Abstract
Both GO (7,8-dihydro-8-oxoguanine) and hoU (5-hydroxyuracil) are highly mutagenic because DNA polymerase frequently misincorporates adenine opposite these damaged bases. In Escherichia coli, MutY DNA glycosylase can remove misincorporated adenine opposite G or GO on the template strand during DNA replication. MutY remains bound to the product that contains an AP (apurinic/apyrimidinic) site. Endo VIII (endonuclease VIII) can remove oxidized pyrimidine and weakly remove GO by its DNA glycosylase and β/δ-elimination activities. In the present paper, we demonstrate that Endo VIII can promote MutY dissociation from AP/G, but not from AP/GO, and can promote β/δ-elimination on the products of MutY. MutY interacts physically with Endo VIII through its C-terminal domain. MutY has a moderate affinity for DNA containing a hoU/A mismatch, which is a substrate of Endo VIII. MutY competes with Endo VIII and inhibits Endo VIII activity on DNA that contains a hoU/A mismatch. Moreover, MutY has a weak adenine glycosylase activity on hoU/A mismatches. These results suggest that MutY may have some role in reducing the mutagenic effects of hoU.
Keywords: 7,8-dihydro-8-oxoguanine (GO); DNA glycosylase; endonuclease VIII; MutY; oxidized pyrimidine; protein–protein interaction
Abbreviations: AP, apurinic/apyrimidinic; BER, base excision repair; CBD, chitin-binding protein; Endo VIII (etc.), endonuclease VIII (etc.); GO, 7,8-dihydro-8-oxoguanine; GST, glutathione S-transferase; hoU, 5-hydroxyuracil; UDG, uracil DNA glycosylase
INTRODUCTION
DNA bases are subjected to damage by ROS (reactive oxygen species) derived from cellular metabolism as well as exogenous stimuli, such as ionizing radiation and various chemical oxidants [1]. Some of this damage, if not repaired, can be bypassed by DNA polymerases to induce mutagenesis and degenerative conditions, including aging and cancer. Living organisms have evolved various defence systems to protect their genomes from oxidative DNA damage. BER (base excision repair) is the major repair pathway for the oxidized DNA bases and is initiated by DNA glycosylases. GO (7,8-dihydro-8-oxoguanine), one of the most stable oxidized purines, has the most deleterious effects because it can mispair with adenine during DNA replication [2,3]. In Escherichia coli, MutM, MutS, MutT, MutY DNA glycosylases and Endo VIII (endonuclease VIII) are involved in defending against the mutagenic effects of GO lesions (reviewed in [4]). The MutT protein eliminates 8-oxo-7,8-dihydro-2′-dGTP from the nucleotide pool using its pyrophosphohydrolase activity [5], whereas the MutM glycosylase (Fpg protein) removes both mutagenic GO adducts and ring-opened purine lesions. When C/GO mismatches are not repaired by MutM, adenines are frequently incorporated opposite GO bases during DNA replication and can subsequently cause G:C→T:A transversions. MutS- and MutY-dependent mismatch repairs can increase replication fidelity by removing the adenine that is misincorporated opposite GO or guanine. The MutS-dependent mismatch repair removes mismatched adenine on the daughter DNA strands (reviewed in [6]). MutY, an adenine and weak guanine DNA glycosylase, is active on A/G, A/GO, A/C and G/GO mismatches [4]. A/GO mismatches are particularly important biological substrates of MutY glycosylase. Endo VIII has been shown to serve as a backup pathway to repair GO in the absence of MutM and MutY [7]. Similar to MutM, Endo VIII can remove GO from GO/C. In addition, Endo VIII can also repair GO when GO is misincorporated opposite adenine during DNA replication [8].
The N-terminal domain of the E. coli MutY protein contains the catalytic activity [9,10], while the C-terminal domain of MutY plays an important role in the recognition of GO lesions [9,11–13]. The binding affinity and reaction rate of a truncated MutY on A/GO-containing DNA are reduced when compared with those of the intact MutY [12,13]. Moreover, deletion of the C-terminal domain of MutY confers a mutator phenotype in vivo [12]. The action of MutY is characterized by its tight binding to the AP (apurinic/apyrimidinic) site produced after the adenine is removed [11,12,14]. The affinity of MutY for AP/GO is particularly strong and is mediated by its C-terminal domain [11–13]. The AP endonucleases, Endo IV and Exo III, have been shown to enhance the turnover of MutY with A/G but not A/GO substrates [15].
HoU (5-hydroxyuracil), a deaminated and oxidized form of cytosine [4], is highly mutagenic because DNA polymerase frequently misincorporates an adenine opposite this damaged base and subsequently causes a G:C→A:T transition [16]. In E. coli, oxidized pyrimidines are mainly repaired by Endo III and Endo VIII, which are encoded by nth and nei genes respectively [17,18]. The substrate preferences of Endo III and Endo VIII overlap, but are not identical [19,20]. Despite similar substrate specificity, Endo III and Endo VIII have no sequence or structure homology [21]. Instead, Endo VIII shares sequence and functional homologies with MutM, and both enzymes have β/δ-elimination activities [20].
In the present study, we show that there is an interaction between pathways that are involved in the repair of oxidized purines and pyrimidines. We demonstrate that Endo VIII interacts physically and functionally with MutY. Endo VIII can promote MutY dissociation from its products and acts further on these products. MutY competes with Endo VIII and inhibits Endo VIII from removing oxidized pyrimidines. Furthermore, MutY can remove adenine from hoU/A mismatches and may have a role in reducing the mutagenic effects of hoU.
MATERIALS AND METHODS
Bacteria
The E. coli strain GM7724 is a ΔmutY::Cam strain derived from AB1157 [ara14 argE3 Δ(gpt-proA)62 galK2 hisG4 kdgK51 leuB6 lacY1 mtl-1 rac− rfbD1 rpsL31 thr-1 tsx-33 supE44 xyl-5] by replacing the mutY gene with the chloramphenicol-resistance gene (Cam). The strain was provided by Dr Michael Volkert and Dr Martin G. Marinus (University of Massachusetts, Worcester, MA, U.S.A.). SW2-8 is Δnei::Cam derived from KL16 (LAM− e14− relA1 spoT1 thi-1) and was provided by Dr Susan Wallace (University of Vermont, Burlington, VT, U.S.A.). GM7724 and SW2-8 with DE3 lysogen were constructed according to the procedures described by Invitrogen.
GST (glutathione S-transferase)-fusion protein constructs
The GST fusions of MutY and its deletion constructs were made using PCR with template pMYW-1 [22] and the primer sets listed in Supplementary Table 1a (http://www.BiochemJ.org/bj/393/bj3930381add.htm). The PCR products were digested with BamHI and XhoI and were ligated into the BamHI/XhoI-digested pGEX-4T-2 vector (GE Healthcare). The GST tag is at the N-termini of all constructs. GST–MutY contains the intact MutY, GST–MutY-(1–226) contains residues 1–226 of MutY, GST–MutY-(216–350) contains residues 216–350 of MutY with one extra methionine residue before MutY, and GST–MutY-(230–350) contains residues 230–350 of MutY.
Expression of CBD (chitin-binding protein)-tagged Endo VIII protein
The plasmid pTYB2[NEI], a gift from Dr Susan Wallace, contains the nei gene cloned into pTYB2 (New England Biolabs). The Endo VIII was fused to the CBD at its C-terminus. SW2-8/DE3 cells containing pTYB2[NEI] were grown in LB (Luria–Bertani) broth containing 100 mg/ml ampicillin at 25 °C. Protein expression was induced at a D590 of 0.6 by the addition of IPTG (isopropyl β-D-thiogalactoside) to a final concentration of 0.2 mM, and the cells were harvested 16 h later. The cell paste, from a 500 ml culture, was resuspended in 9 ml of buffer G (50 mM Tris/HCl, pH 7.4, 150 mM NaCl and 2 mM EDTA) containing 0.5 mM dithiothreitol and 0.1 mM PMSF and treated with lysozyme (1 mg/ml) for 30 min at 4 °C. After sonication, the solution was centrifuged at 10000 g for 20 min, and the supernatant was stored at −80 °C in aliquots.
GST pull-down assay
The GST–MutY constructs were expressed in host E. coli strain GM7724/DE3. The GST constructs (300 ng), which were immobilized on glutathione–Sepharose 4B (Amersham Biosciences) as described previously [23], were incubated overnight at 4 °C with 0.13 mg of cell extracts containing CBD-tagged Endo VIII in 200 μl of buffer G containing 0.1% Nonidet P40. After centrifugation at 1000 g, the pellets were washed four times with 1 ml of buffer G, reconstituted in 1×SDS loading buffer [30 mM Tris/HCl, pH 6.8, 5% (v/v) glycerol, 1% (w/v) SDS, 0.5 mg/ml Bromophenol Blue and 1% (v/v) 2-mercaptoethanol], and resolved on a 10% (w/v) polyacrylamide gel containing SDS [24]. The proteins were transferred on to a membrane and were allowed to react with antibodies against CBD (E8034S; New England Biolabs). Western blotting was detected by the ECL® (enhanced chemiluminescence) analysis system from GE Healthcare.
Enzymes used
Intact MutY and the N-terminal domain MutY-(1–226) were purified as described in [25]. E. coli UDG (uracil DNA glycosylase) and Endo VIII were purchase from Invitrogen and New England Biolabs respectively.
Oligonucleotide substrates
The DNA substrates used in the present study are listed in Supplementary Table 1a at http://www.BiochemJ.org/bj/393/bj3930381add.htm. The 51-mer oligonucleotides were provided by Dr Sankar Mitra (University of Texas Medical Branch, Galveston, TX, U.S.A.). One strand of the duplex was labelled at the 5′ end with [γ-32P]ATP using polynucleotide kinase and then it was annealed to the other strand. The single-stranded overhangs of the 19-mer were filled in with the Klenow fragment of DNA polymerase I and unlabelled dNTPs as described by Lu [26].
MutY gel mobility-shift assay
The MutY binding assay using 1.8 fmol of labelled oligonucleotide substrates in a 20 μl reaction volume was performed as described by Lu [26]. In experiments repeated at least three times, nine different MutY enzyme concentrations were used to bind DNA substrates to determine Kd values. Bands corresponding to enzyme-bound and free DNA were quantified from PhosphorImager images, and Kd values were obtained from analyses by a computer-fitted curve generated by the Enzfitter program [27].
MutY glycosylase assay
The glycosylase assay was carried out in a 10 μl reaction volume containing 1.8 fmol of DNA substrate, 20 mM Tris/HCl (pH 7.6), 1 mM dithiothreitol, 1 mM EDTA, 2.9% (v/v) glycerol and 50 μg/ml BSA. After incubation at 37 °C for 30 min, some reaction mixtures were supplemented with 1 μl of 1 M NaOH and heated at 90 °C for 30 min as indicated. A 5 μl volume of formamide dye (90% formamide, 10 mM EDTA, 0.1% Xylene Cyanol and 0.1% Bromophenol Blue) was added to the sample, which was heated at 90 °C for 2 min, and 5 μl of the mixture was loaded on to a 14% (w/v) polyacrylamide sequencing gel containing 7 M urea.
For time-course studies, samples were heated at 90 °C for 30 min with 0.1 M NaOH immediately after enzyme incubation. Kinetic analyses were performed using DNA concentrations ranging from 0.09 to 11 nM and 14.4 nM of MutY. Bands corresponding to cleavage products and intact DNA were quantified from PhosphorImager images. The Km and Vmax values were obtained from analyses by a computer-fitted curve generated by the Enzfitter program [27].
Endo VIII glycosylase/AP lyase assay
The assay was carried out similarly to the MutY glycosylase assay, except that a DNA substrate containing a hoU/A mismatch was used and NaOH treatment was omitted.
RESULTS
Endo VIII enhances MutY dissociation from the DNA product
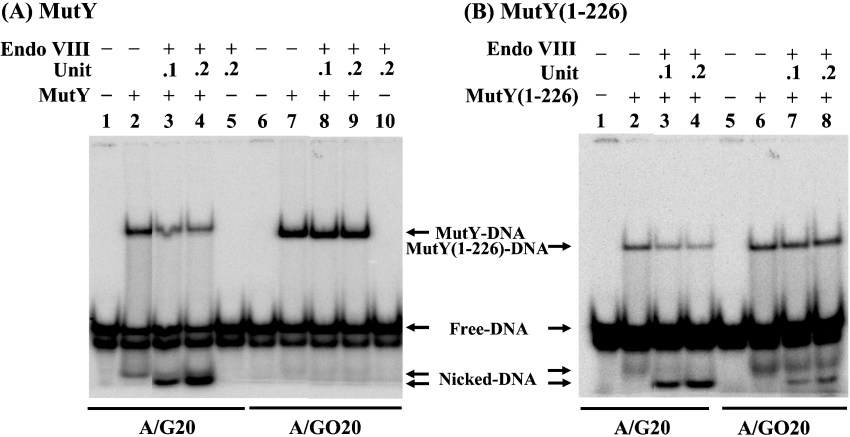
After the MutY glycosylase reaction, the enzyme remained tightly bound to the AP-containing DNA [11,12,14]. In search of enzymes that can enhance the turnover of MutY, we tested DNA glycosylases with β/δ-elimination activities [20]. Using a gel mobility-shift assay, we showed that Endo VIII could affect DNA binding of MutY. The complex formation of MutY with A/G substrate was reduced 3-fold when Endo VIII was added to the binding reaction mixture (Figure 1A, lanes 2–4). A small nicked product was observed with MutY alone (Figure 1A, lane 2) owing to its weak AP lyase activity [9,10,28,29]. Substantial increase in the quantity of nicked product was observed when Endo VIII was added to the MutY binding reaction mixture (Figure 1A, lanes 3 and 4). The mobility of the nicked product in lanes 3 and 4 is slightly faster than that in lane 2 (Figure 1A). We interpret from this that the products have different groups at their 3′ ends: the nicked product in lane 2 of Figure 1(A) has an unsaturated sugar generated by the β-elimination of MutY, while the nicked product in lanes 3 and 4 of Figure 1(A) has a phosphate group due to the β/δ-elimination of Endo VIII (Figure 2). In contrast, the complex formation of MutY with A/GO substrate did not change when Endo VIII was added to the binding reaction mixture (Figure 1A, lanes 7–9). Endo VIII did not bind to either A/G or A/GO substrate (Figure 1A, lanes 5 and 10). Thus Endo VIII can enhance the turnover of MutY with A/G, but not with A/GO substrates.
Figure 1. Effects of Endo VIII on DNA binding by MutY and MutY-(1–226).
(A) Effects of Endo VIII on DNA-binding activity of MutY. Lane 1, 5′-end-labelled A/G-containing 20-mer DNA (A/G20). Lane 2, 1.8 fmol (90 pM) of A/G20 DNA substrate was incubated with MutY (1.8 nM). Lanes 3 and 4, same as lane 2, but with 0.1 unit (0.17 nM) and 0.2 unit (0.34 nM) of Endo VIII respectively. Lane 5, 1.8 fmol (90 pM) of A/G20 DNA substrate was incubated with Endo VIII (0.2 unit or 0.34 nM). Lanes 6–10 are similar to lanes 1–5, except using A/GO-containing 20-mer DNA (A/GO20) and the MutY concentration is 0.11 nM. All reaction mixtures were incubated for 30 min at 37 °C and fractionated on a 6% native gel. (B) Effects of Endo VIII on DNA-binding activity of MutY-(1–226). Lanes 1–4 are similar to lanes 1–4 in (A) and lanes 5–8 are similar to lanes 6–9 in (A), except that MutY-(1–226) was used. Free DNA, nicked DNA, MutY–DNA complex and MutY-(1–226)–DNA complex are indicated.
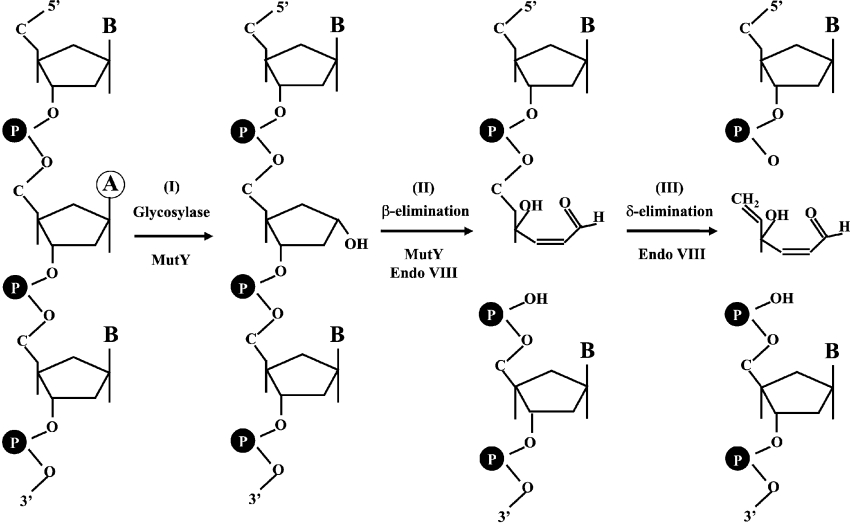
Figure 2. Reactions of MutY DNA glycosylase and β/δ-elimination activities of Endo VIII.
Step I, MutY DNA glycosylase removes a mismatched adenine (circled) which is paired with GO, guanine or cytosine on the other strand (not shown). The MutY product contains an AP site. Step II, the weak AP lyase activity of MutY and the β-elimination activity of Endo VIII cleave the DNA backbone to generate two fragments, one of which contains an unsaturated sugar moiety. Step III, the δ-elimination activity of Endo VIII removes the unsaturated sugar to produce a DNA fragment with a 3′ phosphate group.
Because the C-terminal domain of MutY contributes to the tight binding to AP/GO [11–13], we then tested the effect of Endo VIII on MutY-(1–226) binding to A/G and A/GO. The complex formation of MutY-(1–226) with A/G substrate was reduced 3.5-fold, and the nicked product was enhanced when Endo VIII was added to the binding reaction mixture (Figure 1B, lanes 2–4). The complex formation of MutY-(1–226) with A/GO substrate was slightly reduced by 20%, and the nicked product was observed when Endo VIII was added to the binding reaction (Figure 1B, lanes 7–8). Thus Endo VIII can only slightly enhance the turnover of MutY-(1–226) with A/GO.
Endo VIII can act on MutY products
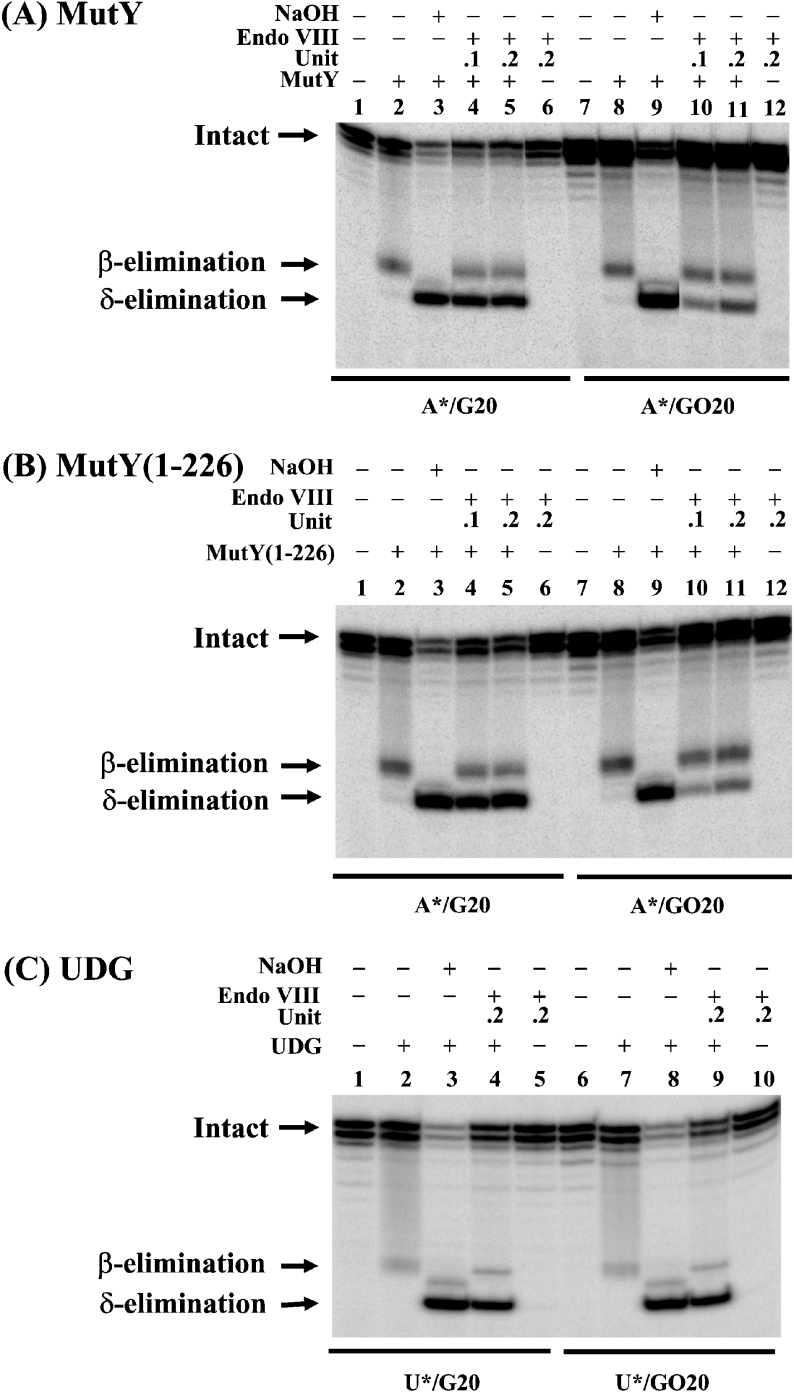
In addition to glycosylase activity on various DNA lesions, Endo VIII has β/δ-elimination activity [20]. To test whether Endo VIII can act on the products of MutY, we added Endo VIII to MutY glycosylase reactions with A/G- and A/GO-containing DNA. The nicked products generated by β- and δ-eliminations (Figure 2) can be easily separated on a sequencing gel. NaOH treatment of MutY product reveals the amount of AP sites generated by MutY glycosylase (Figure 3A, lanes 3 and 9). Endo VIII promoted β/δ-elimination on AP/G, a product of MutY glycosylase (Figure 3A, lanes 4 and 5). Approx. 75% of AP/G products were processed to DNA with 3′ phosphate by the β/δ-elimination of Endo VIII (Figure 3A, compare lanes 3 and 5). Endo VIII also weakly promoted β/δ-elimination on AP/GO after MutY reaction (Figure 3A, lanes 10 and 11). Approx. 17% of AP/GO products were processed to DNA with 3′ phosphate by the β/δ-elimination of Endo VIII (Figure 3A, compare lanes 9 and 11). This nicked product was not detected in the non-denatured gel in Figure 1(A) because of poor dissociation of cleaved product from MutY. Similar results were obtained when Endo VIII was added to MutY-(1–226) glycosylase reactions (Figure 3B), except that slightly more δ-elimination product was observed with A/GO substrate in the presence of Endo VIII (Figure 3B, lanes 10 and 11). Approx. 21% of AP/GO products were processed to DNA with 3′ phosphate by the β/δ-elimination of Endo VIII (Figure 3B, compare lanes 9 and 11). For comparison, Endo VIII equally promoted β/δ-elimination on UDG's products, AP/G and AP/GO (Figure 3C). Thus MutY and MutY-(1–226) binding to AP/GO provides a barrier for the access of Endo VIII.
Figure 3. Endo VIII activity on the DNA products generated by MutY, MutY-(1–226) and UDG.
(A) The activity of Endo VIII on DNA products of MutY. Lanes 1 and 7, 5′-end-labelled A/G- and A/GO-containing DNA respectively. A/G20 DNA substrate (1.8 fmol or 180 pM) was incubated with 7.2 nM MutY (lanes 2–5) and A/GO20 DNA substrate (1.8 fmol or 180 pM) was incubated with 1.8 nM MutY (lanes 8–11) at 37 °C for 30 min. Endo VIII [0.1 unit (0.34 nM) or 0.2 unit (0.68 nM)] was added to reaction mixtures in lanes 4–6 and lanes 10–12 as indicated. Lanes 3 and 9, after glycosylase reaction, the mixtures were supplemented with 1 μl of 1 M NaOH and heated at 90 °C for 30 min. Formamide dye (5 μl) was added to the sample, which was heated at 90 °C for 2 min, and 5 μl of the mixture was loaded on to a 14% (w/v) polyacrylamide sequencing gel containing 7 M urea. The positions of the intact substrate and the products generated by β- and δ-eliminations are marked. (B) The activity of Endo VIII on DNA products of MutY-(1–226). Reactions were similar to (A) except that MutY-(1–226) was used. (C) The activity of Endo VIII on DNA products of UDG DNA glycosylase. Reactions were similar to (A), except 0.01 unit of UDG with U/G20 and U/GO20 DNA substrates were used. Asterisks mark the 32P-labelled DNA strands.
MutY physically interacts with Endo VIII
We tested further whether MutY had any physical interaction with Endo VIII. The GST-tagged MutY was immobilized on glutathione–Sepharose 4B and was used to pull down CBD-tagged Endo VIII. As shown in Figure 4, Endo VIII bound to GST–MutY (lane 1), but did not bind to GST alone (lane 4). By using constructs containing different portions of MutY fused to GST, the regions of MutY engaged in the physical interactions with Endo VIII were localized to residues 230–350, the C-terminal domain of MutY.
Figure 4. Physical interaction of MutY with Endo VIII.
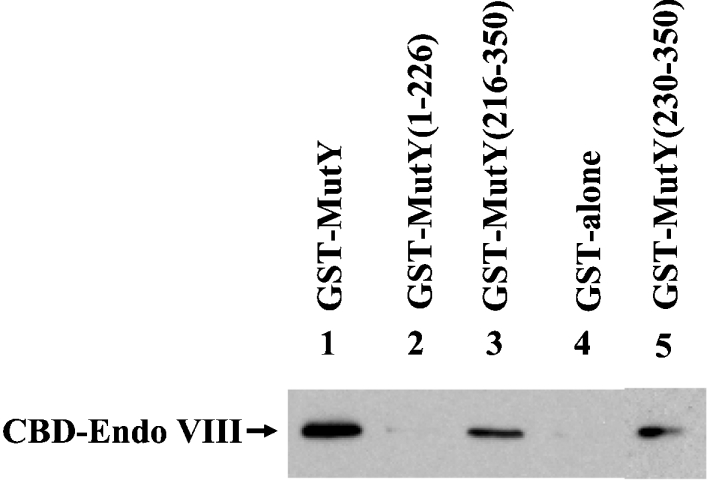
GST alone and several GST–MutY constructs (300 ng) immobilized on glutathione–Sepharose were incubated with 0.13 mg of E. coli extracts expressing CBD-tagged Endo VIII protein as described in the Materials and methods section. The pellets were fractionated on an SDS/10% polyacrylamide gel followed by Western blot analysis using the antibody against CBD. Lane 1, GST-tagged intact MutY (residues 1–350); lane 2, GST–MutY-(1–226); lane 3, GST–MutY-(216–350); lanes 4, GST alone; lane 5, GST–MutY-(230–350).
MutY has binding affinity with hoU/A mismatch
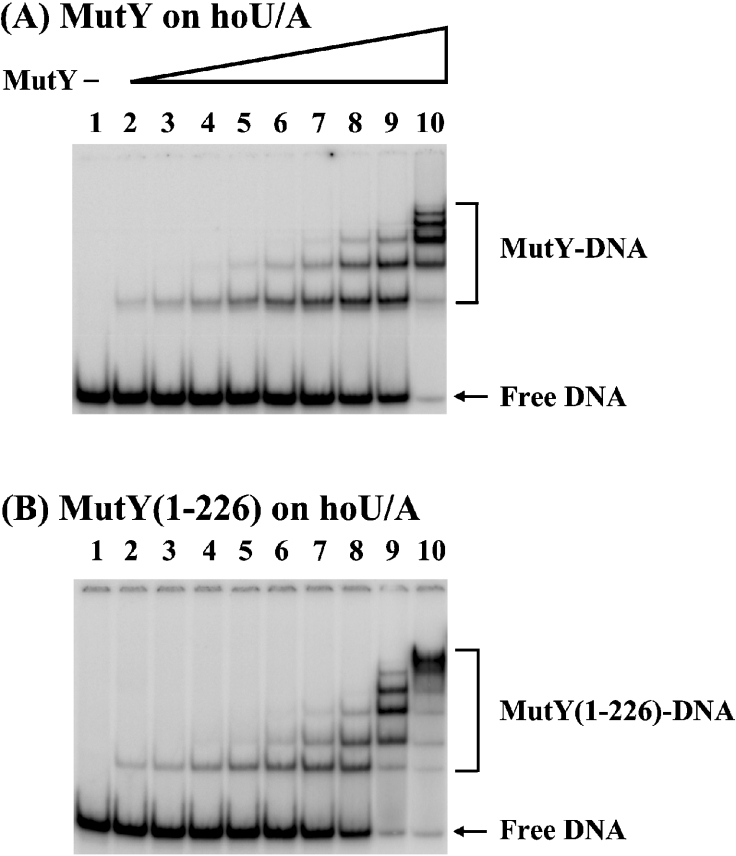
Because MutY interacts with Endo VIII, we performed gel mobility-shift assays to determine whether MutY could bind a 51-mer DNA substrate containing a hoU/A mismatch, a catalytic substrate of Endo VIII. As shown in Figure 5, MutY and MutY-(1–226) were able to bind to this DNA substrate. Up to six protein–DNA complexes were observed (see Figure 5A, lane 10 for example) in a concentration-dependent manner. Because MutY has been shown to be a monomer in solution up to 230 nM by glycerol gradient [30], the multiple protein–DNA complexes detected here are probably not due to protein aggregates at high concentrations of protein. The Kd (app) values of MutY and MutY-(1–226) with hoU/A were 124 and 93 nM respectively (Table 1). The similar Kd values of MutY and MutY-(1–226) with hoU/A imply that the C-terminal domain has little contribution to this binding. The binding affinity of MutY with hoU/A-containing DNA is much weaker than those of MutY with A/G- and A/GO-containing DNA (Kd values of 1.8 and 0.14 nM respectively) [28,31]. However, the affinity of MutY with hoU/A is higher than that of MutY with the homoduplex (a Kd value of 315 nM) [31].
Figure 5. Binding activities of MutY and MutY-(1–226) with DNA containing hoU/A mismatch.
(A) HoU/A-containing DNA was incubated with MutY at various protein concentrations at 37 °C for 30 min. Lane 1, DNA labelled at the 5′ end of the hoU-containing strand. Lanes 2–10, hoU/A-containing DNA was incubated with MutY protein at concentrations of 0.9, 1.8, 3.6, 7.2, 14.4, 28.8, 57.6, 115.2 or 230.4 nM respectively. Products were fractionated on 6% native gels. (B) is similar to (A), except using MutY-(1–226). The positions of the free DNA, MutY–DNA complex and MutY-(1–226)–DNA complex are marked.
Table 1. Kd (app) values of MutY and MutY-(1–226) for hoU/A DNA binding as well as kinetic parameters of MutY on hoU/A.
Kd values are means±S.E.M. for three experiments using eight protein concentrations. ND, not determined.
| Enzyme | Kd (nM) | Km (nM) | Vmax (pM·min−1) | kcat (min−1) | kcat/Km (min−1·μM−1) |
|---|---|---|---|---|---|
| MutY | 124±14 | 224 | 22 | 0.0015 | 0.064 |
| MutY-(1–226) | 93±13 | ND | ND | ND | ND |
MutY can inhibit Endo VIII activities on DNA substrate containing hoU/A mismatch
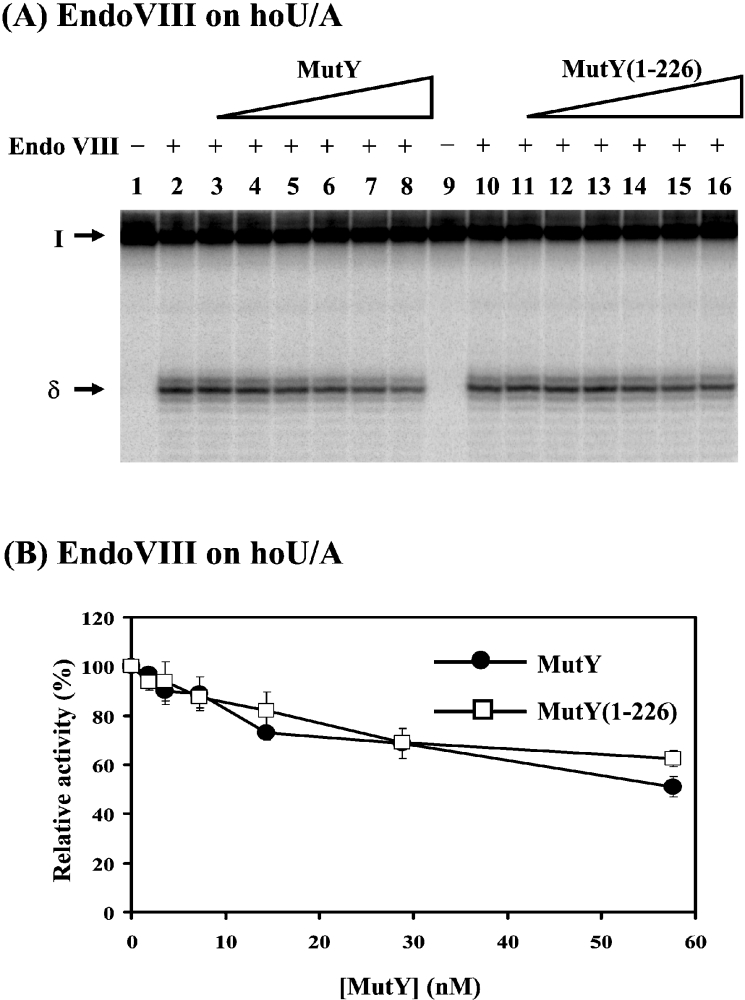
As shown above, MutY has modest binding to Endo VIII's catalytic substrate containing a hoU/A. Next, we tested the effect of MutY on the glycosylase/AP lyase activity of Endo VIII. MutY could inhibit Endo VIII glycosylase/AP lyase activity on DNA substrates containing hoU/A (Figure 6). At a MutY/Endo VIII molar ratio of 50, Endo VIII activity with hoU/A was inhibited by 30%. MutY-(1–226) had a similar effect as MutY on inhibiting Endo VIII activity (Figure 6).
Figure 6. MutY and MutY-(1–226) can inhibit Endo VIII activity.
(A) Activity of Endo VIII on hoU/A substrate can be inhibited by MutY and MutY-(1–226). Lanes 1 and 9, hoU/A-containing DNA labelled at 5′ end of the hoU-containing strand. DNA substrate (1.8 fmol or 180 pM) was incubated with 0.34 nM (0.1 unit) of Endo VIII (lanes 2–8 and 10–16) at 37 °C for 30 min. MutY (1.8, 3.6, 7.2, 14.4, 28.8 or 57.6 nM) was added to reaction mixtures in lanes 3–8. MutY-(1–226) (1.8, 3.6, 7.2, 14.4, 28.8 or 57.6 nM) was added to reaction mixtures in lanes 11–16. After reactions, 5 μl of formamide dye was added to the sample, which was heated at 90 °C for 2 min, and 5 μl of the mixture was loaded on to a 14% (w/v) polyacrylamide sequencing gel containing 7 M urea. The positions of the intact substrate (I) and the products generated by δ-elimination (δ) are marked. (B) Quantitative analyses of MutY and MutY-(1–226) inhibition on Endo VIII activity on hoU/A substrate. Results are means±S.D. from PhosphorImager quantitative analyses of gel images from three experiments as represented in (A).
MutY has glycosylase activity on hoU/A-DNA substrate
Because MutY can bind hoU/A-containing DNA, we tested whether MutY has adenine glycosylase activity on this substrate. Surprisingly, MutY could remove adenine from hoU/A (Figure 7). This glycosylase activity was very weak and required a long incubation time and high molar excess of enzyme over DNA. The kcat/Km of MutY with hoU/A-containing DNA was measured to be 0.064 min−1·μM−1 (Table 1), which is much weaker than those of MutY with 20-mer A/G and A/GO (kcat/Km in the range 3–10 min−1·μM−1) [29].
Figure 7. MutY glycosylase activity on DNA containing a hoU/A mismatch.
(A) Time-course study of MutY glycosylase activity on hoU/A-containing DNA. DNA (1.8 fmol) labelled at the 5′ end of the adenine-containing strand was incubated with 14.4 nM MutY at 37 °C. (B) MutY glycosylase activity on hoU/A-containing DNA at different protein concentrations. hoU/A-containing DNA (1.8 fmol) labelled at the 5′ end of the adenine-containing strand was incubated with MutY protein at 3.6, 7.2, 14.4, 28.8 or 57.6 nM at 37 °C for 1 h. After incubation, the reaction mixtures were supplemented with 1 μl of 1 M NaOH and heated at 90 °C for 30 min. Formamide dye (5 μl) was added to the sample, which was heated at 90 °C for 2 min, and 5 μl of the mixture was loaded on to a 14% (w/v) polyacrylamide sequencing gel containing 7 M urea.
DISCUSSION
AP sites generated by DNA glycosylases are potentially mutagenic owing to lack of base coding information and blockage of DNA synthesis [32,33]. To prevent this toxic effect, MutY, like other DNA glycosylases, binds tightly to its AP site products [12,14] until other components are recruited to carry out the next repair step. It has been suggested that the BER pathway may involve highly co-ordinated processes that are governed by protein–protein and protein–DNA interactions [4,34,35]. In E. coli, Endo IV and Exo III greatly enhance the rate of product release of MutY with a A/G substrate; however, neither AP endonuclease enhances MutY turnover with an A/GO substrate [15]. In the present study, we show that Endo VIII can promote MutY dissociation from AP/G, but not from AP/GO. So far, it remains unclear how MutY dissociates from AP/GO in vivo.
Hazra et al. [8] have demonstrated that Endo VIII does not affect MutY activity on A/GO and G/GO. However, we show that Endo VIII can promote β/δ-elimination on AP/G and weakly promote β/δ-elimination on AP/GO, when AP/G and AP/GO are products of MutY reaction. In contrast, Endo VIII can equally promote β/δ-elimination on AP/G and AP/GO after UDG reaction. Different accessibility of Endo VIII to AP/G and AP/GO may be caused by different binding of MutY to these DNA substrates. It is interesting to note that approx. 17% of AP/GO products generated by MutY were processed by the β/δ-elimination of Endo VIII, but that all AP/GO-containing DNA were still bound by MutY (compare Figures 1A and 3A). This result is due to poor dissociation of the cleaved product from MutY and it suggests that the phosphate groups on the 5′ and 3′ sides of an AP site opposite GO of the MutY-bound DNA are fairly accessible by Endo VIII. How these two enzymes interact with the same DNA remains to be determined.
The C-terminal domain of MutY moderately contributes to preventing MutY dissociation from AP/GO by Endo VIII (Figure 1B). This is somewhat contradictory to this domain's role in binding to the GO strand [9,11–13]. The C-terminal domain of MutY does not limit the accessibility of Endo VIII's β/δ-elimination activity on AP/G and AP/GO (compare Figures 3A and 3B). These results are consistent with the MutY structure, which shows that the C-terminal domain of MutY does not contact the mismatched adenine strand [36]. The physical interaction of Endo VIII with the C-terminal domain of MutY appears to be unnecessary for their functional interaction observed in vitro. The significance of this physical interaction in vivo remains to be determined.
There are interplays between pathways involved in repair oxidized purines and pyrimidines. The GO repair system involves MutT, MutS, MutY, MutM and Endo VIII. Endo VIII serves as a backup pathway to repair GO in the absence of MutM and MutY [7]. However, its major role is to repair oxidized pyrimidines [7,20]. We and others have shown that MutY can inhibit MutM glycosylase activity on GO/C and GO/G mismatches [8,12]. The human MutS homologue (MSH2/MSH6) interacts and enhances hMYH (human MutY homologue) [37]. Similarly, E. coli MutS interacts with MutY (H. Bai and A.-L. Lu, unpublished work). In the present study, we show that Endo VIII glycosylase activity of removing oxidized cytosine from hoU/A can be inhibited by MutY. Similarly, Hazra et al. [38] have shown that the repair of hydantoins (the oxidation product of GO) by Endo VIII is inhibited by MutY. However, Hazra et al. [8] have also shown that Endo VIII GO glycosylase activity on GO/G, GO/A and GO/C substrates is not affected by MutY. This is surprising, in view of the tight binding of MutY to A/GO and AP/GO [9,11–13] and the structures of both MutY and Endo VIII [21,36].
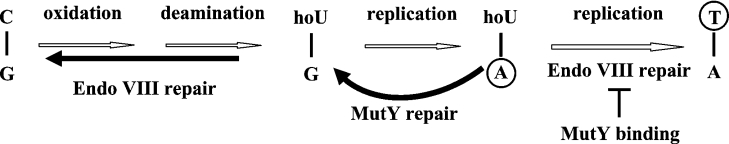
MutY can remove adenine from A/G, A/GO, and A/C mismatches and guanine from G/GO mismatches [12,39–42]. Because MutY activity on A/C and G/GO is very weak, the most frequent mutations observed in mutY mutants are G:C→T:A transversions. It has been reported that MutY has a minor role in reducing the mutagenic effects on G:C→A:T transitions and G:C→C:G transversions [42–44]. In the present paper, we identified a new MutY catalytic substrate: MutY can remove adenine from hoU/A mismatches. hoU is generated readily under oxidative stress from cytosine by oxidation, deamination and dehydration [45]. When hoU/G mismatches are not repaired by Endo VIII, adenines are frequently incorporated opposite hoU bases during DNA replication [16] and can subsequently cause G:C→A:T transitions (Figure 8). The glycosylase activity of MutY on A/hoU may be involved in increasing replication fidelity by removing the adenine misincorporated opposite hoU if a guanine is incorporated opposite template hoU during MutY repair synthesis (Figure 8, solid thick curved arrow). In addition, MutY may inhibit Endo VIII repair on hoU/A when hoU is on the template DNA strand (Figure 8). Thus Endo VIII is restricted to acting on hoU/G substrates (Figure 8, solid thick straight arrow). The MutY glycosylase activity on A/hoU is similar to its weak activity on A/C mismatches [44,46]. The increased G:C→A:T transitions in mutY mutants were interpreted previously as a failure of A/C mismatch repair. Because hoU is an oxidized form of cytosine, the increased G:C→A:T transitions in mutY mutants may be attributed to the deficiency of A/hoU repair. Taken together, our data suggest that MutY may have some role in reducing the mutagenic effects of hoU and in modulating Endo VIII on repair of oxidized pyrimidines.
Figure 8. E. coli MutY may have a role in preventing the mutagenic effects of hoU.
hoU in DNA can be derived from oxidation, deamination and dehydration of cytosine. The Endo VIII glycosylase/AP lyase removes hoU adducts. When hoU/G is not repaired by Endo VIII, adenines are frequently incorporated opposite hoU bases during DNA replication. A/hoU mismatches may be repaired to G/hoU by the MutY pathway. Without repair, the second round of DNA replication of A/hoU can generate G:C→A:T transitions. If Endo VIII repairs an A/hoU mismatch when hoU is on the parental DNA strand, a G:C→A:T mutation occurs. MutY may inhibit Endo VIII repair on A/hoU. Bases on the daughter DNA strands are circled. Repair pathways that can reduce mutagenesis are marked with solid thick lines.
Online Data
Acknowledgments
We thank Dr Michael Volkert, Dr Martin Marinus and Dr Susan Wallace for providing E. coli strains. We offer our special thanks to Dr Susan Wallace and Dr Sankar Mitra for providing plasmid and DNA substrate containing hoU respectively. We are grateful to Min Guo for technical assistance. This work is supported by grant GM 35132 from the National Institutes of General Medical Science, National Institutes of Health.
References
- 1.Halliwell B., Gutteridge J. M. New York: Oxford University Press; 1989. Free Radicals in Biology and Medicine. [Google Scholar]
- 2.Michaels M. L., Miller J. H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tchou J., Grollman A. P. Repair of DNA containing the oxidatively-damaged base 8-hydroxyguanine. Mutat. Res. 1993;299:277–287. doi: 10.1016/0165-1218(93)90104-l. [DOI] [PubMed] [Google Scholar]
- 4.Lu A.-L., Li X., Gu Y., Wright P. M., Chang D.-Y. Repair of oxidative DNA damage. Cell Biochem. Biophys. 2001;35:141–170. doi: 10.1385/CBB:35:2:141. [DOI] [PubMed] [Google Scholar]
- 5.Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature (London) 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 6.Modrich P., Lahue R. S. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 7.Blaisdell J. O., Hatahet Z., Wallace S. S. A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J. Bacteriol. 1999;181:6396–6402. doi: 10.1128/jb.181.20.6396-6402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazra T. K., Hill J. W., Izumi T., Mitra S. Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:193–205. doi: 10.1016/s0079-6603(01)68100-5. [DOI] [PubMed] [Google Scholar]
- 9.Gogos A., Cillo J., Clarke N. D., Lu A.-L. Specific recognition of A/G and A/8-oxoG mismatches by Escherichia coli MutY: removal of the C-terminal domain preferentially affects A/8-oxoG recognition. Biochemistry. 1996;35:16665–16671. doi: 10.1021/bi960843w. [DOI] [PubMed] [Google Scholar]
- 10.Manuel R. C., Lloyd R. S. Cloning, overexpression, and biochemical characterization of the catalytic domain of MutY. Biochemistry. 1997;36:11140–11152. doi: 10.1021/bi9709708. [DOI] [PubMed] [Google Scholar]
- 11.Chmiel N. H., Golinelli M. P., Francis A. W., David S. S. Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Res. 2001;29:553–564. doi: 10.1093/nar/29.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Wright P. M., Lu A.-L. The C-terminal domain of MutY glycosylase determines the 7,8-dihydro-8-oxo-guanine specificity and is crucial for mutation avoidance. J. Biol. Chem. 2000;275:8448–8455. doi: 10.1074/jbc.275.12.8448. [DOI] [PubMed] [Google Scholar]
- 13.Noll D. M., Gogos A., Granek J. A., Clarke N. D. The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine.adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- 14.Porello S. L., Leyes A. E., David S. S. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 15.Pope M. A., Porello S. L., David S. S. Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G:A substrates. J. Biol. Chem. 2002;277:22605–22615. doi: 10.1074/jbc.M203037200. [DOI] [PubMed] [Google Scholar]
- 16.Purmal A. A., Kow Y. W., Wallace S. S. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace S. S., Bandaru V., Kathe S. D., Bond J. P. The enigma of endonuclease VIII. DNA Repair (Amsterdam) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 18.Zharkov D. O., Shoham G., Grollman A. P. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amsterdam) 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 19.Hazra T. K., Izumi T., Venkataraman R., Kow Y. W., Dizdaroglu M., Mitra S. Characterization of a novel 8-oxoguanine-DNA glycosylase activity in Escherichia coli and identification of the enzyme as endonuclease VIII. J. Biol. Chem. 2000;275:27762–27767. doi: 10.1074/jbc.M004052200. [DOI] [PubMed] [Google Scholar]
- 20.Jiang D., Hatahet Z., Melamede R. J., Kow Y. W., Wallace S. S. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 21.Zharkov D. O., Golan G., Gilboa R., Fernandes A. S., Gerchman S. E., Kycia J. H., Rieger R. A., Grollman A. P., Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright P. M., Yu J., Cillo J., Lu A.-L. The active site of the Escherichia coli MutY DNA adenine glycosylase. J. Biol. Chem. 1999;274:29011–29018. doi: 10.1074/jbc.274.41.29011. [DOI] [PubMed] [Google Scholar]
- 23.Parker A., Gu Y., Mahoney W., Lee S.-H., Singh K. K., Lu A.-L. Human homolog of the MutY protein (hMYH) physically interacts with protein involved in long-patch DNA base excision repair. J. Biol. Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U. K. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Li X., Lu A.-L. Intact MutY and its catalytic domain differentially contact with A/8-oxoG-containing DNA. Nucleic Acids Res. 2000;28:4593–4603. doi: 10.1093/nar/28.23.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu A.-L. Repair of A/G and A/8-oxoG mismatches by MutY adenine DNA glycosylase. In: Vaughan P., editor. DNA Repair Protocols: Prokaryotic Systems. Totowa: Humana Press Inc.; 2000. pp. 3–16. [Google Scholar]
- 27.Leatherbarrow R. J. Amsterdam: Elsevier Science Publishers BV; 1987. Enzfitter: a Non-linear Regression Analysis Program for IBM PC. [Google Scholar]
- 28.Lu A.-L., Tsai-Wu J.-J., Cillo J. DNA determinants and substrate specificities of Escherichia coli MutY. J. Biol. Chem. 1995;270:23582–23588. doi: 10.1074/jbc.270.40.23582. [DOI] [PubMed] [Google Scholar]
- 29.Lu A.-L., Yuen D. S., Cillo J. Catalytic mechanism and DNA substrate recognition of Escherichia coli MutY protein. J. Biol. Chem. 1996;271:24138–24143. doi: 10.1074/jbc.271.39.24138. [DOI] [PubMed] [Google Scholar]
- 30.Lee C.-Y., Bai H., Houle R., Wilson G. M., Lu A.-L. An Escherichia coli MutY mutant without the six-helix barrel domain is a dimer in solution and assembles cooperatively into multisubunit complexes with DNA. J. Biol. Chem. 2004;279:52653–52663. doi: 10.1074/jbc.M405271200. [DOI] [PubMed] [Google Scholar]
- 31.Lu A.-L., Fawcett W. P. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from Schizosacchromyces pombe. J. Biol. Chem. 1998;273:25098–25105. doi: 10.1074/jbc.273.39.25098. [DOI] [PubMed] [Google Scholar]
- 32.Barzilay G., Hickson I. D. Structure and function of apurinic/apyrimidinic endonucleases. BioEssays. 1995;17:713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 33.Randall S. K., Eritja R., Kaplan B. E., Petruska J., Goodman M. F. Nucleotide insertion kinetics opposite abasic lesions in DNA. J. Biol. Chem. 1987;262:6864–6870. [PubMed] [Google Scholar]
- 34.Mol C. D., Izumi T., Mitra S., Tainer J. A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature (London) 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 35.Wilson S. H., Kunkel T. A. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 36.Fromme J. C., Banerjee A., Huang S. J., Verdine G. L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature (London) 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 37.Gu Y., Parker A., Wilson T. M., Bai H., Chang D. Y., Lu A.-L. Human MutY homolog (hMYH), a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins hMSH2/hMSH6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 38.Hazra T. K., Muller J. G., Manuel R. C., Burrows C. J., Lloyd R. S., Mitra S. Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Au K. G., Cabrera M., Miller J. H., Modrich P. Escherichia coli mutY gene product is required for specific A–G→C·G mismatch correction. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu A.-L., Chang D.-Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988;54:805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- 41.Michaels M. L., Cruz C., Grollman A. P., Miller J. H. Evidence that MutM and MutY combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q. M., Ishikawa N., Nakahara T., Yonei S. Escherichia coli MutY protein has a guanine-DNA glycosylase that acts on 7,8-dihydro-8-oxoguanine:guanine mispair to prevent spontaneous G:C to C:G transversions. Nucleic Acids Res. 1998;26:4669–4675. doi: 10.1093/nar/26.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nghiem Y., Cabrera M., Cupples C. G., Miller J. H. The mutY gene: a mutator locus in Escherichia coli that generates G:C to T:A transversions. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radicella J. P., Clark E. A., Fox M. S. Some mismatch repair activities in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9674–9678. doi: 10.1073/pnas.85.24.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dizdaroglu M. Characterization of free radical-induced damage to DNA by the combined use of enzymatic hydrolysis and gas chromatography-mass spectrometry. J. Chromatogr. 1986;367:357–366. doi: 10.1016/s0021-9673(00)94856-8. [DOI] [PubMed] [Google Scholar]
- 46.Tsai-Wu J.-J., Liu H.-F., Lu A.-L. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A·C and A·G mispairs. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8779–8783. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.