Abstract
NO• (nitric oxide) is a pleiotropic signalling molecule, with many of its effects on cell function being elicited at the level of the mitochondrion. In addition to the well-characterized binding of NO• to the CuB/haem-a3 site in mitochondrial complex IV, it has been proposed by several laboratories that complex I can be inhibited by S-nitrosation of a cysteine. However, direct molecular evidence for this is lacking. In this investigation we have combined separation techniques for complex I (blue-native gel electrophoresis, Superose 6 column chromatography) with sensitive detection methods for S-nitrosothiols (chemiluminescence, biotin-switch assay), to show that the 75 kDa subunit of complex I is S-nitrosated in mitochondria treated with S-nitrosoglutathione (10 μM–1 mM). The stoichiometry of S-nitrosation was 7:1 (i.e. 7 mol of S-nitrosothiols per mol of complex I) and this resulted in significant inhibition of the complex. Furthermore, S-nitrosothiols were detected in mitochondria isolated from hearts subjected to ischaemic preconditioning. The implications of these results for the physiological regulation of respiration, for reactive oxygen species generation and for a potential role of S-nitrosation in cardioprotection are discussed.
Keywords: complex I, mitochondrion, nitric oxide, reactive oxygen species, S-nitrosothiol
Abbreviations: DETA-NONOate, (Z)-1-[2-(2-aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate; DTPA, diethylenetriaminepenta-acetic acid; GSNO, S-nitrosoglutathione; IPC, ischaemic preconditioning; MALDI-TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS; NO•, nitric oxide; ROS, reactive oxygen species; SNO, S-nitrosothiol(s)
INTRODUCTION
One of the many cellular reactions of NO• (nitric oxide) is S-nitrosation of protein thiols, resulting in the generation of SNO (S-nitrosothiols) [1,2]. Studies of the regulation and biological impact of this modification are technically difficult, due to degradation of SNO in response to light, low-molecular-mass thiols, and metal ions, as well as limited methodology to detect S-nitrosated targets. Nevertheless, SNO formation has been implicated in several physiological and pathological phenomena, including caspase inhibition [3], transport of NO• in the bloodstream [4], and effects on mitochondria [5–7]. Notably, formation of SNO has been found under hypoxic conditions [8] and it is thought that some of the cardioprotective effects of nitrite [9] may be mediated through SNO formation.
The mitochondrion is an essential organelle for normal cellular function, being the chief site of ATP synthesis and an integrator for apoptotic signalling [10,11]. Mitochondria interact with NO• at several levels, and one particularly well characterized example is the inhibition of complex IV (cytochrome c oxidase), via binding of NO• to its binuclear CuB/haem-a3 active site [12,13]. Several studies have suggested that complex IV is not the only site within the respiratory chain that can be inhibited by NO• [6,14,15], thereby alluding to additional NO•-dependent control points within mitochondria.
There are several reasons why S-nitrosation may be an important mitochondrial regulatory mechanism. Mitochondria contain sizeable thiol pools, are abundant in transition metals, and have an internal alkaline pH, all of which are known to modulate SNO biochemistry [16]. In addition, mitochondria are highly membranous and sequester lipophilic molecules such as NO•, and the formation of the putative S-nitrosating intermediate N2O3 is enhanced within membranes [17]. Thus the mitochondrial respiratory chain embedded within the inner membrane would seem to be an ideal target for S-nitrosation.
Previous studies have suggested that SNO may affect various parts of the respiratory chain. In particular, indirect evidence exists for S-nitrosation of complex I, the primary point of electron entry to the chain [5,6,18,19]. In these studies complex I inhibition upon exposure to NO• was reversed by SNO-degrading processes, such as exposure to light, or low molecular mass thiols including GSH and dithiothreitol. Although this provided strong evidence that complex I was a target for S-nitrosation, it is notable that NO• inhibition of complex IV is also sensitive to light [20], and to date there has been no direct measurement of SNO formation within complex I or determination of which peptides are S-nitrosated. Thus the aim of the present study was to develop methodology for the direct detection of SNO within complex I. Through the use of complex I isolating systems (blue native gel electrophoresis [21] and size exclusion chromatography [22]), in combination with SNO detection methods (chemiluminescence [23] and biotin switch [24]), S-nitrosation of complex I was directly measured and a target subunit was identified. In addition, consistent with their potential role in cardioprotection, SNO were detected in mitochondria isolated from perfused hearts subjected to IPC (ischaemic preconditioning).
MATERIALS AND METHODS
Materials
Male Sprague–Dawley rats (Rattus norvegicus), 200 g body mass, were obtained from Harlan. All chemicals were reagent grade and purchased from Sigma–Aldrich, except Superose 6 (GE Biosciences), DETA-NONOate, {(Z)-1-[2-(2-aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate; Alexis} and EZ-link biotin HPDP {(N-[6-(biotinamido)hexyl]-3′-(2′-pyridyl-dithio)propionamide; Pierce}. Peroxynitrite was synthesized via the reaction of NaNO2 with acidified H2O2 as previously described in [25], was quantified spectrophotometrically (302 nm, ϵ=1670 M−1·cm−1), and stock solutions were prepared in 10 mM NaOH. GSNO (S-nitrosoglutathione) was synthesized via the reaction of GSH with acidified NaNO2 as detailed in [26], precipitated using acetone, filtered and washed with ether under vacuum, freeze dried, and quantified using the Saville assay [27]. In addition SNO content was monitored spectrophotometrically (338 nm, ϵ=855 M−1·cm−1).
Mitochondrial isolation, incubations, and assays
Rat heart mitochondria were isolated using differential centrifugation, as previously described in [28]. Protein was determined by the Folin-phenol method [29] against a standard curve constructed using BSA. Mitochondrial incubations and all subsequent steps were performed in the dark. Mitochondrial protein (1 mg) was suspended in 1 ml of mitochondrial respiration buffer [28] containing respiratory substrates glutamate (5 mM), malate (2.5 mM) and succinate (5 mM). Nitric oxide donors (GSNO 1 mM or DETA-NONOate 2 mM) were then added, and the suspension incubated at 37 °C for 40 min with periodic aeration. Samples were centrifuged (5 min, 14000 g), and the supernatants removed. The pellets were then resuspended in 1 ml of respiration buffer and centrifuged again (5 min, 14000 g). Pellets were frozen in liquid N2 for subsequent analysis. For ONOO− treatments, 4 μl of 25 mM ONOO− stock (100 μM final concentration) was added to the cap of the reaction tube, the cap quickly closed and the tube shaken immediately. The process was then repeated twice at 30 s intervals, for a total of 3 additions. Mitochondria were then pelleted, washed, and frozen as described for GSNO samples. Complex I activity was assayed as the rotenone-sensitive oxidation of NADH, at 340 nm in the presence of coenzyme Q1 [5]. Complex IV was assayed by monitoring the cyanide-sensitive first-order oxidation of reduced cytochrome c, at 550 nm as previously described [12].
Blue-native gel electrophoresis and protein extraction
All procedures were performed in the dark. Blue-native gel electrophoresis was carried out as previously described in [21] with minor adjustments, loading 200 μg of mitochondrial protein per well. High-blue cathode buffer contained DTPA (diethylenetriaminepenta-acetic acid; 100 μM) and EDTA (1 mM). In addition DTPA (100 μM) was added to the buffer used for preparing gradient gels. Gels were run at a constant 40 V for 6 h, and then cut into (∼23) 2 mm bands, which were frozen at −80 °C. Prior to chemiluminescent analysis (see below), protein extraction from the gel was performed at 22 °C. Each band was homogenized in 100 μl of gel extraction buffer [6 M urea, 1×PBS (pH 7.2), 100 μM DTPA, 1 mM EDTA, and 1% (w/v) dodecyl maltoside]. The homogenate was then centrifuged at 2000 g at 22 °C for 2 min. The supernatant was removed and placed in a clean tube on ice. Additional buffer was then added to the gel pieces, and the homogenization/centrifugation process repeated three more times, yielding a final extracted sample of approx. 300 μl volume.
Superose 6 column chromatography
A 25 cm×1 cm Bio-Rad glass Econo-Column™ was used, with appropriate flow adaptor (Bio-Rad Laboratories) and a Masterflex™ peristaltic pump (Cole Parmer). Mitochondrial protein (3 mg) was extracted in 300 μl of column running buffer [50 mM Tris (pH 7.7), 50 mM KCl, 100 μM DTPA, and 1% (w/v) lauryl-maltoside]. The sample (250 μl) was loaded on to the column, which was run at a flow rate of 1 ml/min. Fractions (200 μl) were collected, and their A280 measured to determine relative protein content. In addition, the SNO content (see below) and complex I activity (see above) of each fraction were analysed.
Chromatography experiments were performed over several months, with slight variations in the column dimensions. Therefore, for data comparison the relative position of eluted column fractions was expressed as a percentage of the total column bed volume. For example if the column had a bed volume of 20 ml, then a fraction at 1 ml elution volume would correspond to 5% of the bed volume. Within a given data set the SNO, protein, and complex I activity profiles were all internally consistent.
Chemiluminescent and biotin-switch analysis of SNO content
Chemiluminescent analysis of SNO content was performed on samples originating from blue-native gels, intact mitochondria and Superose 6 chromatography fractions, as described in [23]. Blue-native samples (300 μl) were divided among three tubes to undergo preparatory chemistry, (Figure 1) comprising addition of either (i) 40 μl of extraction buffer (see above), (ii) 20 μl of buffer plus 20 μl of 5% (w/v) sulphanilamide, or (iii) 20 μl of 5% (w/v) sulphanilamide plus 20 μl of 50 mM HgCl2. Samples of intact mitochondria (1 mg pellets) were solubilized in 300 μl of extraction buffer (see above) and subjected to the same chemistry. Fractions originating from Superose 6 chromatography (200 μl) were divided among three tubes (50 μl), to which were added either (i) 10 μl column running buffer, (ii) 5 μl of 5% sulphanilamide plus 5μl of buffer, or (iii) 5 μl of 5% sulphanilamide plus 5 μl of 50 mM HgCl2. All samples were reacted for 2 min, then 50 μl of each injected into an argon-fed purge vessel containing tri-iodide reagent [23], connected to a Seivers NOA 280 chemiluminescent NO• analyser (Ionics Instruments). Figure 1 shows a typical trace for a sample of GSNO-treated mitochondria. Each sample was run in duplicate and quantified using a standard curve created with known concentrations of NaNO2.
Figure 1. Chemiluminescent analysis of SNO.
Upper panel shows a schematic of the derivatization chemistry. Each sample was divided into three aliquots. Sulphanilamide reacts with NO2−, HgCl2 reacts with SNO. The lower panel shows a typical chemiluminescent NO analyser (NOA) trace, for injection of samples derived from GSNO-treated isolated rat heart mitochndria.
For biotin-switch analysis, intact mitochondrial pellets (1 mg of protein solubilized in extraction buffer, see above) or chromatography column fractions were analysed according to [24]. Samples were separated in duplicate on non-reducing SDS/12% PAGE gels; half the gel was stained with Coomassie Blue, and the other half blotted and probed with streptavidin–horseradish peroxidase and ECL® detection (GE Biosciences). Blots and stained gels were aligned, and S-nitrosated proteins excised, trypsinized, and identified by peptide mass fingerprinting at the University of Rochester's Proteomics Core facility (Protein/Peptide Facility, University of Rochester Medical Center, Room G-9843, 601 Elmwood Avenue, Rochester, NY 14642, U.S.A.). Mass spectra [MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight)] were analysed using the Mascot™ algorithm (http://www.matrixscience.com).
Heart perfusions and mitochondrial isolation
Langendorff heart perfusions were performed as previously described in [28] in constant flow mode, with the exception that all steps, including subsequent mitochondrial isolations, were performed in the dark. Hearts were preconditioned by subjecting them to three 5 min periods of global ischaemia, with 5 min reperfusions in between. Control hearts were continuously perfused for 30 min. At the end of perfusion protocols, hearts were removed on to ice-cold buffer and mitochondria prepared as described above.
Statistics
Blots, gels and chromatographs are representative of at least three independent experiments. Other data are presented as means±S.E.M. (n>3). Statistical differences between control and GSNO treatment groups were determined by Student's t test.
RESULTS AND DISCUSSION
Mitochondrial complex I has previously been suggested to be inhibited by S-nitrosation [5,6,19]; however, to date there has been no direct measurement of this. The aim of the present study was to apply novel methods to measure S-nitrosation within complex I.
S-nitrosation conditions
Figure 2(A) illustrates the nitrosation capabilities of a variety of NO• donors added to mitochondria. A biotin-switch analysis [24] was performed on the entire mitochondrial protein extract. It should be noted that the bands detected in the controls of all biotin-switch experiments are due to endogenous biotin-containing mitochondrial proteins (e.g. pyruvate carboxylase). These proteins were not removed prior to the assay, due to the danger of losing additional proteins during such purification, and this ensured a more complete picture of mitochondrial SNO content. Figure 2(A) clearly shows that GSNO results in far more S-nitrosation than DETA-NONOate or ONOO−. While ONOO− has previously been hypothesized to inhibit complex I via SNO formation [5], and may indeed form SNO under certain conditions [30], the current data suggest this is not the case in isolated mitochondrial preparations. Factors such as the components of the buffer system, or the ONOO− treatment regimen [bolus addition compared with in situ generation from a donor compound such as SIN-1 (3-morpholinosydnonimine hydrochloride)], may account for discrepancies in the ability of ONOO− to cause S-nitrosation. Overall the results in Figure 2(A), along with previous studies [15,31] including the biological impact of mitochondrial glutathione status on complex I activity [6], suggest that GSNO is a useful S-nitrosating agent in mitochondria.
Figure 2. Characterization of mitochondrial S-nitrosation.
(A) S-nitrosation pattern of rat heart mitochondria, visualized by biotin-switch assay, following treatment with various NO• donors as described in the Materials and methods section. Molecular mass markers (kDa) are shown to the right of the blot. (B) Dose response to GSNO. Mitochondria were treated with various doses of GSNO and analysed as described for (A). (C) SNO content analysed by chemiluminescence. Mitochondria were treated with various doses of GSNO then analysed by chemiluminescence as detailed in the Materials and methods section. (D) NO2− content analysed by chemiluminescence, as described for SNO in (C). In (C) and (D), axes are broken between 500 μM and 1 mM GSNO to enhance detail at lower concentrations. Results are expressed as the means±S.E.M. of at least 3 independent experiments. Ctrl, control; mito′, mitochondrial.
Figure 2(B) shows a dose response of mitochondrial protein SNO formation to GSNO over the range 50–250 μM. Remarkably, increasing the GSNO dose from 100 μM to 250 μM did not appear to cause a significant elevation in protein S-nitrosation. However, since the biotin-switch assay has a limited linear dynamic range, and only detects protein-bound SNO, it was decided to also perform a chemiluminescent analysis over a wider range of GSNO concentrations (10 μM to 1 mM). The results (Figure 2C) show that as GSNO treatment concentration increased, the total SNO (i.e. free plus protein-bound) increased in a classical hyperbolic manner, approaching saturation at approx. 300 μM GSNO. Together these results (Figures 2B and 2C) suggest that formation of protein-bound SNO saturates at approx. 250 μM SNO.
Notably, the results in Figure 2(C) show that at 1 mM GSNO, total SNO formation increases drastically. This may represent the additional formation of non-protein-bound SNO from intra-mitochondrial low molecular mass thiols, or possible carry-over of excess unreacted GSNO into the chemiluminescent assay. However, we consider the latter possibility unlikely since the mitochondrial pellets were washed free of excess GSNO. Furthermore, 1 mM GSNO treatment equates to 1 μmol of SNO/mg of protein, whereas the maximum amount detected was 27 nmol/mg.
The results in Figure 2(D) also show that at GSNO concentrations above 250 μM, NO2− levels climb sharply. Since these samples were prepared from mitochondria that had been centrifuged following GSNO treatment and prior to analysis, this suggests that any NO2− or non-protein-bound SNO detected was either trapped inside the mitochondrial matrix during the centrifugation step, or formed from degradation of protein-bound SNO during the sample work-up period for chemiluminescent analysis.
Fe-nitrosyl was virtually undetectable under all conditions examined and quantitatively never reached more than 5% of the SNO content. Therefore, Fe-nitrosyl values were ignored in subsequent experiments and calculations. Together, the results in Figure 2 highlight that protein S-nitrosation can occur at low concentrations of GSNO, whereas at higher GSNO levels secondary reactions such as free SNO or NO2− formation may occur. These results in isolated mitochondria are similar to those previously observed upon treatment of whole cells with GNSO [32].
Analysis of complex I S-nitrosation by blue-native gel electrophoresis and chemiluminescence
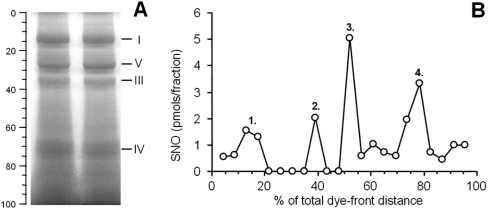
Initial efforts to detect S-nitrosation relied on the isolation of complex I by blue-native gel electrophoresis followed by chemiluminescent SNO analysis. Figure 3(A) shows a typical blue-native gel from the present study, demonstrating that the alterations to the established blue-native method to preserve SNO (metal chelators, running the gel at low voltage in the dark) did not affect the overall pattern of respiratory complex separation (compare with [21]). Excision of bands from the gel permitted the construction of a gel SNO profile, as seen in Figure 3(B). The profile exhibited four major peaks that correlated to gel segments enriched in complex I (peak 1), complex III (peak 2), unknown (peak 3), and complex IV (peak 4). Interestingly, SNO detection in peak 3 suggests the presence of a high-molecular-mass protein other than a respiratory complex that is S-nitrosated, even though it does not appear to be particularly abundant, based on low staining intensity in the corresponding region of the blue-native gel. The protein(s) contained in this band currently await identification.
Figure 3. Blue-native gel electrophoresis and chemiluminescent SNO analysis.
(A) Separation of the mitochondrial respiratory complexes using blue-native gel electrophoresis. The position of the complexes is shown by Roman numerals to the right, and the percentage of total distance down the gel (relative to the dye front) is shown to the left. (B) Chemiluminescent SNO scan of proteins extracted from 2 mm segments of the blue native gel. Each point represents the amount of SNO detected in a given 2 mm gel slice. SNO content data are means of 2 determinations, and overall the data are representative of at least 3 independent experiments.
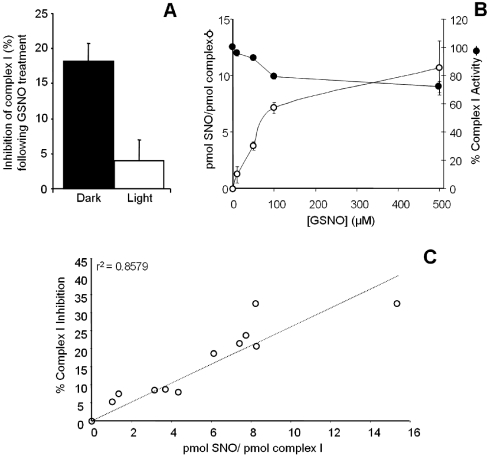
Since the complex I region of the blue native gel contained SNO, it was investigated whether the same S-nitrosating conditions (1 mM GSNO treatment) would also affect complex I activity (rotenone-sensitive NADH/Co-Q1 oxidation/reduction). Figure 4(A) demonstrates that GSNO treatment resulted in similar complex I inhibition to that seen in previous studies [5,6]. This inhibition was significantly reversible after exposure of the mitochondria to light, suggesting the inhibition was an S-nitrosation-dependent mechanism.
Figure 4. Inhibition of complex I following GSNO treatment.
(A) Complex I activity was monitored in mitochondria treated with 1 mM GSNO. For ‘dark’ samples the entire experiment including the assay was performed in the dark. For ‘light’ samples the GSNO treatment was in the dark, but the mitochondrial pellet was exposed to an intense white light source prior to the complex I assay. (B) Complex I activity was monitored using mitochondria treated with 10–500 μM GSNO. Mitochondria from the same suspension were carried over to blue-native gel electrophoresis followed by chemiluminescent SNO detection. Stoichiometric quantification of S-nitrosation was calculated as detailed in the text. (C) Percentage complex I inhibition was plotted against complex I SNO. The line fit and r2 value were obtained by regression analysis.
In addition to this single GSNO concentration, samples were also prepared at a variety of GSNO concentrations (10 μM–500 μM). Complex I activity was then measured, and the samples were run on blue-native gels for quantification of complex I S-nitrosation. Assuming that heart mitochondria contain approx. 50 pmol of complex I per mg of protein [33], and therefore known amounts of complex I were loaded into each lane of the gel, the degree of complex I S-nitrosation could be calculated from the SNO measured in each complex I excised band and the results expressed as pmol of SNO/pmol of complex I. Figure 4(B) shows both the stoichiometric S-nitrosation and inhibition of complex I, as a function of GSNO concentration. S-nitrosation reached a plateau at approx. 7:1 mol/mol ratio, whereas inhibition reached a plateau at approx. 20–25%. This apparent mis-match between the degree of S-nitrosation (7:1) and the degree of inhibition (25%) may reflect S-nitrosylation of complex I on multiple subunits and this possibility was subsequently investigated (see below). Alternatively, since the complex I assay is performed at an analytical wavelength of 340 nm, which is very close to the absorption maxima of SNO at 335 nm, it is possible that the degree of inhibition by S-nitrosation is underestimated, as the S-nitrosation itself may be destroyed during the complex I activity assay. Nevertheless, support for a link between S-nitrosation and inhibition is provided by the data in Figure 4(C), showing a linear correlation (r2=0.86) between these parameters.
In addition, the stoichiometric data in Figure 4 are quantitatively compatible with the data in Figures 2 and 3. At the 100 μM GSNO treatment point, complex I SNO (from Figure 4B) is 7 pmol/pmol. If 1 mg of mitochondrial protein contains 50 pmol of complex I [33], then it would contain 350 pmol of complex I SNO (i.e. 7×50). Simultaneously, Figure 2(C) shows that total SNO at the 100 μM GSNO treatment point is approx. 1700 pmol/mg, and Figure 3(B) suggests that approx. 20% of this total (integrated area under the curve) originates from complex I. Since 20% of 1700 is 340 pmol of complex I SNO, these numbers are internally consistent.
Complex IV activity was also measured utilizing the same mitochondrial samples used for the complex I SNO detection and activity assays. Complex IV activity was inhibited by approx. 5% after treatment with GSNO (results not shown), although we did not pursue this further since the inhibition of complex IV by NO• binding to its binuclear centre has already been well established [12,14,34,35].
Analysis of SNO by Superose 6 chromatography and chemiluminescence
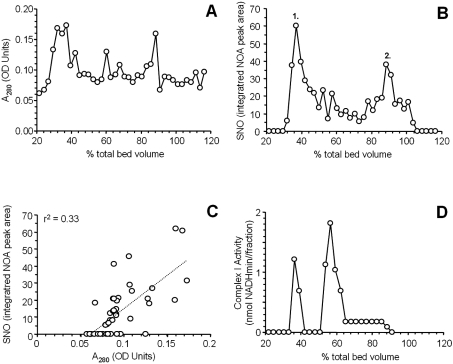
Owing to the technical problems associated with excising multiple bands from a blue-native gel, extracting the proteins, and performing derivative chemistry prior to NO• chemiluminescent analysis, it was decided to seek a liquid phase high-throughput separation method for mitochondrial respiratory complexes. Superose 6 chromatography has been previously used for such separation of high molecular mass proteins [22], and Figure 5(A) shows the protein profile (A280) of mitochondrial proteins separated on a Superose 6 column. Chemiluminescent analysis of each fraction from the column was then used to construct an SNO profile, as shown in Figure 5(B). Notably, most of the SNO appeared to be concentrated within two regions of the chromatogram, eluting at approx. 35% and 85% of the column bed volume (labelled as peaks 1 and 2 respectively). Comparison of the SNO content and protein content of each fraction (Figure 1C) revealed no significant correlation (r2=0.33, Figure 5C), indicating that the peaks of SNO content are not simply those fractions which contain the most protein.
Figure 5. Superose 6 column chromatography and chemiluminescent SNO analysis.
(A) Profile of the protein content in each fraction collected from the column. The relative position of each fraction in the collection profile is expressed as a percentage of the total column bed volume. (B) Chemiluminescent SNO scan of the column fractions. (C) Plot of protein content against SNO content of column fractions. The fit line and r2 value are for a linear regression analysis. (D) Complex I activity of the column fractions. All results are representative of at least three independent experiments. Abbreviation: NOA, NO analyser.
Figure 5(D) shows the complex I content of the column fractions, indicating that the fraction with most SNO content (peak 1 of Figure 5B) correlates with a peak of complex I activity. Whereas another peak of complex I activity also appeared further down the chromatogram, this peak did not appear to have a high SNO content. Complex I is a labile molecule that can break down from its usual approx. 900 kDa L-shaped form into sub-complexes of 400–500 kDa [36]. Consistent with such breakdown either in the column itself or during the preceding preparatory steps, approximate calibration of the column with high-range molecular mass markers (results not shown) showed that the second peak of complex I activity corresponded to a mass of approx. 450 kDa.
Biotin-switch analysis of complex I enriched fraction
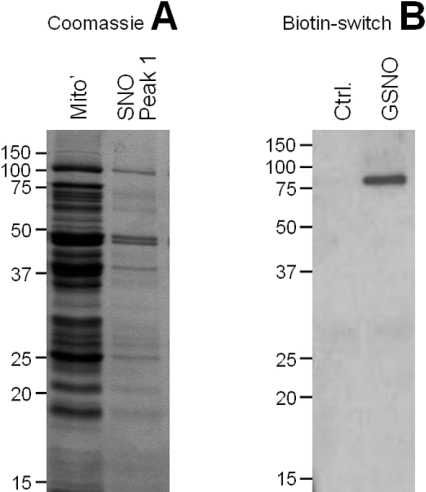
In order to identify S-nitrosated peptides, peak 1 in Figure 5(B), containing the highest SNO content and corresponding to intact complex I, was selected for further analysis by the biotin switch assay. Figure 6(A) shows that this fraction contained significantly fewer proteins than the whole mitochondrial extract, making it possible to perform a one-dimensional gel analysis and obtain sufficient resolution to excise bands from the gel for identification by peptide mass fingerprinting (MALDI-TOF-MS). Figure 6(B) shows the results of the biotin switch assay on this fraction, from both control and GSNO-treated mitochondria, indicating that a single peptide within the fraction is S-nitrosated. Peptide mass fingerprint analysis of the corresponding band in the gel revealed the identity of this protein as the 75 kDa subunit of complex I (accession number 51858651). The MOWSE score for the excised protein was 76 (>58 significant, P<0.05) with 33% sequence coverage.
Figure 6. Biotin-switch analysis of SNO-enriched peak 1.
(A) Coomassie Blue stained gel, comparing the proteins found in a total mitochondrial protein extract with those found in SNO peak 1 from Superose 6 column chromatography (Figure 5B). (B) Streptavidin–horseradish peroxidase Western blot of SNO peak 1 from Superose 6 column chromatography (Figure 5B) subjected to biotin-switch derivatization. Results are shown for control and GSNO-treated mitochondria, and are representative of at least three independent experiments. Mito′, mitochondrial; Ctrl, control.
Together the data in Figure 3(B) and Figure 5(B) (SNO content), and Figure 6(B) (SNO target identification) reveal that approx. 20% of the total protein-bound SNO content of GSNO-treated mitochondria was contained within a single S-nitrosated peptide, the 75 kDa subunit of complex I. There are approx. 2000 proteins within the mitochondrial proteome, and >95% of all proteins have at least one cysteine residue. Therefore the presence of such a large fraction of SNO within a single peptide suggests that the 75 kDa complex I subunit is a highly specific S-nitrosation target. In addition, the data from Figure 4(B), which indicate an approx. 7:1 stoichiometry of S-nitrosation, suggest that multiple cysteine residues within this peptide may be S-nitrosated. There are 11 cysteines within the 75 kDa subunit and which ones are more prone to S-nitrosation is not known. However, a previous study suggested that Cys137 is a likely target for S-nitrosation [6], based on its residence within a putative S-nitrosation consensus sequence.
Physiological and pathological implications of complex I S-nitrosation
Reversible inhibition of complex I by S-nitrosation may represent an additional mechanism for NO•-dependent control of the mitochondrial respiratory chain. Previous studies from this and other laboratories have suggested that sites other than complex IV within the chain may be targets for NO• [14,37]. The relative contributions of complex IV haem-nitrosylation compared with complex I S-nitrosation remain to be determined, and the balance between these two events may shift depending on the intra-mitochondrial conditions (pH, O2 tension etc.)
Notably, although maximum complex I inhibition was approx. 25%, such inhibition is expected to have significant implications for mitochondrial function. This is because unlike complex IV, which has a high ‘threshold’ for inhibition (i.e. a high degree of complex IV inhibition is required before any effect on respiration is seen), complex I has a relatively low threshold, such that a small degree of complex I inhibition can lead to significant inhibition of respiration [14].
The results from the present study also have important implications for the effect of NO• on mitochondrial ROS (reactive oxygen species) generation. Previously, it has been shown that inhibition of complex IV by NO• can cause back-up of electrons in the respiratory chain and increases ROS generation at complex III [15,38]. However, because complex I is an entry point for electrons into the chain, reversibly inhibition of it by S-nitrosation would lower the electron flux through the chain, thereby lowering ROS generation. Whereas complex I inhibition may increase ROS at the complex itself [39], it should be noted that complex I is quantitatively a much smaller source of ROS than complex III [40]. Thus, overall, a small SNO-induced increase in complex I ROS may be beneficial if it inhibits large-scale ROS generation at complex III.
In order to probe a potential beneficial role for mitochondrial S-nitrosation, mitochondria were isolated from hearts subjected to IPC. In IPC, short periods of non-lethal ischaemia have been shown to protect the heart against subsequent ischaemia–reperfusion injury [41]. In addition, significant roles for both NO• and mitochondria in IPC have been proposed [42]. Figure 7 shows that SNO were undetectable (limit 0.5–1 pmol) in control mitochondria, but easily detected in IPC mitochondria. Although this SNO signal has not yet been assigned to complex I, the results of the present study, shown in Figures 1–6, suggest that complex I S-nitrosation is significant whenever mitochondrial SNO are present. In addition, although the exact amount of mitochondrial SNO detected in IPC is much lower than that seen following treatment of mitochondria with GSNO (cf. Figure 2C), significant SNO degradation during the >1 h mitochondrial isolation procedure, especially at the homogenization step, was seen (results not shown). The source of mitochondrial SNO during IPC is not known, but it may originate from nitrite [9], which raises the possibility that complex I S-nitrosation may underlie some of the cardioprotection mediated by nitrite.
Figure 7. SNO detection in IPC.
Rat hearts were subjected to IPC, mitochondria isolated, and the amount of mitochondrial SNO measured using chemiluminescent analysis, all in the dark, as detailed in the Materials and methods section. N.D., not detectable (limit of detection 0.5–1 pmol of SNO).
In contrast to a potentially beneficial role, complex I inhibition has also been implicated in a number of disease pathologies, including Parkinson's disease [6,43] and ischaemia-reperfusion injury [28]. Both ROS and reactive nitrogen species have also been implicated in these diseases [44–46], although exactly how these species inhibit complex I under disease conditions is not well understood. Putative mechanisms include S-nitrosation [6] or nitration of tyrosine and tryptophan residues within the complex [47]. Overall, it is evident that S-nitrosation of complex I may play important roles in mitochondrial physiology and pathology, and that further work is required to elucidate the upstream mechanisms that regulate this important event.
Acknowledgments
A patent relating to the work described in this paper is currently pending. We thank Dr Gurrinder Bedi of the University of Rochester's Proteomics Core Facility for help with MALDI-TOF peptide mass fingerprinting. This work was funded by grant #HL71158 from the National Institutes of Health.
References
- 1.Stamler J. S., Lamas S., Fang F. C. Nitrosylation: the prototypic redox-based signalling mechanism. Cell (Cambridge, Mass.) 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 2.Miersch S., Mutus B. Protein S-nitrosation: biochemistry and characterization of protein thiol-NO interactions as cellular signals. Clin. Biochem. 2005;38:777–791. doi: 10.1016/j.clinbiochem.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Mannick J. B., Hausladen A., Liu L., Hess D. T., Zeng M., Miao Q. X., Kane L. S., Gow A. J., Stamler J. S. Fas-induced caspase denitrosylation. Science (Washington, D.C.) 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 4.Stamler J. S., Jaraki O., Osborne J., Simon D. I., Keaney J., Vita J., Singel D., Valeri C. R., Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borutaite V., Budriunaite A., Brown G. C. Reversal of nitric oxide-, peroxynitrite- and S-nitrosothiol-induced inhibition of mitochondrial respiration or complex I activity by light and thiols. Biochim. Biophys. Acta. 2000;1459:405–412. doi: 10.1016/s0005-2728(00)00178-x. [DOI] [PubMed] [Google Scholar]
- 6.Hsu M., Srinivas B., Kumar J., Subramanian R., Andersen J. Glutathione depletion resulting in selective mitochondrial complex I inhibition in dopaminergic cells is via an NO-mediated pathway not involving peroxynitrite: implications for Parkinson's disease. J. Neurochem. 2005;92:1091–1103. doi: 10.1111/j.1471-4159.2004.02929.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Jin B., Li L., Block E. R., Patel J. M. Nitric oxide-induced persistent inhibition and nitrosylation of active site cysteine residues of mitochondrial cytochrome-c oxidase in lung endothelial cells. Am. J. Physiol. Cell Physiol. 2005;288:C840–C849. doi: 10.1152/ajpcell.00325.2004. [DOI] [PubMed] [Google Scholar]
- 8.Ng E. S., Jourd'heuil D., McCord J. M., Hernandez D., Yasui M., Knight D., Kubes P. Enhanced S-nitroso-albumin formation from inhaled NO during ischemia/reperfusion. Circ. Res. 2004;94:559–565. doi: 10.1161/01.RES.0000117771.63140.D6. [DOI] [PubMed] [Google Scholar]
- 9.Duranski M. R., Greer J. J., Dejam A., Jaganmohan S., Hogg N., Langston W., Patel R. P., Yet S. F., Wang X., Kevil C. G., et al. Cytoprotective effects of nitrite during in vivo ischemia-reperfusion of the heart and liver. J. Clin. Invest. 2005;115:1232–1240. doi: 10.1172/JCI22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duchen M. R. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol. Aspects Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Skulachev V. P. Mitochondrial physiology and pathology; concepts of programmed death of organelles, cells and organisms. Mol. Aspects. Med. 1999;20:139–184. doi: 10.1016/s0098-2997(99)00008-4. [DOI] [PubMed] [Google Scholar]
- 12.Cleeter M. W., Cooper J. M., Darley-Usmar V. M., Moncada S., Schapira A. H. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 13.Palacios-Callender M., Quintero M., Hollis V. S., Springett R. J., Moncada S. Endogenous NO regulates superoxide production at low oxygen concentrations by modifying the redox state of cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7630–7635. doi: 10.1073/pnas.0401723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brookes P. S., Shiva S., Patel R. P., Darley-Usmar V. M. Measurement of mitochondrial respiratory thresholds and the control of respiration by nitric oxide. Methods Enzymol. 2002;359:305–319. doi: 10.1016/s0076-6879(02)59194-1. [DOI] [PubMed] [Google Scholar]
- 15.Poderoso J. J., Carreras M. C., Lisdero C., Riobo N., Schopfer F., Boveris A. Nitric oxide inhibits electron transfer and increases superoxide radical production in rat heart mitochondria and submitochondrial particles. Arch. Biochem. Biophys. 1996;328:85–92. doi: 10.1006/abbi.1996.0146. [DOI] [PubMed] [Google Scholar]
- 16.Foster M. W., Stamler J. S. New insights into protein S-nitrosylation. Mitochondria as a model system. J. Biol. Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 17.Bruckdorfer R. The basics about nitric oxide. Mol. Aspects Med. 2005;26:3–31. doi: 10.1016/j.mam.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Brown G. C., Borutaite V. Inhibition of mitochondrial respiratory complex I by nitric oxide, peroxynitrite and S-nitrosothiols. Biochim. Biophys. Acta. 2004;1658:44–49. doi: 10.1016/j.bbabio.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Clementi E., Brown G. C., Feelisch M., Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarti P., Giuffre A., Barone M. C., Forte E., Mastronicola D., Brunori M. Nitric oxide and cytochrome oxidase: reaction mechanisms from the enzyme to the cell. Free Radical Biol. Med. 2003;34:509–520. doi: 10.1016/s0891-5849(02)01326-6. [DOI] [PubMed] [Google Scholar]
- 21.Brookes P. S., Pinner A., Ramachandran A., Coward L., Barnes S., Kim H., Darley-Usmar V. M. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signalling complexes. Proteomics. 2002;2:969–977. doi: 10.1002/1615-9861(200208)2:8<969::AID-PROT969>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Danial N. N., Gramm C. F., Scorrano L., Zhang C. Y., Krauss S., Ranger A. M., Datta S. R., Greenberg M. E., Licklider L. J., Lowell B. B., et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature (London) 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 23.Yang B. K., Vivas E. X., Reiter C. D., Gladwin M. T. Methodologies for the sensitive and specific measurement of S-nitrosothiols, iron-nitrosyls, and nitrite in biological samples. Free Radical Res. 2003;37:1–10. doi: 10.1080/1071576021000033112. [DOI] [PubMed] [Google Scholar]
- 24.Jaffrey S. R., Snyder S. H. The biotin switch method for the detection of S-nitrosylated proteins. Science STKE. 2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 25.Moro M. A., Darley-Usmar V. M., Goodwin D. A., Read N. G., Zamora-Pino R., Feelisch M., Radomski M. W., Moncada S. Paradoxical fate and biological action of peroxynitrite on human platelets. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6702–6706. doi: 10.1073/pnas.91.14.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank S., Stallmeyer B., Kampfer H., Schaffner C., Pfeilschifter J. Differential regulation of vascular endothelial growth factor and its receptor fms-like-tyrosine kinase is mediated by nitric oxide in rat renal mesangial cells. Biochem. J. 1999;338:367–374. [PMC free article] [PubMed] [Google Scholar]
- 27.Tarpey M. M., Wink D. A., Grisham M. B. Methods for detection of reactive metabolites of oxygen and nitrogen: in vitro and in vivo considerations. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R431–R444. doi: 10.1152/ajpregu.00361.2003. [DOI] [PubMed] [Google Scholar]
- 28.Tompkins A., Burwell L., Digerness S., Zaragoza C., Holman W., Brookes P. S. Mitochondrial dysfunction in cardiac ischemia-reperfusion injury: ROS from complex I, without inhibition. Biochim. Biophys. Acta. 2005;1762:223–231. doi: 10.1016/j.bbadis.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Uppu R. M., Lemercier J. N., Squadrito G. L., Zhang H., Bolzan R. M., Pryor W. A. Nitrosation by peroxynitrite: use of phenol as a probe. Arch. Biochem. Biophys. 1998;358:1–16. doi: 10.1006/abbi.1998.0825. [DOI] [PubMed] [Google Scholar]
- 31.Steffen M., Sarkela T. M., Gybina A. A., Steele T. W., Trasseth N. J., Kuehl D., Giulivi C. Metabolism of S-nitrosoglutathione in intact mitochondria 1. Biochem. J. 2001;356:395–402. doi: 10.1042/0264-6021:3560395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordge M. P., Addis P., Noronha-Dutra A. A., Hothersall J. S. Cell-mediated biotransformation of S-nitrosoglutathione. Biochem. Pharmacol. 1998;55:657–665. doi: 10.1016/s0006-2952(97)00498-x. [DOI] [PubMed] [Google Scholar]
- 33.Schagger H., Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 34.Brunori M., Giuffre A., Forte E., Mastronicola D., Barone M. C., Sarti P. Control of cytochrome c oxidase activity by nitric oxide 18. Biochim. Biophys. Acta. 2004;1655:365–371. doi: 10.1016/j.bbabio.2003.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Sarti P., Arese M., Bacchi A., Barone M. C., Forte E., Mastronicola D., Brunori M., Giuffre A. Nitric oxide and mitochondrial complex IV 3. IUBMB Life. 2003;55:605–611. doi: 10.1080/15216540310001628726. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich T., Bottcher B. The gross structure of the respiratory complex I: a Lego System. Biochim. Biophys. Acta. 2004;1608:1–9. doi: 10.1016/j.bbabio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Poderoso J. J., Lisdero C., Schopfer F., Riobo N., Carreras M. C., Cadenas E., Boveris A. The regulation of mitochondrial oxygen uptake by redox reactions involving nitric oxide and ubiquinol. J. Biol. Chem. 1999;274:37709–37716. doi: 10.1074/jbc.274.53.37709. [DOI] [PubMed] [Google Scholar]
- 38.Brookes P., Darley-Usmar V. M. Hypothesis: the mitochondrial NO(*) signalling pathway, and the transduction of nitrosative to oxidative cell signals: an alternative function for cytochrome c oxidase. Free Radical Biol. Med. 2002;32:370–374. doi: 10.1016/s0891-5849(01)00805-x. [DOI] [PubMed] [Google Scholar]
- 39.Taylor E. R., Hurrell F., Shannon R. J., Lin T. K., Hirst J., Murphy M. P. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 2003;278:19603–19610. doi: 10.1074/jbc.M209359200. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q., Vazquez E. J., Moghaddas S., Hoppel C. L., Lesnefsky E. J. Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 41.Murry C. E., Jennings R. B., Reimer K. A. Preconditioning with ischemia: a delay of lethal cell injury in ischaemic myocardium 2. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 42.Zaugg M., Schaub M. C. Signalling and cellular mechanisms in cardiac protection by ischaemic and pharmacological preconditioning. J. Muscle Res. Cell Motil. 2003;24:219–249. doi: 10.1023/a:1026021430091. [DOI] [PubMed] [Google Scholar]
- 43.Orth M., Schapira A. H. Mitochondrial involvement in Parkinson's disease. Neurochem. Int. 2002;40:533–541. doi: 10.1016/s0197-0186(01)00124-3. [DOI] [PubMed] [Google Scholar]
- 44.Dawson T. M., Dawson V. L. Molecular pathways of neurodegeneration in Parkinson's disease. Science (Washington, D.C.) 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 45.Lalu M. M., Wang W., Schulz R. Peroxynitrite in myocardial ischemia-reperfusion injury. Heart Fail. Rev. 2002;7:359–369. doi: 10.1023/a:1020766502316. [DOI] [PubMed] [Google Scholar]
- 46.Zweier J. L., Flaherty J. T., Weisfeldt M. L. Direct measurement of free radical generation following reperfusion of ischaemic myocardium. Proc. Natl. Acad. Sci. U.S.A. 1987;84:1404–1407. doi: 10.1073/pnas.84.5.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray J., Taylor S. W., Zhang B., Ghosh S. S., Capaldi R. A. Oxidative damage to mitochondrial complex I due to peroxynitrite: identification of reactive tyrosines by mass spectrometry. J. Biol. Chem. 2003;278:37223–37230. doi: 10.1074/jbc.M305694200. [DOI] [PubMed] [Google Scholar]