Abstract
Recent work indicates that the LKB1 tumour suppressor protein kinase, which is mutated in Peutz–Jeghers cancer syndrome, phosphorylates and activates a group of protein kinases that are related to AMPK (AMP-activated protein kinase). Ten of the 14 AMPK-related protein kinases activated by LKB1, including SIK (salt-induced kinase), MARK (microtubule-affinity-regulating kinase) and BRSK (brain-specific kinase) isoforms, possess a ubiquitin-associated (UBA) domain immediately C-terminal to the kinase catalytic domain. These are the only protein kinases in the human genome known to possess a UBA domain, but their roles in regulating AMPK-related kinases are unknown. We have investigated the roles that the UBA domain may play in regulating these enzymes. Limited proteolysis of MARK2 revealed that the kinase and UBA domains were contained within a fragment that was resistant to trypsin proteolysis. SAXS (small-angle X-ray scattering) analysis of inactive and active LKB1-phosphorylated MARK2 revealed that activation of MARK2 is accompanied by a significant conformational change that alters the orientation of the UBA domain with respect to the catalytic domain. Our results indicate that none of the UBA domains found in AMPK-related kinases interact with polyubiquitin or other ubiquitin-like molecules. Instead, the UBA domains appear to play an essential conformational role and are required for the LKB1-mediated phosphorylation and activation of AMPK-related kinases. This is based on the findings that mutation or removal of the UBA domains of several AMPK-related kinases, including isoforms of MARK, SIK and BRSK, markedly impaired the catalytic activity and LKB1-mediated phosphorylation of these enzymes. We also provide evidence that the UBA domains do not function as LKB1–STRAD (STE20-related adaptor)–MO25 (mouse protein 25) docking/interacting sites and that mutations in the UBA domain of SIK suppressed the ability of SIK to localize within punctate regions of the nucleus. Taken together, these findings suggest that the UBA domains of AMPK-related kinases play an important role in regulating the conformation, activation and localization of these enzymes.
Keywords: AMP-activated protein kinase (AMPK), LKB1 tumour suppressor protein kinase, Peutz–Jeghers syndrome (PJS), small-angle X-ray scattering analysis (SAXS analysis), ubiquitination
Abbreviations: AMPK, AMP-activated protein kinase; BRSK, brain-specific kinase; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, haemagglutinin; MARK, microtubule-affinity-regulating kinase; MO25, mouse protein 25; NEDD8, neural precursor cell-expressed developmentally down-regulated 8; NSD, normalized spatial discrepancy; NUAK1 kinase, ARK5 (AMPK-related kinase 1); PJS, Peutz–Jegher's syndrome; QIK, Qin-induced kinase; Rhp23, a functional homologue of the human excision repair enzyme Hhr23A; SAXS, small-angle X-ray scattering; SAKS1, stress-activated-kinase substrate-1; SIK, salt-induced kinase; SNF1, sucrose-non-fermenting kinase-1; SNRK, sucrose-non-fermenting-related kinase; STRAD, STE20-related adaptor; SUMO1, small ubiquitin-related modifier-1; TAP, tandem affinity purification; UBA, ubiquitin-associated domain
INTRODUCTION
LKB1 has been identified as a gene mutated in the inherited PJS (Peutz–Jeghers syndrome), in which subjects are predisposed to developing benign and malignant tumours [1,2]. Subsequent work demonstrated that overexpression of LKB1 in various cell lines induce a G1 cell-cycle arrest [3,4] and that mice lacking one allele of the LKB1 gene develop benign polyps similar to those found in PJS in humans (reviewed in [5]). In addition to regulating cell growth, studies in nematode-worm (Caenorhabditis elegans) [6], fruitfly (Drosophila melanogaster) [7], South-African-clawed-frog (Xenopus laevis) [8] and mammalian cells [9] suggested that LKB1 also plays an important role in regulating cell polarity, which may contribute to tumour formation in PJS patients.
In vivo, LKB1 is complexed to STRAD (STE20-related adaptor), an inactive pseudokinase [10], as well as an Armadillo-repeat-domain scaffolding protein MO25 (mouse protein 25) [11,12]. The binding of LKB1 to STRAD–MO25, activates LKB1 and also anchors it in the cell cytosol [10,11]. The first identified physiological substrate of LKB1 was AMPK (AMP-activated protein kinase) [13–15], which is a master regulator of cellular energy charge [16]. The LKB1–STRAD–MO25 complex phosphorylates the T-loop threonine residue of AMPK (Thr172), leading to activation of this enzyme. Further evidence that LKB1 is a key regulator of AMPK and energy levels comes from the finding that, in mice lacking LKB1 in either skeletal muscle [17] or liver [18], AMPK is not activated in response to a variety of stimuli. Recent studies have also indicated that, in addition to the two isoforms of AMPK, 13 other less studied kinases that are related to AMPK {MARK1 (microtubule-affinity-regulating kinase 1), MARK2, MARK3, MARK4, QIK (Qin-induced kinase), QSK kinase, SIK (salt-induced kinase), NUAK1 kinase [ARK5 (AMPK-related kinase 5)], NUAK2, BRSK1 (brain-specific kinase 1), BRSK2 and SNRK (sucrose-non-fermenting-related kinase)} are also phosphorylated and activated by LKB1 in vitro and possess markedly lower activity in LKB1-deficient cell lines compared with LKB1-expressing cells [19,20]. Interestingly, many of the AMPK-related kinases that are activated by LKB1 possess a UBA (ubiquitin-associated) domain immediately succeeding the kinase domain. As the function of this domain within these enzymes is unknown, we undertook, in the present study, experiments to explore the roles that it may play.
MATERIALS AND METHODS
Materials
Complete™ protease-inhibitor cocktail tablets were obtained from Roche; P81 phosphocellulose paper was from Whatman; [32P]ATP and glutathione–Sepharose were purchased from Amersham Biosciences. Precast SDS/polyacrylamide/Bis-Tris gels were from Invitrogen; tissue-culture reagents were from Life Technologies; tetra-ubiquitin and K48 (Lys48)- and K63 (Lys63)-linked polyubiquitin were purchased from Affiniti Research Products (Exeter, U.K.) and ubiquitin was purchased from Sigma. All peptides were synthesized by Dr Graham Bloomberg (Department of Biochemistry, School of Medical Science, University of Bristol, Bristol, U.K.). Purified LKB1–STRAD–MO25 complex obtained from an insect-cell baculovirus expression system was kindly provided by Dr Gursant Kular (MRC Protein Phosphorylation Unit, MSI/WTB Complex, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.).
Antibodies
The following antibodies were raised in sheep and affinity-purified on the appropriate antigen: phospho-anti-T-loop QIK/SIK (residues 175–189 of human SIK, KSGEPLSpTWCGSPPY phosphorylated at Thr182, used for immunoblotting), phospho-anti-T-loop MARK (residues 204-218 of human MARK3, TVGGKLDpTFCGSPPY phosphorylated at Thr211, used for immunoblotting), phospho-anti-T-loop BRSK1/BRSK2 (residues 238–252 of human BRSK1, VGDSLLEpTSCGSPHY phosphorylated at Thr245, used for immunoblotting) and the anti-GST [anti-(glutathione S-transferase) protein, used for immunoblotting]. Anti-QSK (residues 1349–1369 of human QSK, TDILLSYKHPEVSFSMEQAGV, used for immunoblotting and immunoprecipitation), anti-SIK (residues 1–20 of human SIK, MVIMSEFSADPAGQGQGQQK, used for immunoblotting and immunoprecipitation). The antibody recognizing both MARK2 and MARK3 isoforms (anti-c-TAK #05-680) was from Upstate Biotech, Lake Placid, NY, U.S.A.. The anti-ubiquitin antibody was purchased from Dako, and the monoclonal antibody recognizing the HA (haemagglutinin) epitope tag was purchased from Roche. The secondary antibodies coupled to horseradish peroxidase used for immunoblotting were obtained from Pierce.
General methods
Tissue culture, transfection, immunoblotting, restriction-enzyme digests, DNA ligations and other recombinant DNA procedures were performed using standard protocols. All mutagenesis was carried out using the QuikChange® site-directed mutagenesis method (Stratagene). DNA constructs used for transfection were purified from Escherichia coli DH5α cells using Qiagen plasmid Mega or Maxi kit according to the manufacturer's protocol. All DNA constructs were verified by DNA sequencing, which was performed by The Sequencing Service, School of Life Sciences, University of Dundee, Scotland, U.K., using DYEnamic ET terminator chemistry (Amersham Biosciences) on Applied Biosystems automated DNA sequencers.
Buffers
Lysis Buffer contained 50 mM Tris/HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% (w/v) Triton-X 100, 1 mM sodium orthovana-date, 10 mM sodium β-glycerophosphate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 0.1% (v/v) 2-mercaptoethanol and Complete™ proteinase-inhibitor cocktail (one tablet/50 ml). Buffer A contained 50 mM Tris/HCl, pH 7.5, 0.1 mM EGTA and 0.1% (v/v) 2-mercaptoethanol. Buffer B comprised 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.27 M sucrose, 0.1% (v/v) 2-mercaptoethanol, 1 mg/ml lysozyme, 1 mg/ml DNAse and Complete™ proteinase-inhibitor mixture (one tablet/25 ml). Buffer C contained 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA and 0.1% (v/v) 2-mercaptoethanol, and Buffer D contained 25 mM Tris/HCl, pH 7.5, and 1 mM dithiothreitol. Buffer E contained 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl, 0.27 M sucrose, 1% (w/v) Nonidet P40 and 1 mM dithiothreitol. Buffer F contained 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl, 1 mM MgCl2, 1 mM imidazole, 2 mM CaCl2, 0.27 M sucrose and 1 mM dithiothreitol.
Plasmids
The generation of wild-type and mutant epitope-tagged AMPK-related kinases in the pEBG2T vector encoding for the expression of GST-fusion proteins in eukaryotic cells or pGEX for expression in E. coli have been described previously [19,20]. The wild-type and mutant cDNA clones of SIK and MARK4 were subcloned into a NotI-modified pEGFP-C1 vector (Clontech) as BamHI–NotI fragments. For the SAXS (small-angle X-ray scattering) analysis, MARK2 from amino acid 6 to amino acid 350 [MARK2-(6–350)-peptide] was amplified by PCR from existing cDNA using the following primers: 5-agatctaactcagccacctctgctgatgagcag-3 and 5-gctgaaggccggggtttcagggtgatggtgtcg-3. The resulting PCR product was ligated into pCR 2.1 TOPO vector (Invitrogen), sequenced, subcloned as a BgII–NotI fragment into the BamHI–NotI site of pGEX-6P-1 expression vector (Amersham). The UBA domains of the AMPK-related kinases with an N-terminal HA tag were amplified by PCR, subcloned and sequenced in pCR 2.1 vector, and further subcloned into various expression vectors. The isolated UBA domains encompassed the indicated residues: BRSK1, amino acids 304–366; BRSK2, amino acids 287–349; MARK1, amino acids 319–380; MARK2, amino acids 289–329; MARK3, amino acids 317–376; MARK4, amino acids 341–400; SIK, amino acids 293–353; QSK, amino acids 334–394; QIK, amino acids 285–345; SNRK, amino acids 281–344. The UBA domains of AMPK-related kinases were subcloned from the above EBG vector into the pEBGFP-C2-TAP vector described previously [21], as BamHI–NotI inserts. Dr Colin Gordon (MRC Human Genetics Unit, Western General Hospital, Edinburgh, Scotland, U.K.) kindly provided the bacterial pGEX GST-Rhp23 expression vector [22] (Rhp23 is a functional homologue of the human excision repair enzyme Hhr23A). The bacterial pGEX GST fusion expression vectors for expression of NEDD8 (neural precursor cell-expressed developmentally down-regulated 8), SUMO1 (small ubiquitin-related modifier-1), SUMO2 and SUMO3 were obtained for Professor Ron T. Hay, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K. The expression vector employed to express full-length GST–SAKS1 (GST–stress-activated-kinase substrate-1) in the pGEX vector has been described [23] and was subcloned into the KpnI sites of pEBG6P in order to express GST–SAKS in mammalian cells.
Expression and purification of inactive MARK2 kinase for SAXS analysis
The pGEX expression construct encoding the kinase and UBA domains of MARK2-(6–350)-peptide with an N-terminal GST tag was transformed in E. coli BL21 cells. Cultures (4 litres) were grown at 37 °C in Luria broth containing 100 μg/ml ampicillin until the attenuance (D600) reached 0.7. Induction of protein expression was carried out by adding 200 μM isopropyl β-D-galactoside and the cells were cultured for a further 16 h at 21 °C. Cells were isolated by centrifugation, resuspended in 200 ml of ice-cold Buffer B and lysed in one round of freeze–thawing, followed by sonication to fragment DNA (six cycles of 30 s each). The lysate was centrifuged at 4 °C for 30 min at 26000 g and the supernatant was incubated for 1 h at 4 °C with 4 ml of glutathione–Sepharose. The beads were then washed three times with 40 ml of Buffer B containing 0.5 M NaCl, followed by three washes with Buffer C. The resin was then resuspended to a final volume of 15 ml in Buffer C. The amount of protein bound to the glutathione–Sepharose was quantified, and 50 μg of GST-tagged-PreScission™ protease (Amersham)/mg of protein bound to the resin was added. The resin was gently mixed for 16 h at 4 °C, and eluted protein collected and subjected to gel filtration (without concentration) on a Superdex-200-26/60 gel-filtration column equilibrated against 25 mM Tris/HCl (pH 7.5)/150 mM NaCl/1 mM dithiothreitol. The fractions containing homogenous inactive MARK2-(6–350)-peptide were pooled and concentrated, using a Viva-spin column (VivaScience, Lincoln, U.K.), to 8.2 mg/ml for the SAXS analysis.
Expression and purification of activated MARK2 kinase for SAXS analysis
MARK2-(6–350)-peptide was expressed and purified on glutathione–Sepharose as described above. After elution from glutathione–Sepharose with the GST-tagged-PreScission protease, MARK2-(6–350)-peptide was activated in a 15 ml reaction mixture containing 15 mg of MARK2-(6–350)-peptide, 3 mg of active LKB1–STRAD–MO25 complex, 10 mM MgCl2, 0.1 mM ATP, 50 mM Tris/HCl pH 7.5, 0.1 mM EGTA, and 0.1% (v/v) 2-mercaptoethanol. After incubation for 90 min at 30 °C, the specific activity of MARK2-(6–350)-peptide increased over 80-fold when assayed using the AMARA (AMARAASAAALARRR) peptide substrate [19]. The activated MARK2-(6–350)-peptide was applied to a Superdex-200-26/60 gel-filtration column equilibrated against 25 mM Tris/HCl (pH 7.5)/50 mM NaCl/1 mM dithiothreitol. The purified fraction from gel filtration containing MARK2 (which separated from the high-molecular-mass LKB1–STRAD–MO25 complex) was pooled and diluted in Buffer D in order to decrease the ionic strength to 15 mM NaCl. The mixture was applied to a Mono-S HR 10/10 column (Amersham Pharmacia Biotech) equilibrated in Buffer D at a flow rate of 2 ml/min. The column was washed in Buffer D, and the inactive and active forms of MARK2-(6–350)-peptide were separated using a linear 120 ml gradient of 0–0.5 M NaCl in Buffer D; fractions of size 4 ml were collected. The first-to-be-eluted peak that contained activated MARK2-(6–350)-peptide was pooled and concentrated to 5.8 mg/ml.
SAXS analysis
The inactive and LKB1-activated MARK2-(6–350)-peptide, were dialysed in 25 mM Tris/HCl (pH 7.5)/200 mM NaCl/1 mM dithiothreitol prior to the SAXS analysis. Immediately before SAXS data measurement, fresh dithiothreitol was added to a final concentration of 10 mM. SAXS data were collected on the EMBL (European Molecular Biology Laboratory) beamline X33 at DESY (Deutschen Elektronen-Synchrotron), Hamburg, Germany, and beamline 2.1 at SRS (Synchroton Radition Source), Daresbury Laboratory, Warrington, Cheshire, U.K.) employing a wavelength of 1.5 Å (1 Å=0.1 nm). Data were collected, and scattering intensities were recorded with 5 min exposure on a MAR-345 image plate detector. Data were corrected for buffer scattering using PRIMUS [24] and XOTOKO software [25] and checked for radiation damage by comparing subsequent measurements on the sample. Transformation to the real space p(r) function was performed with GNOM [26]. Ten individual reciprocal space dummy atom refinement were performed with GASBOR [27] and analysed with the DAMAVER suite [28] showing good consistency [inactive MARK2 NSD (normalized spatial discrepancy) 0.94±0.02 and active MARK2 NSD 1.03±0.01]. MARK2 kinase domain and UBA domain models were generated using MODELLER [29], using as templates cAMP-dependent protein kinase A (Protein Data Bank code 1CTP; 35.3% identity) and the Hhr23A UBA domain (Protein Data Bank code 1IFY; 48.5% identity). These models were then superposed on the dummy atom models using SUPCOMB [30]. All the Figures were made using Pymol software (DeLano Scientific LLC, South San Francisco, CA, U.S.A.; www.pymol.org).
Expression and purification of UBA domains and AMPK-related kinases in E. coli
The pGEX expression constructs encoding fragments of the UBA domain of ten AMPK-related kinases and the UBA domain of Rhp23 or AMPK-related protein kinases were transformed into E. coli BL21 cells. Cultures (1 litre) were grown at 37 °C in Luria broth containing 100 μg/ml ampicillin until the D600 was 0.8. Induction of protein expression was carried out by adding 100 μM isopropyl β-D-galactoside and the cells were cultured for a further 16 h at 26 °C. Cells were isolated by centrifugation, resuspended in 15 ml of ice-cold Lysis Buffer and lysed in one round of freeze/thawing, followed by sonication to fragment DNA. The lysates were centrifuged at 4 °C for 30 min at 26000 g, and the recombinant proteins were affinity-purified on glutathione–Sepharose beads. For ubiquitin-binding assays the protein bound to the beads was used and for kinase and phosphorylation assays the bound protein was eluted in Buffer A containing 20 mM glutathione.
Multi-ubiquitin binding assays
Multi-ubiquitin binding assays were performed as described in [22]. Binding assays were performed in a 0.4 ml reaction mixture containing Lysis Buffer, 50 mM NaCl, 0.1 mg/ml BSA, 5–20 μl glutathione–Sepharose beads conjugated to 1–2 μg of GST-fusion protein and 0.5 μg of tetra-ubiquitin or 1 μg of either K48- or K63-linked polyubiquitin chains (Affiniti Research Products) or 1 μg of ubiquitin (Sigma). After incubation at 4 °C for 2 h, the glutathione–Sepharose beads were isolated, washed three times with ice-cold Lysis Buffer containing 0.5 M NaCl, followed by three washes with Buffer A and resuspended in a 50 μl of Sample Buffer [6.8 mM Tris/0.1% (w/v) lithium dodecyl sulphate/0.5% (v/v) glycerol, containing 1 mM dithiothreitol, final pH 8.5]. The sample was subjected to PAGE and either subjected to Coomassie Brilliant Blue staining, to quantify the levels of GST–UBA domain present, or to immunoblot analysis with the anti-ubiquitin antibody, to detect associated ubiquitin.
In order to monitor binding of isolated UBA domains and AMPK-related kinases to NEDD8 and SUMO1, SUMO2 and SUMO3, the isolated UBA/AMPK-related kinases were expressed in HEK-293 cells from a pCMV5 vector with a HA tag, and 36 h after transfection the cells were lysed as described below. A 1 mg portion of cell lysates was incubated with 1–2 μg of purified GST-fusion protein of NEDD8 or SUMO isoforms bound to 20 μl of glutathione–Sepharose in a total volume of 1 ml of Lysis Buffer containing 0.1 mg/ml BSA. After incubation with gentle agitation for 2 h at 4 °C, the beads were isolated and washed three times with cold Lysis Buffer containing 0.5 M NaCl, followed by three washes with Buffer A. After the final wash the pellet was resuspended in 50 μl of Sample Buffer and subjected to immunoblot analysis with anti-HA antibodies.
Expression and purification of AMPK-related kinases
Ten 10-cm-diameter dishes of HEK-293 cells were cultured and cells in each dish was transfected with 10 μg of the pEBG-2T construct encoding wild-type or indicated mutant forms of AMPK-related kinase using the polyethyleneimine method [31]. The cells were cultured for a further 36 h and lysed in 0.5 ml of ice-cold Lysis Buffer, and the lysates the pooled and centrifuged at 4 °C for 10 min at 26 000 g. The GST-fusion proteins were purified by affinity chromatography on glutathione–Sepharose and were either not eluted from the resin (for ubiquitin-binding assays described above) or eluted in Buffer A containing 20 mM glutathione (for kinase and phosphorylation measurements).
Measurement of AMPK-related kinase catalytic activity
The activity of purified AMPK-related kinases was quantified by measurement of phosphorylation of the AMARA peptide substrate [19]. For the peptide assay, 20–100 ng of AMPK-related kinase was incubated in a 50 μl mixture containing 50 mM Tris/HCl, pH 7.5, 0.1% (v/v) 2-mercaptoethanol, 10 mM MgCl2, 0.1 mM EGTA, 0.1 mM [α-32P]ATP (300 c.p.m./pmol) and 200 μM AMARA peptide for 20 min at 30 °C. Incorporation of [32P]Pi into the peptide substrate was determined by applying 40 μl of the reaction mixture to P81 phosphocellulose paper and liquid-scintillation counting after washing the papers in H3PO4. One unit of activity was defined as that which catalysed the incorporation of 1 nmol of 32P into the substrate.
Activation of AMPK-related kinases by LKB1
A 30–100 ng portion of full-length wild-type or indicated mutant forms or fragments was incubated with or without 0.5 μg LKB1–STRAD–MO25 complex in Buffer A containing 5 mM magnesium acetate and 0.1 mM ATP in a final volume of 20 μl. After incubation at 30 °C for 30 min, 5 μl of the incubation mixture was quenched in 25 μl of ice-cold Buffer A containing 2 mM EDTA. The AMARA peptide kinase activities of the quenched AMPK-related kinase were determined as described above.
In order to monitor the effect of isolated UBA domain on activation of full-length wild-type SIK in vitro by the LKB1 complex, kinase assays were performed in the presence or absence of 4 μg of SIK UBA domain or Rhp23 UBA domain protein as described above.
Generation of stable cell lines and TAP (tandem affinity purification)
HEK-293 cells were cultured in 10-cm-diameter dishes to 50–70% confluence and transfected with 2 μg of pEGFP-C2-TAP construct encoding the UBA domain of SIK, BRSK2 or MARK3 using FuGENE™ reagent (Roche) according to the manufacturer's instructions. After 24 h, G418 (geneticin) was added to the medium to a final concentration of 3 mg/ml, and the medium was changed every 24 h, maintaining G418. After 14–20 days, individual surviving colonies expressing GFP (green fluorescent protein) fluorescence were selected and expanded. FACS analysis was also performed to ensure uniform expression of GFP in the selected cell lines and anti-HA immunoblotting analysis of lysed cells was performed to ensure that the expressed proteins migrated as a single-molecular-mass species at the expected apparent molecular mass (the isolated GFP-TAP tag adds 50 kDa to the molecular mass of a protein). The TAP-tagged UBA domains were affinity-purified and analysed as described previously [21].
Immunoprecipitation and kinase assay of endogenous AMPK-related kinase
Clarified HEK-293 cell extract containing 1 mg of total cell protein (stable cell lines expressing UBA domain of SIK, MARK3 or BRSK2) were incubated at 4 °C for 1 h on a shaking platform with 5 μg of the corresponding antibody, which had been previously conjugated to 5 μl of Protein G–Sepharose. The immunoprecipitates were washed twice with Lysis Buffer containing 0.5 M NaCl and twice with Buffer A. The immunoprecipitates were either subjected to protein kinase activity assay towards the AMARA peptide in a total assay volume of 50 μl as described above or analysed by immunoblotting.
Immunoblotting
Samples were heated at 70 °C for 5 min in 1×SDS Sample Buffer and subjected to PAGE and electrotransfer on to nitrocellulose membrane. Membranes were blocked for 1 h in 50 mM Tris/HCl (pH 7.5)/0.15 M NaCl/0.2% (v/v) Tween (TBST Buffer) containing 10% (w/v) dried skimmed milk. The membranes were probed with 1 μg/ml of indicated antibodies or 1-in-1000 dilution of commercial antibodies for 16 h at 4 °C in TBST Buffer containing 5% (w/v) dried skimmed milk. Detection was performed using horseradish-peroxidase-conjugated secondary antibodies and enhanced-chemiluminescence reagent.
Localization studies
The HEK-293 cells were cultured to 50% confluence on glass coverslips (number 1.5) in 60-mm-diameter dishes and transfected with 0.5 μg of pEGFP construct encoding wild-type or indicated SIK and MARK4 mutants, using FuGENE™ reagent according to the manufacturer's protocol. A duplicate set of dishes was used for each condition. The cells were then washed twice with PBS and permeabilized for 10 min with 1% (v/v) Nonidet P40 in PBS, followed by 1% (v/v) Nonidet P40 containing 0.5 μg/ml DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride; purchased from Fluka). The cells were imaged using a Zeiss LSM 510 META or Leica Sp2 AOBS confocal microscope.
RESULTS
UBA domains in AMPK-related kinases
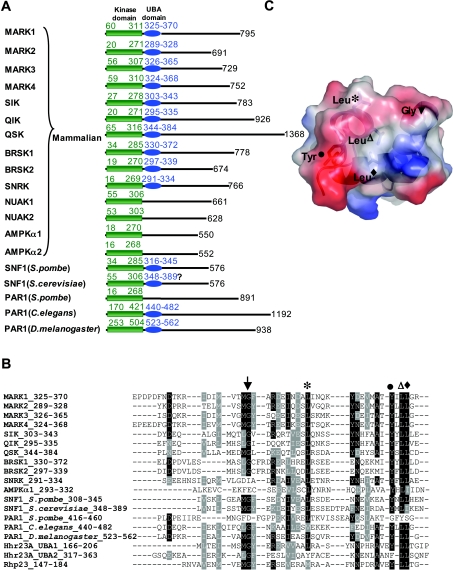
Sequence analysis indicates that ten of the 14 mammalian AMPK-related kinases activated by LKB1, possess a UBA domain that is located immediately after the kinase catalytic domain (Figure 1A). UBA domains comprise ≈45 amino acid residues, are found in diverse proteins and can function as ubiquitin- or polyubiquitin-interacting motifs [32]. The AMPK-related kinases are the only protein kinases in the human genome that possess a UBA domain [33]. C. elegans and Drosophila homologues of the MARK kinases (sometimes termed ‘Par1 kinases’) also possess a UBA domain. Although mammalian AMPKα1 or AMPKα2 catalytic subunits do not possess an obvious UBA domain, SNF1 (sucrose-non-fermenting kinase-1) homologues of AMPK in the fission yeast Schizosaccharomyces pombe and baker's yeast (Saccharomyces cerevisiae) possess a UBA domain (Figure 1A). Structural analysis has revealed that, although UBA domains of different proteins have a low level of sequence conservation, they nevertheless fold into a similar compact three-helix bundle fold that has a largely hydrophobic surface patch that can interact with ubiquitin or polyubiquitin chains [34–37]. Sequence alignments demonstrate that the residues located in this exposed region of UBA domains are conserved in the UBA domains of AMPK-related kinases, including the glycine residue found in most UBA domains (Figures 1B and 1C).
Figure 1. Presence of the UBA domain in AMPK-related kinases.
(A) Schematic representation of the 14 mammalian AMPK subfamily kinases that are activated by LKB1 as well as the homologues of these enzymes in the indicated species. The presence or absence of the UBA domain was determined employing the ScanProsite (http://us.expasy.org/tools/scanprosite/) and SMART (Simple Modular Architecture Research Tool) programs (http://smart.embl-heidelberg.de/). The residues encompassing the catalytic and UBA domains are in green and blue respectively. The number of residues present in each kinase is also indicated in black. Neither SMART or ScanProsite program recognizes the putative UBA domain indicated in S. cerevisiae SNF1, which is, therefore, indicated with ‘?’. (B) The alignment of the UBA domain sequences of AMPK subfamily protein kinases with UBA domains of Hhr23A and Rhp23. The conserved residues that lie on the exposed ubiquitin-binding surface of Hhr23A shown in (C) are indicated with different symbols. The identical residues are black and the conserved residues are in grey (C). Electrostatic potential surface of the Hhr23A UBA domain structure (PDB 1IFY [34]) showing the location of the conserved amino acid residues indicated in (B). Basic charged residues are displayed in blue and acidic charged residues are shown in red. Cartoon representation of the three helices bundle structure and sticks representation of the conserved residues are also shown in the Figure.
SAXS analysis of MARK2
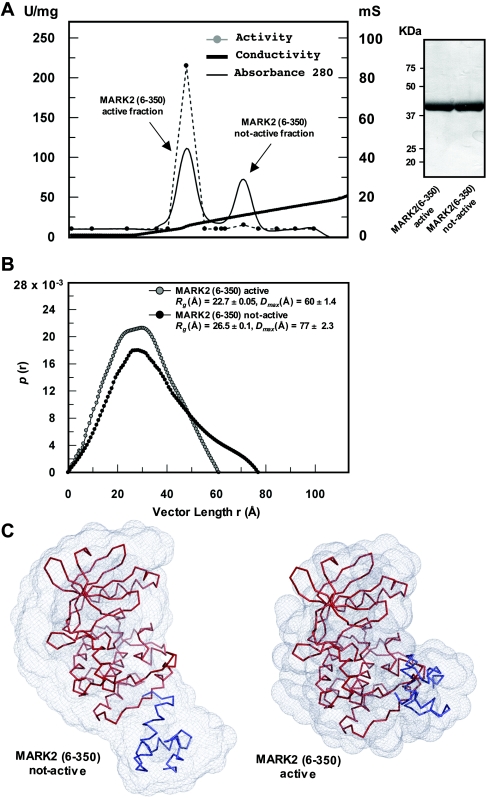
Limited tryptic proteolysis of full-length MARK2 and MARK3, expressed as active forms in HEK-293 cells, revealed the presence of a proteolyticlly resistant domain comprising residues 6–350 of MARK2 and residues 41–393 of MARK3, that encompass both the catalytic domain and UBA domain (F.Villa, unpublished work). To explore whether the UBA and catalytic domains in MARK isoforms might associate with each other, we undertook a conformational analysis of the MARK2-(6–350)-protein fragment employing SAXS analysis, a method that permits the visualization of the overall conformation of proteins [38]. MARK2-(6–350)-protein expressed in E. coli was inactive and could be activated following phosphorylation with LKB1 in vitro and separated from inactive MARK2 on cation-exchange chromatography as an earlier-to-be-eluted peak consistent with it being more phosphorylated (Figure 2A). The earlier-to-be-eluted active peak was only observed when MARK2-(6–350)-protein was phosphorylated with LKB1 complex, and MS confirmed the presence of the phosphorylated T-loop MARK2 peptide only in the earlier-to-be-eluted active fractions (results not shown). SAXS analysis revealed that activated MARK2 was more compact than inactive MARK2, as the Dmax value (the longest dimension of the molecule) and average radius of gyration (Rg) of activated MARK2-(6–350)-protein were significantly lower than that of inactive MARK2-(6–350)-protein (inactive MARK2: Rg=26.5 Å and Dmax=77 Å, compared with active MARK2: Rg=22.7 Å and Dmax=60 Å; Figure 2B). From the scattering data, the ATSAS suite of programs was used to generate ‘dummy atom’ models for both the active and inactive forms of MARK2-(6–350)-protein as described in the Materials and methods section. The inactive MARK2-(6–350)-protein was more elongated, with the UBA domain visible as domain separated from the larger catalytic domain (Figure 2C, left panel). In contrast, the activated MARK2-(6–350)-protein was more compact (Figure 2C, right panel) and the UBA domain was more closely associated with the large lobe of the catalytic domain than in inactive MARK2-(6–350)-protein.
Figure 2. SAXS analysis of inactive and LKB1 activated MARK2-(6–350)-peptide.
(A) Inactive recombinant MARK2-(6–350)-peptide was phosphorylated with the LKB1–STRAD–MO25 complex as described in the Materials and methods section and was subjected to chromatography on a Mono-S cation-exchange column. A280 (solid thin black line), conductivity (solid thick black line) and AMARA peptide kinase activity (broken grey line and filled circles) are plotted against eluted fractions. (B) Pair-distance distribution functions computed from scattering data of inactive non-LKB1 phosphorylated MARK2-(6–350)-protein (black circles) and LKB1-phosphorylated activated MARK2-(6–350)-protein (grey circles). Pair distances are shown as arbitrary units. (C) Comparison of the scattering solution shape of the model for MARK2-(6–350)-protein in the inactive conformation (left panel) and for MARK2-(6–350)-protein in the active conformation (right panel). MARK2 kinase domain model (red) and UBA domain (blue) are represented in ribbon format. The results of the scattering solution shape are representative of five independently run experiments.
UBA domains of AMPK-related kinases do not interact with ubiquitin-like molecules
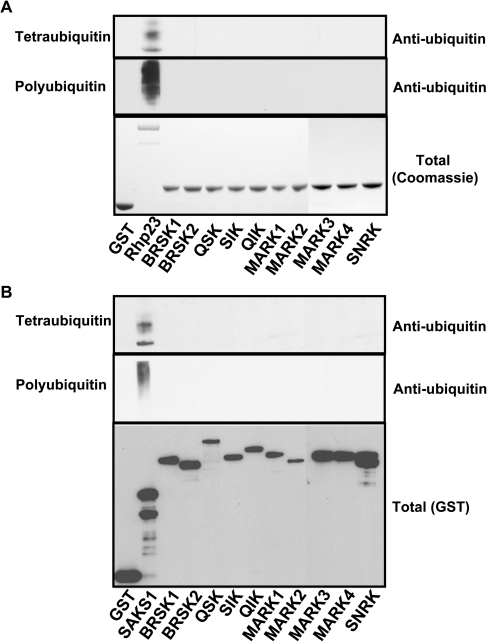
We next tested whether the isolated UBA domains of each of the ten AMPK-related kinases interacted with tetra-ubiquitin (Figure 3A, upper panel), K48-linked polyubiquitin (Figure 3A, middle panel) or K63-linked polyubiquitin (M. Jaleel, unpublished work). Employing standard binding assay that has been previously utilized to monitor binding of UBA domains to ubiquitin-like molecules, we failed to observe a significant interaction of the UBA domains of AMPK-related kinases with the ubiquitin species tested. In a parallel assays, the UBA-domain-containing protein Rhp23, interacted with tetra-ubiquitin, K48-linked polyubiquitin (Figure 3A) and K63-linked polyubiquitin (M. Jaleel, unpublished work), as reported previously [22]. We also found that the full-length UBA-domain containing AMPK-related kinases failed to interact with tetra-ubiquitin or K48-linked polyubiquitin under conditions which the full-length UBA-domain containing SAKS1 [23] protein bound to these ligands (Figure 3B). We also tested whether isolated UBA domains or full-length AMPK-related kinases could interact with mono-ubiquitin or the ubiquitin-like molecules NEDD8, SUMO1, SUMO2 or SUMO3 as described in the Materials and methods section, but observed no evidence of any significant interaction occurring (M. Jaleel, unpublished work).
Figure 3. UBA domain of AMPK-related kinases do not bind tetra-ubiquitin or polyubiquitin.
(A) GST-fusion proteins of the indicated UBA domains of AMPK subfamily kinases or the tandem UBA domains of Rhp23 bound to glutathione–Sepharose were incubated with either tetra-ubiquitin or K48-linked polyubiquitin. The beads were washed and the associated protein was eluted and subjected to PAGE. The gels were either stained with Coomassie Blue (lower panel) to visualize levels of GST-fusion proteins or immunoblotted with anti-ubiquitin antibody to detect associated ubiquitin forms (upper panels). (B) As above, except that full-length GST-fusion proteins of AMPK subfamily kinases were employed rather than isolated UBA domains. The full-length SAKS1 ubiquitin-binding UBA domain was employed as a control [23]. The level of GST-fusion proteins associated with the glutathione–Sepharose was assessed by immunoblot analysis employing a anti-GST antibody (lower panel). The results are representative of two or three separate experiments.
Role of UBA domain in controlling the activity and phosphorylation of AMPK-related kinases
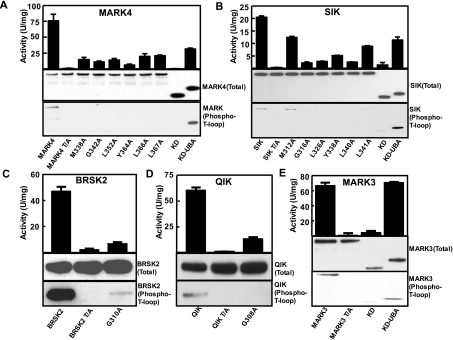
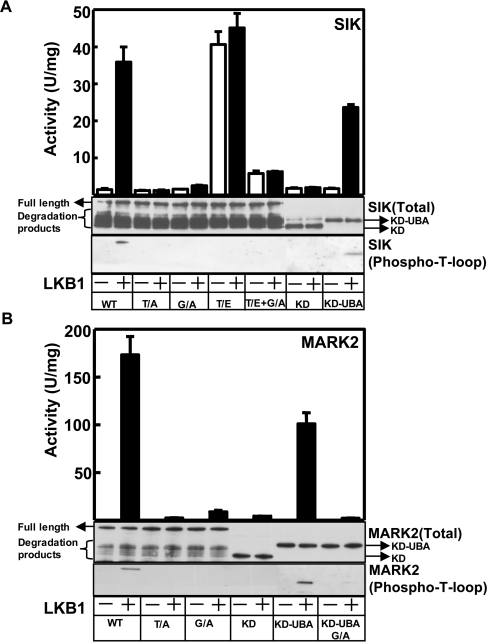
We next assessed how mutation of several of the conserved residues on the UBA domain (Figure 1B), affected catalytic activity and phosphorylation of the T-loop of MARK4 (Figure 4A) or SIK (Figure 4B). Wild-type MARK4 and SIK, when transfected in mammalian HEK-293 cells that express endogenous LKB1, were substantially active and phosphorylated at their T-loop. As expected, mutation of the T-loop threonine residue that is phosphorylated by LKB1 to alanine, abolished MARK4 or SIK activity and recognition of these enzymes with T-loop phosphospecific antibodies (Figures 4A and 4B). Interestingly, mutation of most of the conserved residues in the UBA domain of MARK4 (Figure 4A) or SIK (Figure 4B), including the glycine residue, markedly decreased catalytic activity and T-loop phosphorylation of these enzymes. Moreover, a fragment of MARK4 and SIK encompassing only the isolated catalytic domain lacking the UBA domain was almost devoid of activity and T-loop phosphorylation. In contrast, longer fragments of MARK4 and SIK that included the UBA domain were active and phosphorylated at their T-loop residue (Figures 4A and 4B). We also investigated how mutation of the conserved glycine residue in the UBA domain of BRSK2 (Figure 4C) and QIK (Figure 4D) affected the activity and T-loop phosphorylation of these enzymes. Similar to the findings with SIK and MARK4, mutation of the conserved glycine residue markedly suppressed BRSK2 and QIK catalytic activity as well as the T-loop phosphorylation. We also found that the isolated catalytic domain of MARK3 lacking the UBA domain was inactive and was not phosphorylated at the T-loop residue when expressed in HEK-293 cells, in contrast with the longer UBA-domain-containing fragment that was active and phosphorylated at the T-loop (Figure 4E). It should be noted that the correlation between the activity of AMPK-related kinases and their T-loop phosphorylation is not linear (Figure 4). This is likely to arise as a result of mutations that we introduced influencing catalytic activity independently of T-loop phosphorylation.
Figure 4. Role of the UBA domain in regulating AMPK-related kinase activity and T-loop phosphorylation.
(A–E) HEK-293 cells were transfected with constructs encoding GST-fusion proteins of full-length wild type (WT) or indicated mutants of the AMPK-related kinases. ‘T/A’ indicates mutants in which the LKB1 T-loop threonine residue is changed to alanine. KD is a fragment of the enzyme encompassing only the kinase domain (MARK4 residues 2–329; SIK residues 2–305, MARK3 residues 2–326) and KD-UBA is a fragment that encompasses the kinase domain and UBA domains MARK4 residues 2–400, SIK residues 2–350 and MARK3 residues 2–383). At 36 h post-transfection, the AMPK-related kinases were affinity-purified from the cell lysates using glutathione–Sepharose and assayed for AMARA-peptide kinase activity (top panel) or subjected to immunoblot analysis with an anti-GST antibody to quantify relative protein levels (Total) or immunoblotted with T-loop phosphoantibodies recognizing the LKB1 phosphorylated forms of these enzymes (Phospho-T-loop). Results of the kinase catalytic assays are presented as the mean catalytic activities±S.D. for assays carried out in triplicate. The results presented are representative of two or three independent experiments.
UBA domains are required for phosphorylation and activation of SIK and MARK2 by LKB1 in vitro
We next investigated how mutation of the UBA domains in SIK and MARK2 affected their phosphorylation and activation by LKB1 in vitro. These enzymes were expressed in a non-phosphorylated form in E. coli. Incubation of wild-type inactive SIK (Figure 5A) or MARK2 (Figure 5B) with LKB1 and MgATP markedly increased the T-loop phosphorylation and catalytic activity of these enzymes (Figure 5A). In contrast, mutants of these enzymes in which the conserved glycine residue was changed to alanine were barely phosphorylated or activated following incubation with LKB1 and MgATP. A fragment of SIK or MARK2 encompassing only the kinase domain and lacking the UBA domain were also not significantly activated or phosphorylated by LKB1. In contrast, a longer fragment of SIK (Figure 5A) and MARK2 (Figure 5B) that encompasses both the kinase and UBA domains was phosphorylated and activated by LKB1. Mutation of the conserved glycine residue in the UBA domain prevented phosphorylation and activation of the MARK2 kinase UBA-domain fragment by LKB1 (Figure 5B). Mutation of the T-loop threonine residue of SIK that is phosphorylated by LKB1 to a glutamic acid residue to mimic phosphorylation, significantly activated SIK (Figure 5A), as reported previously [19]. Interestingly, mutation of the conserved glycine residue within this threonine→glutamic acid mutant of SIK, markedly decreased catalytic activity, indicating that the UBA domain is required for maximal activity of the phosphorylated form of SIK.
Figure 5. Role of UBA domain in enabling SIK and MARK2 to be phosphorylated and activated by the LKB1 complex in vitro.
The indicated wild-type and mutant forms of SIK (A) and MARK2 (B) were expressed in E. coli and incubated in the presence (+) or absence (−) of purified LKB1–STRAD–MO25 complex in the presence of MgATP. The AMPK-related kinases were then assayed for AMARA peptide kinase activity (top panel, activity) or subjected to immunoblot analysis with an anti-GST antibody to quantify relative protein levels (Total) or immunoblotted with T-loop phosphoantibodies recognizing the LKB1 phosphorylated forms of these enzymes (Phospho-T-loop). Results of the kinase catalytic assays are presented as the mean catalytic activities±S.D. for assays carried out in triplicate. The results presented are representative of two or three independent experiments. T/A indicates mutants in which the LKB1 T-loop threonine residue is changed to alanine; G/A indicates mutants in which the conserved glycine residue in the UBA domain (see Figure 1B) is mutated to alanine; T/E indicates mutants in which the LKB1 T-loop threonine residue is changed to glutamic acid; KD indicates a fragment of the enzyme encompassing only the kinase domain (SIK residues 2–305, MARK2 residues 6–281) or KD-UBA, which comprises a fragment that encompasses the kinase domain and UBA domains (SIK residues 2–350, MARK2 residues 6–350). As indicated, a significant proportion of purified full-length GST-SIK and GST-MARK2 expressed in E. coli is degraded, but only the full-length non-proteolysed form is phosphorylated by LKB1.
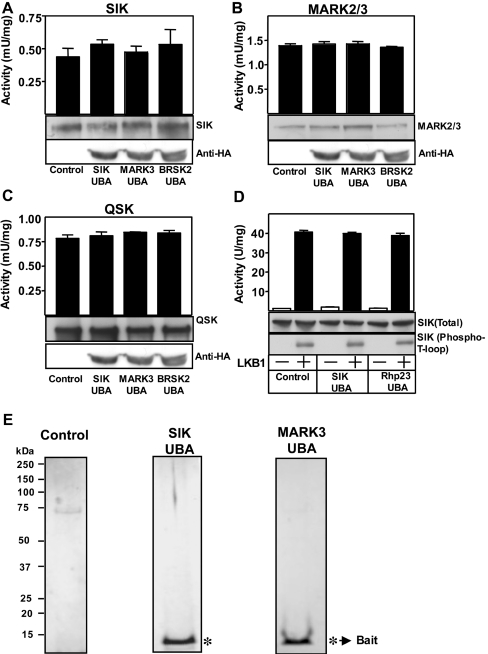
UBA domain does not act as a docking site for LKB1
To investigate whether the UBA domain of AMPK-related kinases was acting as a docking site for the LKB1–STRAD–MO25 complex, we stably overexpressed the isolated UBA domain of SIK, MARK3 and BRSK2 in HEK-293 cells and found that this did not influence the activity of endogenously expressed SIK (Figure 6A), MARK2/3 (Figure 6B) or QSK (Figure 6C). We also found that phosphorylation and activation of SIK by LKB1 in vitro was not affected by the addition of an excess of the UBA domain of SIK (Figure 6D). Moreover, tandem affinity purification of the isolated UBA domains of SIK and MARK3 UBA domains from HEK-293 cells, did not reveal any associated protein of significance (Figure 6E).
Figure 6. Evidence that the UBA domain of AMPK-related kinases do not interact with the LKB1–STRAD–MO25 complex.
The indicated endogenously expressed AMPK-related kinases, SIK (A), MARK2/3 (B) and QSK (C) were immunoprecipitated from HEK-293 cells lysates stably expressing HA-epitope-tagged UBA domain of SIK, MARK3, BRSK2 or control cell lines that express the empty vector. The SIK, MARK2/3 and QSK immunoprecipitates were subjected to in vitro kinase activity towards the AMARA peptide (top panel) and immunoblot analysis with indicated antibodies (middle panel) to quantify relative protein levels. The cell lysates prior to immunoprecipitation were also subjected to immunoblot analysis with an anti-HA antibody to quantify the expression of the UBA domain (lower panel). (D) Wild-type full-length SIK (150 ng) was expressed in E. coli and incubated in the presence (+) or absence (−) of purified LKB1–STRAD–MO25 complex (0.5 μg) and UBA domain of SIK or Rhp23 (4 μg) in the presence of MgATP. The activity of SIK towards the AMARA peptide substrate was measured (top panel, activity) or subjected to immunoblot analysis with an anti-GST antibody to quantify relative protein levels (Total) or immunoblotted with T-loop phosphoantibodies recognizing the LKB1 phosphorylated forms of these enzymes (Phospho-T-loop). Results of the kinase catalytic assays are presented as the mean catalytic activity±S.D. of assays carried out in triplicate. (E) The UBA domain of SIK and MARK3 were subjected to tandem affinity purification from HEK-293 cells stably expressing these domains, as described in the Materials and methods section. These were electrophoresed on a polyacrylamide gel and the protein bands were visualized following colloidal Coomassie Blue staining. * Indicates the bands identified as the bait UBA domain.
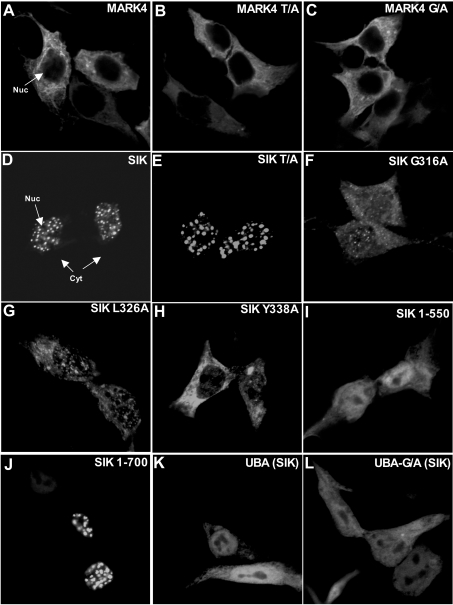
Effect of the UBA domain on the localization of MARK4 and SIK
MARK4 was largely localized in the cytoplasm and mutation of either the conserved UBA-domain glycine residue or the threonine residue in the T-loop that LKB1 phosphorylated did not significantly alter MARK4 cytosolic localization (Figures 7A–7C). Consistent with previous studies [21], we found that SIK was mainly localized in punctate regions of the nucleus (Figure 7D). Mutation of the T-loop threonine residue to alanine, which inactivates SIK and also abolishes its ability to interact with 14-3-3 protein isoforms [21], did not alter the ability of SIK to localize within punctate regions of the nucleus (Figure 7E). Strikingly, however, mutation of the conserved UBA-domain Gly316 (Figure 7F), Leu326 (Figure 7G) or Tyr338 (Figure 7H) residues markedly affected SIK localization, so that the mutant enzyme distributed throughout the cytoplasm and nucleus of cells. Although the UBA domain is required for the punctate nuclear localization of SIK, it does not directly confer the ability of SIK to localize within punctate nuclear regions, as we observed that a fragment of SIK comprising the kinase and UBA domains (residues 1–550) (Figure 7I) or the isolated UBA domain (Figure 7K) was localized throughout the cells, whereas a longer fragment (residues 1–700) had the same punctate localization as the wild-type enzyme (Figure 7J). Thus other non-catalytic residues of SIK, in addition to an intact UBA domain, are also required for the localization of SIK to punctate nuclear regions.
Figure 7. Role of the UBA domain in regulating the localization of MARK4 and SIK.
HEK-293 cells were transfected with the indicated constructs encoding for the expression of GFP-MARK4 (panels A–C) or GFP-SIK (panels D–L). At 24 h post-transfection the cells were fixed in 3% (v/v) paraformaldehyde and GFP localization was visualized directly by observing GFP fluorescence. The cells were imaged using Zeiss LSM 510 META or Leica Sp2 AOBS confocal microscopes. The cells shown are representative images obtained in two separate experiments. UBA indicates the isolated UBA domain of SIK (residues 293–353) and UBA-G/A indicates the isolated UBA domain of SIK in which the conserved glycine residue (Figure 1B) is mutated to alanine.
DISCUSSION
Our findings suggest that, rather than binding to polyubiquitin, the UBA domains of AMPK-related kinases may play an intrinsic structural role and are required for efficient phosphorylation and activation of these enzymes by LKB1 (Figures 4 and 5). In line with this hypothesis, SAXS analysis reveals that the phosphorylation of MARK2 by LKB1 results in a conformational change in which the UBA domain becomes more closely associated with the large lobe of the catalytic domain (Figure 2). This may explain why limited trypsin proteolysis of full-length active MARK2 and MARK3 yielded a protease-resistant fragment encompassing the catalytic and UBA domains. It is possible that phosphorylation of the T-loop threonine residue of MARK2 by LKB1 induces a conformational change within the large lobe of the kinase domain that promotes its association with the UBA domain. The SAXS analysis of the active/inactive forms of MARK2 suggest that the UBA domain, after LKB1 phosphorylation, moves towards the activation loop of the kinase. This might also protect the T-loop threonine residue from becoming dephosphorylated by protein phosphatases in vivo. The finding in Figure 5(A) that mutation of the conserved glycine residue in the UBA domain of the active threonine→glutamic acid mutant of SIK markedly inhibited enzyme activity, suggests that binding of the UBA domain to the kinase domain is required for the activation of the LKB1-phosphorylated enzyme. Further detailed structural studies are required in order to determine the molecular mechanism by which the UBA domain of AMPK-related kinases interacts and regulates the activity/conformation of the catalytic domain. Our findings suggest that, for future crystallographic studies on AMPK-related kinases, the conformation of the catalytic domains of these enzymes should preferably be studied with the presence of the UBA domain rather than in isolation.
Our results indicate that the low activity of UBA-mutant forms of AMPK-related kinases, when expressed in mammalian HEK-293 cells, is likely to result from the reduced T-loop phosphorylation of these enzymes (Figure 4). These observations could be explained if the UBA domain of AMPK-related kinases functioned as a docking site for the LKB1–STRAD–MO25 complex. We have investigated this possibility, but our analysis suggests that this is not the case. First, we were unable to demonstrate significant interaction between isolated UBA domains of AMPK-related kinases and any of the components of the LKB1 complex in overexpression binding studies in cells (M. Jaleel, unpublished work). Secondly, we generated stable HEK-293 cell lines expressing high levels of the UBA domains of MARK3 and SIK and failed to observe association of these domains with LKB1 or any other protein (Figure 6E). Thirdly, the activity of AMPK-related kinases in cell lines overexpressing UBA domains was not affected, which might be expected if these domains comprised an LKB1 docking site (Figures 6A–6C). Finally, we also observed that phosphorylation of SIK in vitro by the LKB1–STRAD–MO25 complex was not inhibited by the addition of a large excess of UBA domain (Figure 6D). As mutation or truncation of the UBA domain of AMPK-related kinases prevented phosphorylation by LKB1, we favour the notion that the UBA domain is likely to influence the structure of the catalytic domain in a manner that enables it to be efficiently recognized and phosphorylated by LKB1.
UBA domains are found on many of the mammalian AMPK-related kinases as well as homologues of these enzymes in C. elegans and Drosophila (Figure 1A), indicating that they are likely to perform a fundamental role. Although the mammalian AMPKα1 or AMPKα2 catalytic subunits do not possess an obvious UBA domain, the yeast Schiz. pombe SNF1 AMPKα counterpart does possesses a UBA domain in the same position of the catalytic domain as found in the mammalian AMPK-related kinases that is clearly recognized as a UBA domain by the SMART [39] and ScanProsite [40] motif searching programs (Figure 1). Analysis of the S. cerevisiae SNF1 sequence with the SMART and ScanProsite software did not detect a UBA domain, but sequence alignment of the S. cerevisiae and S. pombe SNF1 sequences demonstrate that the conserved residues making UBA domains are nevertheless conserved between these species (Figure 1B), suggesting that S. cerevisiae SNF1 does possess a UBA domain. Only 15 Schiz. pombe proteins possess a UBA domain and SNF1 is the only protein kinase possessing such a domain [41]. The crystal structure of the catalytic domain of S. cerevisiae SNF1, lacking the region which bears similarity with the UBA domain, has recently been reported [42]. It would be interesting to determine the structure of a longer fragment of SNF1 that encompasses the region bearing similarity to the UBA domain and how it might regulate catalytic function. These findings indicate that, during the evolutionary process, mammalian AMPKα1 and AMPKα2 may have lost their UBA domain, whereas this domain may have been maintained in mammalian AMPK-related kinases. Although sequence alignment of AMPKα1 and AMPKα2 with other AMPK-related kinases within the UBA domain region clearly demonstrate that most of the conserved residues that make up a UBA domain, including the conserved glycine residue, are lacking in AMPKα1 and AMPKα2, there is nevertheless some weak sequence conservation within this region (Figure 1B). Previous studies have shown that a fragment of AMPKα1 encompassing residues 1–312 lacking the UBA-domain homologous region, in contrast with the AMPK-related kinases investigated in the present study (Figure 4), is fully active and phosphorylated at its T-loop when expressed in mammalian cells [43]. Moreover, the 1–312 fragment of AMPKα1 is efficiently phosphorylated and activated by the LKB1–STRAD–MO25 complex in vitro [13], in contrast with mutants of MARK2 and SIK that lack the UBA domain (Figure 5). Interestingly, a longer fragment of AMPKα1 encompassing residues 1–392 that comprises the region showing weak homology with the UBA domain was markedly less active when expressed in mammalian cell lines than the 1–312 fragment [43], suggesting that the 312–392 region of AMPKα1 comprises an auto-inhibitory domain. These results indicate that the region of AMPKα1/AMPKα2 encompassing the residues showing low similarity in sequence to a UBA domain, play a distinct role on AMPKα1/AMPKα2 compared with AMPK-related kinases. It is possible that sequences on the regulatory AMPKβ and AMPKγ subunits may substitute for the role that the UBA domain of AMPK-related kinases plays.
In conclusion, we have for the first time investigated the role of the conserved UBA domain that is present on many of the AMPK-related kinases that are activated by LKB1. Our results indicate that the UBA domain on these enzymes functions in a different manner to UBA domains characterized to date and do not interact with ubiquitin or ubiquitin-like molecules. Instead these domains play a crucial role in enabling AMPK-related kinases such as MARK and SIK isoforms to be phosphorylated and activated by the LKB1–STRAD–MO25 complex. This effect is not mediated by the ability of the UBA domains to interact with the LKB1–STRAD–MO25 complex, but instead our findings suggest that the UBA domain interact directly with the catalytic domain of these enzymes, permitting them to be in a conformation that can be readily phosphorylated and activated by the LKB1 complex.
Acknowledgments
We thank the DESY EMBL staff Dr Dmitri Svergun and Dr Peter Konarev and SRS Daresbury staff Dr Günter Grossmann and Dr Kalotina Geraki for SAXS time and their help. We also thank the protein-production and antibody-purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] co-ordinated by Dr Hilary McLauchlan and Dr James Hastie for expression and purification of GST-Precision Protease and affinity purification of antibodies and the Sequencing Service (School of Life Sciences, University of Dundee, Scotland, U.K.) for DNA sequencing, as well as Dr Mark Peggie for subcloning of SAKS1 cDNA. D. M. F. van A. was supported by a Wellcome Trust Senior Research Fellowship and the European Molecular Biology Laboratory Young Investigator Programme (EMBO YIP). We are grateful to the Association for International Cancer Research, Diabetes UK, the Medical Research Council, the Moffat Charitable Trust and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck and Co. Inc., Merck KgaA and Pfizer) for financial support.
References
- 1.Hemminki A., Markie D., Tomlinson I., Avizienyte E., Roth S., Loukola A., Bignell G., Warren W., Aminoff M., Hoglund P., et al. A serine/threonine kinase gene defective in Peutz–Jeghers syndrome. Nature (London) 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 2.Jenne D. E., Reimann H., Nezu J., Friedel W., Loff S., Jeschke R., Muller O., Back W., Zimmer M. Peutz–Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat. Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 3.Tiainen M., Vaahtomeri K., Ylikorkala A., Makela T. P. Growth arrest by the LKB1 tumor suppressor: induction of p21(WAF1/CIP1) Hum. Mol. Genet. 2002;11:1497–1504. doi: 10.1093/hmg/11.13.1497. [DOI] [PubMed] [Google Scholar]
- 4.Tiainen M., Ylikorkala A., Makela T. P. Growth suppression by Lkb1 is mediated by a G1 cell cycle arrest. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9248–9251. doi: 10.1073/pnas.96.16.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudeau J., Sapkota G., Alessi D. R. LKB1, a protein kinase regulating cell proliferation and polarity. FEBS Lett. 2003;546:159–165. doi: 10.1016/s0014-5793(03)00642-2. [DOI] [PubMed] [Google Scholar]
- 6.Watts J. L., Morton D. G., Bestman J., Kemphues K. J. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467–1475. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
- 7.Martin S. G., St Johnston D. A role for Drosophila LKB1 in anterior–posterior axis formation and epithelial polarity. Nature (London) 2003;421:379–384. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- 8.Ossipova O., Bardeesy N., DePinho R. A., Green J. B. LKB1 (XEEK1) regulates Wnt signalling in vertebrate development. Nat. Cell Biol. 2003;5:889–894. doi: 10.1038/ncb1048. [DOI] [PubMed] [Google Scholar]
- 9.Baas A. F., Kuipers J., van der Wel N. N., Batlle E., Koerten H. K., Peters P. J., Clevers H. C. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–466. doi: 10.1016/s0092-8674(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 10.Baas A. F., Boudeau J., Sapkota G. P., Smit L., Medema R., Morrice N. A., Alessi D. R., Clevers H. C. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boudeau J., Baas A. F., Deak M., Morrice N. A., Kieloch A., Schutkowski M., Prescott A. R., Clevers H. C., Alessi D. R. MO25α/β interact with STRADα/β enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 2003;22:5102–5114. doi: 10.1093/emboj/cdg490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milburn C. C., Boudeau J., Deak M., Alessi D. R., van Aalten D. M. Crystal structure of MO25α in complex with the C terminus of the pseudo kinase STE20-related adaptor. Nat. Struct. Mol. Biol. 2004;11:193–200. doi: 10.1038/nsmb716. [DOI] [PubMed] [Google Scholar]
- 13.Hawley S. A., Boudeau J., Reid J. L., Mustard K. J., Udd L., Makela T. P., Alessi D. R., Hardie D. G. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods A., Johnstone S. R., Dickerson K., Leiper F. C., Fryer L. G., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003;13:2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 15.Shaw R. J., Kosmatka M., Bardeesy N., Hurley R. L., Witters L. A., DePinho R. A., Cantley L. C. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie D. G. The AMP-activated protein kinase pathway – new players upstream and downstream. J. Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto K., McCarthy A., Smith D., Green K. A., Hardie D. G., Ashworth A., Alessi D. R. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 2005;24:1810–1820. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw R. J., Lamia K. A., Vasquez D., Koo S. H., Bardeesy N., DePinho R. A., Montminy M., Cantley L. C. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lizcano J. M., Goransson O., Toth R., Deak M., Morrice N. A., Boudeau J., Hawley S. A., Udd L., Makela T. P., Hardie D. G., Alessi D. R. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaleel M., McBride A., Lizcano J. M., Deak M., Toth R., Morrice N. A., Alessi D. R. Identification of the sucrose non-fermenting related kinase SNRK, as a novel LKB1 substrate. FEBS Lett. 2005;579:1417–1423. doi: 10.1016/j.febslet.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 21.Al-Hakim A. K., Goransson O., Deak M., Toth R., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J. Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 23.McNeill H., Knebel A., Arthur J. S., Cuenda A., Cohen P. A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs. Biochem. J. 2004;384:391–400. doi: 10.1042/BJ20041498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 25.Boulin C. J., Kempf R., Koch M. H. J., McLaughlin S. M. Data appraisal, evaluation and display for synchroton radiation experiments: hardware and software. Nucl. Instrum. Methods. 1986;A249:399–407. [Google Scholar]
- 26.Semenyuk a. V., Svergun D. I. Gnom – a program package for small-angle scattering data-processing. J. Appl. Crystallogr. 1991;24:537–540. [Google Scholar]
- 27.Petoukhov M. V., Svergun D. I. New methods for domain structure determination of proteins from solution scattering data. J. Appl. Crystallogr. 2003;36:540–544. [Google Scholar]
- 28.Volkov V. V., Svergun D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A., Blundell T. L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Kozin M. B., Svergun D. I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 31.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicke L., Schubert H. L., Hill C. P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 33.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 34.Mueller T. D., Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein–protein interactions. J. Mol. Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 35.Varadan R., Assfalg M., Raasi S., Pickart C., Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Ohno A., Jee J., Fujiwara K., Tenno T., Goda N., Tochio H., Kobayashi H., Hiroaki H., Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure (Cambridge) 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Trempe J. F., Brown N. R., Lowe E. D., Gordon C., Campbell I. D., Noble M. E., Endicott J. A. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vachette P., Koch M. H., Svergun D. I. Looking behind the beamstop: X-ray solution scattering studies of structure and conformational changes of biological macromolecules. Methods Enzymol. 2003;374:584–615. doi: 10.1016/S0076-6879(03)74024-5. [DOI] [PubMed] [Google Scholar]
- 39.Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., Ponting C. P., Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gattiker A., Gasteiger E., Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformat. 2002;1:107–108. [PubMed] [Google Scholar]
- 41.Hartmann-Petersen R., Semple C. A., Ponting C. P., Hendil K. B., Gordon C. UBA domain containing proteins in fission yeast. Int. J. Biochem. Cell Biol. 2003;35:629–636. doi: 10.1016/s1357-2725(02)00393-x. [DOI] [PubMed] [Google Scholar]
- 42.Rudolph M. J., Amodeo G. A., Bai Y., Tong L. Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1. Biochem. Biophys. Res. Commun. 2005;337:1224–1228. doi: 10.1016/j.bbrc.2005.09.181. [DOI] [PubMed] [Google Scholar]
- 43.Crute B. E., Seefeld K., Gamble J., Kemp B. E., Witters L. A. Functional domains of the α1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 1998;273:35347–35354. doi: 10.1074/jbc.273.52.35347. [DOI] [PubMed] [Google Scholar]