Abstract
YZGD from Paenibacillus thiaminolyticus is a novel bifunctional enzyme with both PLPase (pyridoxal phosphatase) and Nudix (nucleoside diphosphate x) hydrolase activities. The PLPase activity is catalysed by the HAD (haloacid dehalogenase) superfamily motif of the enzyme, and the Nudix hydrolase activity is catalysed by the conserved Nudix signature sequence within a separate portion of the enzyme, as confirmed by site-directed mutagenesis. YZGD's phosphatase activity is very specific, with pyridoxal phosphate being the only natural substrate, while YZGD's Nudix activity is just the opposite, with YZGD being the most versatile Nudix hydrolase characterized to date. YZGD's Nudix substrates include the CDP-alcohols (CDP-ethanol, CDP-choline and CDP-glycerol), the ADP-coenzymes (NADH, NAD and FAD), ADP-sugars, TDP-glucose and, to a lesser extent, UDP- and GDP-sugars. Regardless of the Nudix substrate, one of the products is always a nucleoside monophosphate, suggesting a role in nucleotide salvage. Both the PLPase and Nudix hydrolase activities require a bivalent metal cation, but while PLPase activity is supported by Co2+, Mg2+, Zn2+ and Mn2+, the Nudix hydrolase activity is Mn2+-specific. YZGD's phosphatase activity is optimal at an acidic pH (pH 5), while YZGD's Nudix activities are optimal at an alkaline pH (pH 8.5). YZGD is the first enzyme reported to be a member of both the HAD and Nudix hydrolase superfamilies, the first PLPase to be recognized as a member of the HAD superfamily and the first Nudix hydrolase capable of hydrolysing ADP-x, CDP-x and TDP-x substrates with comparable substrate specificity.
Keywords: ADP-coenzyme, ADP-sugar, CDP-alcohol, haloacid dehalogenase (HAD) superfamily, Nudix hydrolase, pyridoxal phosphatase
Abbreviations: HAD, haloacid dehalogenase; MDP-1, magnesium-dependent phosphatase-1; NMP, nucleoside monophosphate; Nudix, nucleoside diphosphate x; PLP, pyridoxal 5-phosphate; PLPase, pyridoxal phosphatase
INTRODUCTION
We became interested in YZGD from Paenibacillus thiaminolyticus when we discovered, through BLAST searches [1], that YZGD belongs to two separate and distinct enzyme families; the first approx. 250 amino acids show homology to the ‘nitrophenyl phosphatase’ family of the HAD (haloacid dehalogenase) superfamily [2], and the last approx. 160 amino acids contain the signature sequence of the Nudix (nucleoside diphosphate x) hydrolase superfamily [3]. YZGD is the first enzyme found to belong to both of these enzyme superfamilies.
The HAD superfamily was discovered by computer analysis of bacterial HADs [2]. Using an iterative approach to sequence database searching, a number of known enzymes and open reading frames were found to have several conserved amino acids within three motifs [2]. These conserved amino acids have been defined most recently as: an aspartate nucleophile in motif I, a serine or threonine for phosphoryl binding in motif II, a lysine or arginine for phosphoryl binding, and carboxylate residues forming a metal-binding pocket in motif III [4]. These motifs compose the core domain. Many family members have an additional cap domain involved in substrate recognition and positioning [5].
The HAD superfamily is a fairly large superfamily, containing over 3000 members [4]. The characterized enzymes in this superfamily catalyse group transfer and fall within one of the following classifications: HADs, phosphonatases, phosphate monoesterases, ATPases and phosphomutases [4]. The phosphate monoesterases can be further divided into a number of distinct phosphatase families including phosphoglycolate phosphatases, phosphoserine phosphatases, histidinol phosphatases, trehalose phosphatases [2], the MDP-1 (magnesium-dependent phosphatase-1) family [6] and the phospho 1 ethanolamine/choline phosphatase family [7].
One of the phosphatase families that was delineated by cluster analysis based on BLAST scores includes ‘the nitrophenyl phosphatases and related putative proteins’ [2]. Although the nitrophenyl phosphatase family of the HAD superfamily contains over 400 open reading frames, the function of most of these putative proteins remains unknown. When this family was discovered [2], only two enzymes of the family had been identified, the p-nitrophenyl phosphate-specific phosphatases from Saccharomyces cerevisiae [8–10] and Schizosaccharomyces pombe [11]. Though the enzymes were identified as phosphatases, no biologically relevant substrate could be determined, no obvious phenotype was revealed, and both enzymes appeared dispensable for vegetative growth and sporulation [10,11]. A phosphatase with protein phosphatase activity on histone II-A and casein was suggested to be the p-nitrophenyl phosphatase from S. cerevisiae [12], though protein phosphatase activity was sought, but could not be detected in the earlier studies [10,11].
The only other enzyme in this family that has been previously characterized is a human PLPase (pyridoxal phosphatase). This enzyme was initially isolated and purified from human erythrocytes [13] and a 20 amino acid peptide sequence was determined [14]. From this peptide sequence, human ESTs (expressed sequence tags) were uncovered and the corresponding cDNA clones were identified from the brain [15]. Jang et al. [15] identified a number of similar protein sequences, but failed to recognize this human PLPase or these other proteins as members of the HAD superfamily.
The Nudix hydrolases are a family of enzymes that hydrolyse a phosphoanhydride bond in substrates consisting of a nucleoside diphosphate linked to some moiety, x, hence the acronym ‘Nudix’, and are defined by the signature sequence GX5EX7-REUXEEXGU, where U represents the bulky aliphatic amino acids leucine, isoleucine or valine [3]. Members of the Nudix hydrolase family are ubiquitous throughout all three domains of life with over 1800 putative proteins [16]. With each new enzyme uncovered, the substrates predominantly fall into the categories of nucleoside triphosphates, nucleotide sugars, dinucleoside polyphosphates and nucleotide coenzymes [3], with few exceptions [18,19]. Though the substrate specificity is varied, it is believed that a general role of the Nudix hydrolases is to regulate the level of metabolites that would be detrimental to the cell at elevated levels [3].
In the present paper, we describe the cloning and expression of the yzgd gene, as well as the purification and characterization of the YZGD enzyme. We characterize YZGD as both a PLPase of the nitrophenyl phosphatase family of the HAD superfamily and a versatile member of the Nudix hydrolase superfamily with activity on CDP-alcohols, ADP-coenzymes, ADP-sugars, TDP-glucose, and to a lesser extent on UDP- and GDP-sugars. We use site-directed mutagenesis to show that the phosphatase activity and Nudix hydrolase activity are separate and distinct. We also use bioinformatics to define better the nitrophenyl phosphatase family of the HAD superfamily and show that this family is not a PLPase family, but rather a family of enzymes that so far appears to act upon phosphorylated substrates containing an aromatic ring.
EXPERIMENTAL
Materials
Oligodeoxynucleotides were from Integrated DNA Technologies, the plasmid pET11b from Novagen, and the plasmid pAN13 [20] containing the yzgd gene was kindly provided by Dr Tadhg Begley (Cornell University, Department of Chemistry and Chemical Biology, Ithaca, NY, U.S.A.). Escherichia coli DH5α competent cells were from Invitrogen and E. coli HMS174(DE3) competent cells were from Novagen. Pfu DNA polymerase was from Stratagene, restriction enzymes NdeI and BlpI, T4 DNA ligase and calf intestinal alkaline phosphatase were from Invitrogen and New England Biolabs and inorganic pyrophosphatase was from Sigma. Media components were from Difco, isopropyl β-D-thiogalactoside was from Research Organics, resins and substrates were from Sigma and general chemicals were from Sigma and J. T. Baker. Thiazole l-phosphate was a gift from Dr Tadhg Begley. The QuikChange® Site-Directed Mutagenesis kit was from Stratagene.
Cloning
The yzgd gene was subcloned from the plasmid pAN13 [20]. The yzgd gene was amplified using PCR, where an NdeI restriction site was incorporated at the start of the gene and a BlpI site was incorporated at the end of the gene by using primers containing these sites. The amplified gene was gel-purified, digested with NdeI and BlpI and ligated into plasmid pET11b, which had also been digested with NdeI and BlpI. This put the yzgd gene under the control of a T7 lac promoter for overexpression. The resultant plasmid, pETyzgd, was transformed into E. coli DH5α for storage and E. coli HMS174(DE3) for expression. The sequence of yzgd in pETyzgd was confirmed by the fluorescent dideoxy terminator method on a PerkinElmer ABI 377 automated DNA sequencer at the Johns Hopkins University core facility.
Expression and purification of the enzyme
E. coli HMS174(DE3) containing pETyzgd was grown at 37 °C in 2 litres of Luria–Bertani medium containing 100 μg/ml ampicillin. When growth reached an A600 (absorbance) of 0.75, the culture was induced with 1 mM isopropyl β-D-thiogalactoside. After an additional 2 h of growth, the cells (4.4 g) were harvested, washed with buffered iso-osmotic saline (50 mM Tris/HCl, pH 7.5, 0.5% KCl and 0.5% NaCl), re-centrifuged and stored at −80 °C. A negative control of E. coli HMS174(DE3) containing pET11b was treated in an identical manner.
The cells (4.4 g) were resuspended in 8.8 ml of buffer A (50 mM Tris/HCl, pH 7.5, 1 mM EDTA and 0.1 mM dithiothreitol), sonicated using a Branson sonifier and centrifuged. The supernatant was separated from the pellet and the protein concentration of the supernatant was adjusted to 10 mg/ml with buffer A (fraction I). Fraction I contained 130 mg of total protein with a PLPase specific activity of 1.2 units/mg. Streptomycin sulphate (10%, w/v) was added to fraction I to give a final concentration of 1% streptomycin sulphate and incubated on ice for 15 min, and the precipitate containing nucleic acids was discarded. Most of the enzymatic activity remained in the supernatant (fraction II). Saturated ammonium sulphate was added to fraction II to give a final concentration of 55% ammonium sulphate. The precipitant, containing YZGD, was resuspended in 5 ml of buffer A (fraction III) and loaded on to a 2.5 cm×60 cm Sephadex G-100 column calibrated with molecular mass protein standards. Protein was eluted from the column with buffer A containing 200 mM NaCl. Fractions containing YZGD were pooled and concentrated with 80% ammonium sulphate, and the precipitant containing YZGD was resuspended in l ml of buffer A (fraction IV). Fraction IV was dialysed against 50 mM Tris/HCl (pH 7.6) and 1 mM EDTA (buffer B). The dialysed sample was loaded on to a 1.5 cm×6 cm DEAE-Sepharose column equilibrated with buffer B and proteins were eluted by rinsing the column with 2 column volumes of buffer B containing 200, 300 and 400 mM NaCl sequentially. YZGD eluted with buffer B containing 300 mM NaCl. Fractions containing YZGD were pooled and concentrated using Centriprep-10 concentrators from Millipore (fraction V). The final purified enzyme contained approx. 15 mg of protein with a PLPase specific activity of 7.0 units/mg.
Enzyme assays
For all assays, 1 unit of YZGD hydrolyses 1 μmol of substrate per min.
Phosphatase assay
A standard reaction mixture consisted of 50 mM Tris/maleate (pH 5), 5 mM CoCl2, 1 mM dithiothreitol, 4 mM PLP (pyridoxal 5-phosphate) and 0.5–4.0 m-units of YZGD in 50 μl. Negative controls replaced YZGD with diluent, YZGDD12N, or crude extract of pET11b in HMS174(DE3). The reactions were carried out for 30 min at 37 °C, quenched by the addition of 50 μl of 4:1 Norit (20% packed volume)/HClO4 (7%) and centrifuged, and 50 μl of the supernatant was analysed for inorganic orthophosphate by the colorimetric method of Ames and Dubin [21]. Other phosphorylated substrates were analysed by the colorimetric method of Ames and Dubin [21] or Fiske and Subbarow [22].
Protein phosphatase assay
A 10 mg/ml casein solution was prepared and dialysed against Tris/HCl (pH 7.5), 10% (v/v) glycerol, 1 mM dithiothreitol and 1 mM EDTA to remove any free inorganic phosphate. The reaction mixture contained 50 mM Tris/maleate (pH 5) or Tris/HCl (pH 7.2), 5 mM MgCl2 or MnCl2, 1 mM dithiothreitol, 50 μg of casein and 0.5 or 30 m-units of YZGD. A positive control contained 0.5 mg of protein phosphatase 1 instead of YZGD. The reactions were incubated at 37 °C for 1 h or overnight and further treated as described under the subsection Phosphatase assay.
Nudix hydrolase assays
A standard reaction mixture consisted of 50 mM Tris/HCl (pH 8.5), 10 mM MnCl2, 1 mM dithiothreitol, 0.6 unit of calf intestinal alkaline phosphatase, 4 mM substrate and 0.5–4.0 m-units of YZGD in 50 μl. Negative controls replaced YZGD with diluent, YZGDE324Q, or crude extract of pET11b in HMS174(DE3). The reactions were carried out for 15–30 min at 37 °C and quenched with excess EDTA. Inorganic phosphate was quantified using the colorimetric method of Ames and Dubin [21]. All Nudix substrates were analysed in this way except (d)NTPs, which were analysed as described by O'Handley et al. [23], and CoA, which was analysed by HPLC as described below.
Product determination
The products of the reactions were determined by HPLC. Standard assay conditions were used except that calf intestinal alkaline phosphatase was omitted, and the EDTA-quenched reaction mixtures were passed through Microcon-10 filters (Millipore). A YMC ODS-AQ column was used with a mobile phase consisting of 100% buffer (12.5 mM citric acid, 25 mM sodium acetate and 10 mM acetic acid, pH 6.3) for 15 min followed by a gradient over 5 min to 100% methanol. Standards of NMPs (nucleoside monophosphates) and nucleoside diphosphates were analysed, as well as substrates incubated in the reaction mixture, but without enzyme.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange® kit from Stratagene. pETYZGD was the template in the PCR. For YZGDD12N, Asp12 was changed to an asparagine residue by converting GAT into AAT with complementary primers containing single-point mutations. For YZGDE324Q, Glu324 was changed to a glutamine residue by converting GAG into CAG with complementary primers containing single-point mutations. pETyzgd was degraded with DpnI, and pETYZGDE324Q and pETYZGDD12N were each transformed separately, into E. coli DH5α for storage and E. coli HMS174(DE3) for expression. The mutants were sequenced (University of Iowa, Iowa, IA, U.S.A.), expressed and purified as described above for the wild-type protein.
Bioinformatics
BLAST searches were carried out using the standard protein–protein BLAST (blastp) [1] or PSI-BLAST [24]. Sequence alignment of BLAST results was performed using CLUSTAL W [25] or CLUSTAL X [26]. HMMER 2.3.2 [27] was used to build a hidden Markov model for the nitrophenyl phosphatase family of the HAD superfamily. The model was calibrated using ‘hmm-calibrate’ to determine statistical significance parameters before using the model as a search tool. This model was used in ‘hmmsearch’ against known proteins of other families of the HAD superfamily, members of the nitrophenyl phosphatase family as a positive control, and completely unrelated proteins as a negative control.
RESULTS AND DISCUSSION
Subcloning, expression and purification
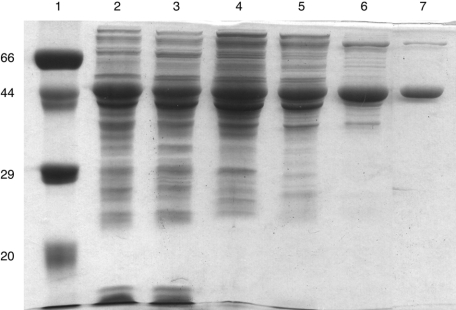
The sequence of the subcloned yzgd from P. thiaminolyticus agrees with the sequence deposited by Costello et al. [28]. Overexpression in E. coli produced a protein product, which appears as a major band at 45 kDa on a denaturing polyacrylamide gel (Figure 1). Additionally, the crude extract of YZGD contained ≥1000-fold more PLPase activity than a corresponding extract lacking YZGD. Purification of YZGD resulted in a highly purified protein (Figure 1). Sonication was necessary to extract soluble YZGD from the cells. Streptomycin sulphate was necessary to remove nucleic acids. YZGD eluted from a G100 column as expected for a 90 kDa protein, indicating that YZGD, with a monomeric molecular mass of 45 kDa, is a dimer in solution. YZGD was predicted to have a net charge of −22 and a pI of 4.9 (DNAid). We took advantage of YZGD's negative charge to purify it over a DEAE anion-exchange column, where YZGD eluted with a NaCl concentration of 0.3 M. This purification method yielded approx. 15 mg of highly purified YZGD from 2 litres of culture with a 6-fold purification and a 67% yield.
Figure 1. Expression and purification of the YZGD PLPase.
A 15% polyacrylamide gel with 1% SDS contained the following: lane 1, reference proteins with molecular masses of 66, 45, 29 and 20 kDa; lane 2, crude extract from E. coli cells containing pETyzgd; lane 3, fraction I, supernatant obtained from sonication of the cells; lane 4, fraction II, supernatant obtained from fractionation with streptomycin; lane 5, fraction III obtained by fractionation with ammonium sulphate; lane 6, fraction IV obtained by purification over a G100 column; lane 7, fraction V obtained by purification over a DEAE-Sepharose column. Lane 7 contains 5 μg of purified YZGD. Lanes 2–6 contain approximately the same number of PLPase units as present in lane 7.
Substrate specificity
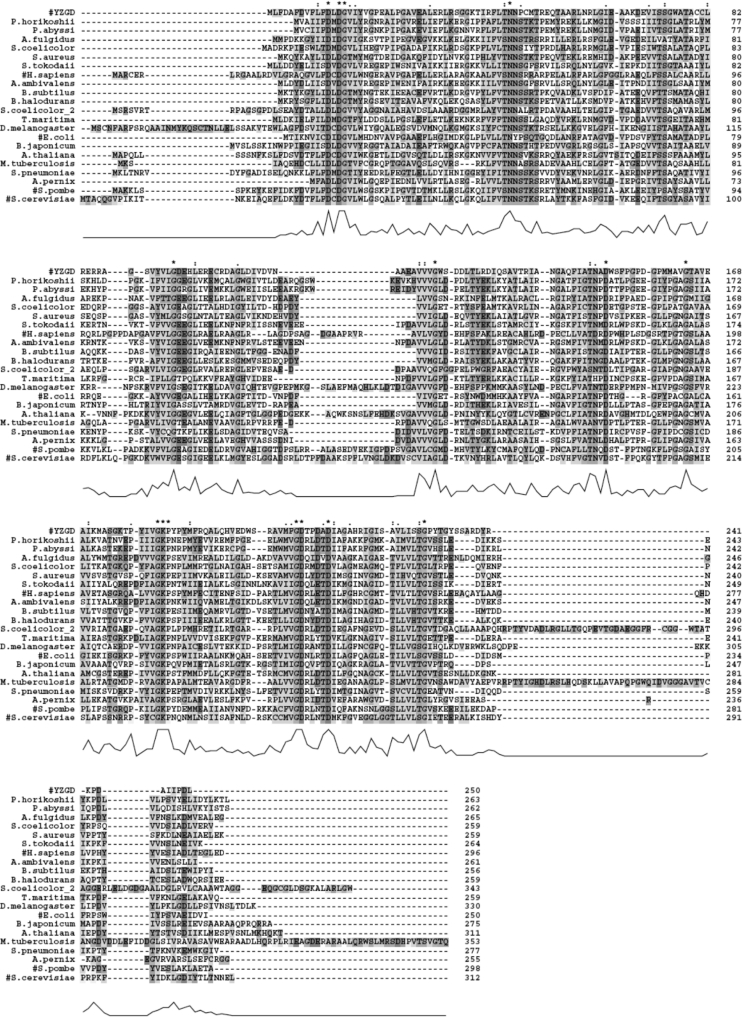
Since YZGD was found through BLAST searches to belong to two separate and distinct protein families, the ‘nitrophenyl phosphatase’ family of the HAD superfamily [2] (Figure 2) and the Nudix hydrolase family [3] (Figure 3), we predicted that YZGD would possess both phosphatase activity and Nudix hydrolase activity.
Figure 2. BLAST search results for the HAD domain of YZGD.
A BLAST search of non-redundant databases using the first 250 amino acids of YZGD uncovered 390 hits with E values of <1. The 20 best matches along with S. pombe and S. cerevisiae p-nitrophenyl phosphatases, hits 23 and 29 respectively, were aligned using CLUSTAL X [26]. Most of the matches were unknown open reading frames. Proteins with characterized phosphatase activity are indicated by a ‘#’, and aligned separately in Figure 6. Absolutely conserved identical amino acids are noted with a ‘*’. Conserved similar amino acids and highly conserved identical amino acids are identified by the symbols ‘:’ and ‘●’ respectively. The amino acids are also highlighted when there is sequence identity or similarity. Similar amino acids are grouped as follows: (1) G; (2) P; (3) I, L, V, A, M, F, W; (4) T, S, N, Q; (5) D, E; R, K; and (6) Y, H. Gaps in the amino acid sequences, inserted by CLUSTAL to optimize alignment, are indicated with a ‘-’. The height of the peaks below the sequence alignment indicate the degree of overall homology; the higher the peak, the greater the overall homology.
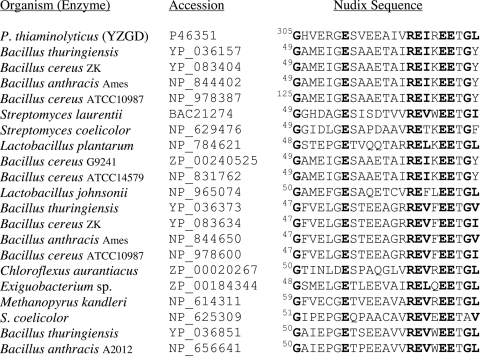
Figure 3. BLAST search results for the Nudix sequence of YZGD.
A BLAST search of non-redundant databases using amino acids 251–413 of YZGD uncovered 280 hits with E values of <1 and the best matches contain the Nudix signature sequence. The Nudix signature sequences of the 20 best matches are shown in order of overall total similarity to YZGD. Twelve of the top 20 hits are from B. thuringiensis, B. cereus or B. anthracis, suggesting the evolutionary relatedness of these organisms to P. thiaminolyticus.
When Koonin and Tatusov [2] delineated the HAD superfamily into subfamilies, the only identified and characterized enzymes of the nitrophenyl phosphatase family of the HAD superfamily were the two yeast p-nitrophenyl phosphatases. YZGD also hydrolyses p-nitrophenyl phosphate; however, p-nitrophenyl phosphate is not a biologically relevant compound. The only biologically significant phosphorylated substrate identified for YZGD is the essential cofactor PLP, with a specific activity of 7 units/mg (Table 1), a kcat of 19.4 s−1, a Km of 0.33 mM and a kcat/Km of 5.8×104 M−1·s−1 (Table 3). Even the related pyridoxamine 5-phosphate is not a substrate for YZGD (Table 1). Once we discovered this PLPase activity, we identified a human PLPase as a member of this family as well. This PLPase activity designates a novel activity for the HAD superfamily. However, this is not a PLPase family, as the yeast nitrophenyl phosphatases do not hydrolyse pyridoxal phosphate [9,11] and we have discovered that another member of this family, NAGD from E. coli, is a UMP hydrolase (I. M. Tirrell, A. T. Nguyen, R. J. Mentz, E. J. Slivka and S. F. O'Handley, unpublished work).
Table 1. Substrate specificity of YZGD for phosphorylated substrates.
All substrates except casein were present at a concentration of 4 mM. All substrates were assayed using the standard phosphatase assay as described in the Experimental section, except for casein, which was assayed using the protein phosphatase assay as described in the Experimental section. One unit of enzyme activity hydrolyses 1 μmol of substrate/min. There was no detectable activity with the following substrates: tyrosine phosphate, serine phosphate, threonine phosphate, histidine phosphate, arginine phosphate, casein (phosphorylated at serine and threonine residues), glycolate 2-phosphate, trehalose 6-phosphate, N-acetylglucosamine 6-phosphate, glucosamine 6-phosphate, fructose 6-phosphate, thiamine phosphate, thiamine pyrophosphate, thiazole phosphate, NADP+, NADPH, FMN, ribose 5-phosphate, glucose 6-phosphate, glucose 1-phosphate, (d)AMP, (d)GMP, (d)CMP, UMP, dTMP, ADP, ATP, inositol phosphates, phosphoenolpyruvate, choline 3-phosphate, ethanolamine 3-phosphate and glycerol 3-phosphate.
| Substrate | Specific activity (units/mg) |
|---|---|
| PLP | 7 |
| Pyridoxamine 5-phosphate | <0.1 |
| p-Nitrophenyl phosphate | 20 |
Table 3. Kinetic parameters for YZGD.
For pyridoxal phosphate, the standard phosphatase assay was used with 0.1–4.0 mM substrate. For the other eight substrates, the standard Nudix hydrolase assay was used with substrate concentrations ranging from 0.5–20 mM for TDP-glucose to 2–80 mM for CDP-choline and CDP-ethanolamine; the other Nudix substrate concentrations were within these two ranges. Vmax and Km were determined from a nonlinear regression analysis and kcat was calculated from Vmax assuming that YZGD is a dimer with a molecular mass of 90.8 kDa. One unit of enzyme hydrolyses 1 μmol of substrate per min.
| Substrate | Vmax (units/mg) | kcat (s−1) | Km (mM) | kcat/Km (×103 M−1·s−1) |
|---|---|---|---|---|
| Pyridoxal phosphate | 12±1 | 19±1 | 0.33±0.07 | 57±16 |
| FAD | 15±1 | 22±1 | 3.7±0.6 | 6.0±2.3 |
| ADP-ribose | 11±1 | 17±2 | 6.9±2.5 | 2.4±0.8 |
| NADH | 5.2±0.3 | 7.8±0.5 | 4.7±1.0 | 1.7±0.5 |
| NAD | 5.3±0.5 | 8.0±0.8 | 8.5±1.3 | 0.9±0.6 |
| CDP-glycerol | 19±1 | 29±2 | 7.1±1.0 | 4.1±1.5 |
| CDP-ethanolamine | 33±2 | 50±3 | 22±3 | 2.3±0.9 |
| CDP-choline | 14±1 | 22±1 | 22±3 | 1.0±0.4 |
| TDP-glucose | 4.1±0.2 | 6.3±0.2 | 1.3±0.2 | 4.9±1.1 |
Although the original studies on the yeast p-nitrophenyl phosphatases showed that these enzymes were not protein phosphatases [8,11], a later study indicated that a protein phosphatase activity might be associated with the p-nitrophenyl phosphatase from S. cerevisiae [12]. Therefore we analysed YZGD for protein phosphatase activity. YZGD is not active on the naturally phosphorylated milk protein casein (Table 1). A positive control of protein phosphatase 1 acting on casein demonstrated that enough free phosphate could be released to detect readily in our assays. Likewise, YZGD is not active on serine phosphate, threonine phosphate, tyrosine phosphate, histidine phosphate or arginine phosphate (Table 1). Thus we have no evidence that YZGD is a protein phosphatase. This is not surprising, as the HAD superfamily consists of a wide variety of phosphatases [4].
Substrates of other members of the HAD superfamily were analysed including glycolate 2-phosphate, trehalose 6-phosphate, serine phosphate, phosphoethanolamine and phosphocholine. No activity was observed for YZGD with these substrates (Table 1). We have determined that NAGD from E. coli is active on several substrates besides UMP, including N-acetylglucosamine 6-phosphate, glucosamine 6-phosphate and fructose 6-phosphate (I. M. Tirrell, A. T. Nguyen, R. J. Mentz, E. J. Slivka, and S. F. O'Handley, unpublished work). Conversely, YZGD is not active on these substrates (Table 1). Because yzgd is just upstream of the thiaminase I gene yzgc [28], thiamine phosphate, thiamine pyrophosphate and a thiamine precursor, thiazole phosphate, were analysed for activity. However, none of these are substrates for YZGD (Table 1), and the relationship of yzgd and yzgc, if any, remains unknown (T. P. Begley, personal communication). Coenzymes, sugar phosphates, NMPs, inositol phosphates and other biologically significant phosphorylated compounds were analysed (Table 1) and we especially took note of the phosphorylated substrates that would be products of YZGD's Nudix hydrolase activity, but none of these compounds were substrates for YZGD's phosphatase activity.
Whereas YZGD's phosphatase activity is highly specific, its Nudix hydrolase activity is extremely broad (Table 2). YZGD displays comparable activity on CDP-alcohols, ADP-coenzymes, ADP-ribose and TDP-glucose and some activity on UDP-sugars and GDP-sugars. YZGD is the first Nudix hydrolase reported to have comparable ADP-x, CDP-x and TDP-x hydrolase activities, the first Nudix hydrolase reported to have a TDP-sugar hydrolase activity and the second Nudix hydrolase reported to have CDP-alcohol hydrolase activity (a CDP-choline hydrolase from Bacillus cereus has been described [16]). There is a rat liver enzyme with substrate specificity similar to the Nudix hydrolase activity of YZGD [29], but its sequence has not been determined, and thus it has not yet been classified as a Nudix hydrolase.
Table 2. Substrate specificity of YZGD for Nudix substrates.
All substrates except CoA were present at a concentration of 10 mM, and were assayed using the standard Nudix hydrolase assay as described in the Experimental section. The reaction of YZGD with CoA was analysed by HPLC as described in the Experimental section. NAD, nicotinamide–adenine dinucleotide; Ap2A–Ap6A, diadenosine diphosphate–diadenosine hexaphosphate.
| Substrate | Specific activity (units*/mg) | Relative activity (%) |
|---|---|---|
| CDP-glycerol | 11.4 | 100 |
| CDP-ethanolamine | 10.5 | 92 |
| CDP-choline | 7.0 | 61 |
| TDP-glucose | 5.5 | 48 |
| FAD | 9.1 | 80 |
| ADP-ribose | 9.1 | 80 |
| NADH | 6.7 | 59 |
| NAD | 6.6 | 58 |
| ADP-mannose | 1.6 | 14 |
| ADP-glucose | 0.8 | 7 |
| Ap2A | 0.7 | 6 |
| UDP-glucose | 4.2 | 37 |
| UDP-mannose | 1.5 | 13 |
| UDP-N-acetylglucosamine | 0.8 | 7 |
| UDP-galactose | 0.3 | 3 |
| GDP-mannose | 0.2 | 2 |
| GDP-glucose | <0.1 | <1 |
| CoA | <0.1 | <1 |
| Ap3A, Ap4A, Ap5A, Ap6A | <0.1 | <1 |
| (d)NTPs | <0.1 | <1 |
* One unit of enzyme hydrolyses 1 μmol of substrate per min.
The only common Nudix substrates for which YZGD shows no activity are the (d)NTPs, the diadenosine polyphosphates and CoA. The (d)NTPs are relatively small, while the diadenosine polyphosphates and CoA are considerably larger compared with YZGD's substrates; thus (d)NTPs, diadenosine polyphosphates and CoA may not fit YZGD's active site properly.
When kinetic assays were carried out, the kcat/Km values were within a factor of six of one another for the eight best Nudix substrates (Table 3). The specific activities for YZGD's Nudix activities are comparable with those of other Nudix hydrolases [30,31]. However, the Km values for YZGD's Nudix substrates are higher than those of other Nudix hydrolases [30–32]. Considering YZGD's broad substrate specificity, the higher Km values are not unexpected. Proof that all of these Nudix hydrolase activities can be attributed to YZGD include the observations that the specific activities are comparable with one another and well above the activities of an E. coli extract lacking YZGD and are affected similarly by bivalent metal cations, pH and site-directed mutagenesis.
With its broad substrate specificity, YZGD is the most versatile Nudix hydrolase to date, capable of hydrolysing derivatives of all of the nucleotides. Thus it is interesting to speculate that YZGD is representative of an evolutionarily ancient Nudix hydrolase with rather non-specific activity from which evolved the more specific Nudix hydrolases seen today.
Products of the reaction
YZGD hydrolyses phosphate from pyridoxal phosphate and the reaction can be written as:
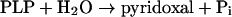 |
(reaction 1) |
YZGD also catalyses the hydrolysis of Nudix substrates as follows: ADP-ribose and NADH yield AMP and ribose 5-phosphate or nicotinamide mononucleotide respectively. CDP-ethanolamine, CDP-choline and CDP-glycerol yield CMP, and phosphoethanolamine, phosphocholine or phosphoglycerol respectively. UDP-mannose and GDP-mannose yield UMP or GMP respectively and mannose 1-phosphate. TDP-glucose yields TMP and glucose 1-phosphate. Regardless of the substrate, one of the products is always an NMP. Thus a general reaction can be written as:
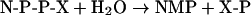 |
(reaction 2) |
where N is any of the canonical nucleosides, P is phosphate and X is any moiety. Most other Nudix hydrolases are pyrophosphohydrolases [3,33], catalysing nucleophilic attack of phosphorous, and YZGD follows suit.
Properties of the enzyme
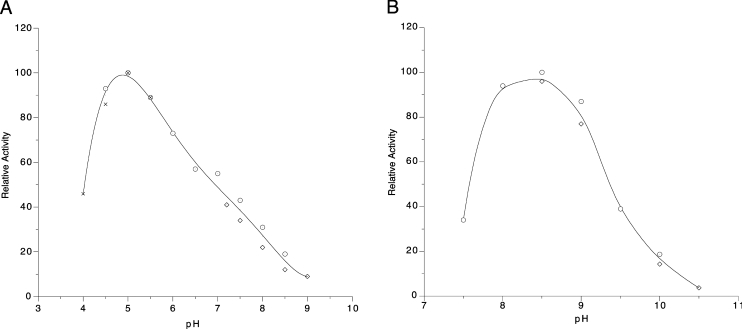
YZGD's PLPase activity is characterized as an acid phosphatase with optimal activity at pH 5 (Figure 4A). The same pH optimum was obtained for YZGD with p-nitrophenyl phosphate. The pH optimum for YZGD's Nudix hydrolase activity is pH 8.5 (Figure 4B). This held true for the three substrates tested: CDP-ethanolamine, ADP-ribose and TDP-glucose. This alkaline pH optimum is characteristic of the Nudix hydrolase family with pH optima typically ranging from pH 8.5 to 9.5 [23].
Figure 4. Effect of pH on YZGD activity.
Reactions were carried out at 37 °C with (A) 4 mM PLP, 5 mM Co2+, 1 mM dithiothreitol, 0.5 m-unit of YZGD and 50 mM Tris/maleate (○), 50 mM Tris/HCl (◇) or 50 mM acetate (×) buffers at various pH values, or (B) 4 mM ADP-ribose, 10 mM Mn2+, 1 mM dithiothreitol, 0.5 m-unit of YZGD and 50 mM Tris/HCl (○) or 50 mM glycine (◇) buffers at various pH values.
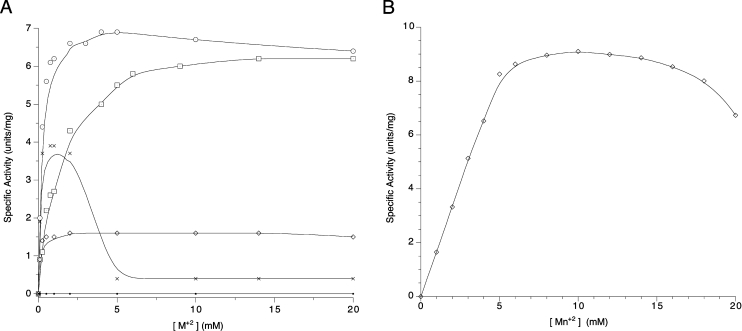
YZGD absolutely requires a bivalent metal ion for both its phosphatase activity and its Nudix hydrolase activity; however, the metal ion requirements differ for the two activities. For YZGD's phosphatase activity, Co2+ is most effective. Mg2+ is almost as effective as Co2+, while Zn2+ and to a lesser extent Mn2+ can partially substitute for Co2+ (Figure 5A). Ni2+ and Fe2+ support catalytic activity with activities reaching 40 and 20% maximal activity respectively, while Ca2+ and Cu2+ do not support activity.
Figure 5. Effect of bivalent metal ion concentration on YZGD activity.
Reactions were carried out at 37 °C with (A) 4 mM PLP, 50 mM Tris/maleate (pH 5), 1 mM dithiothreitol, 0.5 m-unit of YZGD and various concentrations of Co2+ (○), Mg2+ (□), Zn2+ (×), Mn2+ (◇) or Ca2+ (●), or (B) 4 mM ADP-ribose, 50 mM Tris/HCl (pH 8.5), 1 mM dithiothreitol, 0.5 m-unit of YZGD and various concentrations of Mn2+ (◇).
The Nudix hydrolases absolutely require a bivalent metal cation for activity and YZGD is no exception. However, while most Nudix hydrolases are maximally activated by Mg2+ [23,30,32–35], YZGD is absolutely dependent on Mn2+ for its Nudix hydrolase activity (Figure 5B). No other bivalent metal ion tested (Mg2+, Co2+, Zn2+ or Ca2+) supported hydrolysis of Nudix substrates by YZGD. Several Nudix hydrolases from B. cereus and the rat liver enzyme with similar Nudix substrate specificity to YZGD are also Mn2+-dependent enzymes [16,29].
We also investigated the effect of two metals at equal concentrations. There was no enhancement of either activity with two metals. Co2+ and Zn2+ nearly completely inhibited the Mn2+-supported Nudix hydrolase activity, whereas Mg2+ and Ca2+ had little effect. Mn2+ lowered the activity of the Co2+-supported phosphatase activity, but did not eliminate the activity, since Mn2+ supports phosphatase activity itself, albeit at a lower level than does Co2+.
Site-directed mutagenesis studies
The PLPase activity exhibited by YZGD is consistent with YZGD being a member of the HAD superfamily as there are a number of phosphatases within this superfamily [4]. Likewise, the hydrolysis of CDP-alcohols, ADP-coenzymes and nucleotide sugars is consistent with YZGD being a member of the Nudix hydrolase superfamily [3]. Conversely, the Nudix hydrolases are not expected to be phosphatases nor are HAD superfamily members expected to be pyrophosphohydrolases [3,4]. To establish unequivocally that for YZGD, it is the HAD superfamily motif that is responsible for PLPase activity, and the Nudix signature sequence that is responsible for the Nudix hydrolase activity, site-directed mutagenesis studies were carried out.
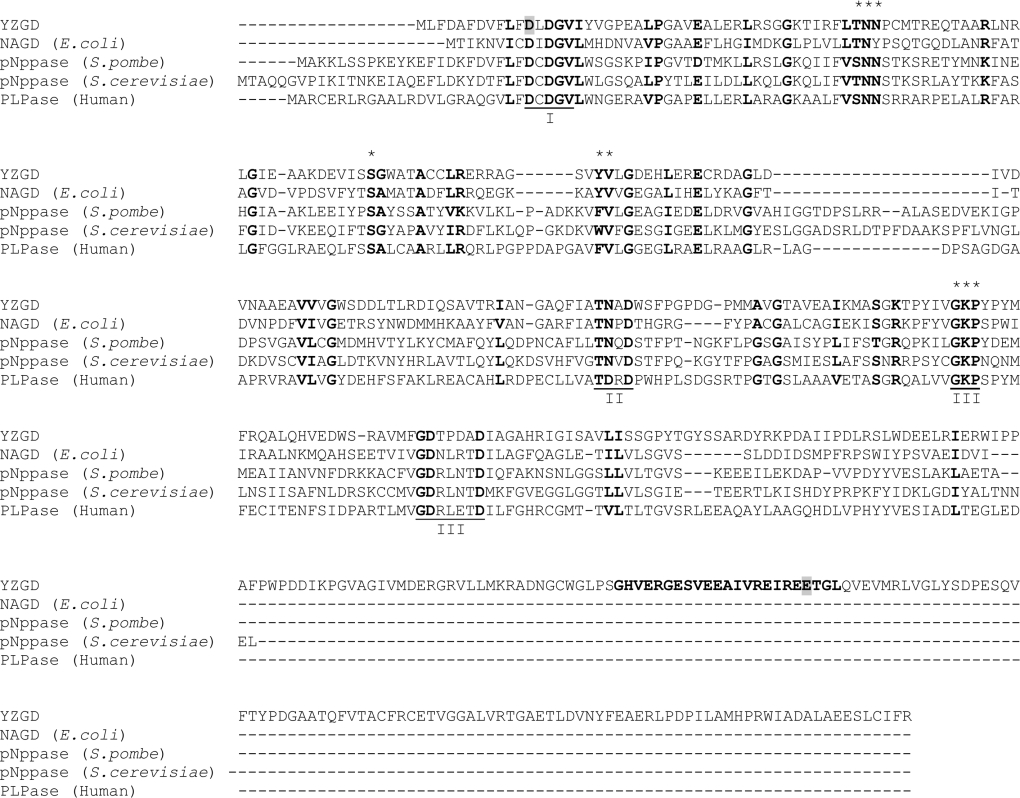
In the first mutant, YZGDD12N, Asp12 was changed to an asparagine residue (Figure 6). The corresponding aspartate residue in other HAD superfamily members has been shown to be within the active site, to co-ordinate the bivalent metal cation necessary for catalysis, and to be essential for catalysis [4]. As expected, there was no change in the expression, solubility or Nudix hydrolase activity of YZGDD12N when compared with wild-type YZGD, but there was a complete loss of PLPase activity.
Figure 6. Alignment of YZGD with other members of the nitrophenyl phosphatase family.
The amino acid sequences of YZGD, NAGD from E. coli, the p-nitrophenyl phosphatases (pNppase) from S. pombe and S. cerevisiae and the human PLPase were aligned using CLUSTAL X [26]. Similar and identical amino acids are in boldface. Gaps in the amino acid sequences inserted to optimize alignment are indicated by a ‘-’. Motifs I, II and III of the HAD superfamily are underlined and labelled. The 23 amino acid Nudix signature sequence in YZGD is in boldface. The amino acids Asp12 (within the HAD domain of YZGD) and Glu324 (within the Nudix signature sequence of YZGD), which were modified by site-directed mutagenesis, are highlighted in grey. The [S/T]NN and [F/W/Y][I/L/V] motifs, which appear to be predictive markers for the nitrophenyl phosphatase family are denoted by a ‘*’, as are the conserved serine between motifs I and II and the conserved GKP sequence of motif III.
For the second mutant, YZGDE324Q, Glu324 was changed to a glutamine residue (Figure 6). This amino acid is one of the four glutamates conserved among Nudix hydrolases [3] and essential for Nudix hydrolase activity [36,37]. When this corresponding glutamate was mutated in MutT, the activity of MutT decreased more than 100000-fold [36]. Upon NMR analysis, Lin et al. [36] discovered that this glutamate is necessary for co-ordinating the bivalent metal cation required for catalysis. When YZGDE324Q was analysed, there was no difference in the expression, solubility or PLPase activity between the mutant or the wild-type enzyme; however, there was complete loss of all Nudix hydrolase activity for all Nudix substrates.
Potential role of YZGD
The only other purified and characterized PLP-specific phosphatase is the human PLP-specific phosphatase [13,14,38,39]. In many cases, alkaline phosphatase is responsible for PLPase activity [40,41]. Although the importance of PLPase activity has been established [42], the exact nature of its role in the cell has not. Fonda and Zhang [39] described how the human PLP phosphatase might be important in vitamin B6 metabolism. Erythrocytes can take up pyridoxine, convert it into PLP, and hydrolyse the PLP to pyridoxal. Pyridoxal is then transported and rephosphorylated to PLP in other tissues, which cannot synthesize PLP by other means.
In a study by Yamada et al. [43], an E. coli vitamin B6 auxotroph could utilize the unphosphorylated forms of vitamin B6, but not the phosphorylated forms if alkaline phosphatase was repressed. The phosphorylated forms could only be taken up and utilized if they were first dephosphorylated. YZGD might perform a similar function in P. thiaminolyticus. It has been suggested that enzymatic hydrolysis of PLP may help control the intracellular level of this coenzyme as well [42]. PLP is an essential coenzyme for many enzymes, particularly those involved in amino acid metabolism [44], and YZGD may be involved in the regulation of these pathways by modulating the levels of intracellular PLP through hydrolysis.
Because YZGD hydrolyses CDP-alcohols, ADP-coenzymes, ADP, TDP, UDP and GDP sugars and one of the products of the reaction is always an NMP, YZGD has the ability to produce CMP, AMP, TMP, UMP and GMP from a wide array of nucleotide derivatives. YZGD's ability to generate all of the ribonucleoside monophosphates from a number of sources suggests a role for the enzyme in nucleotide salvage, designating a new function for a Nudix hydrolase.
Bioinformatics and comparison of YZGD to other family members
We initially uncovered YZGD from P. thiaminolyticus through BLAST searches of Nudix hydrolases, since YZGD contains the Nudix signature sequence GX5EX7REUXEEXGU (where U=I, L or V). A subsequent BLAST search of YZGD uncovered the two yeast p-nitrophenyl phosphatases and a number of open reading frames that compose the nitrophenyl phosphatase family of the HAD superfamily, in addition to members of the Nudix hydrolase family. Therefore we performed two separate BLAST searches, one on the first 250 amino acids of YZGD with sequence similarity to nitrophenyl phosphatase family members, and another on the last 163 amino acids containing the Nudix signature sequence. Results from these two BLAST searches can be seen in Figures 2 and 3 respectively.
Although the number of identified and characterized enzymes of the nitrophenyl phosphatase family is currently limited (Figure 6), there are several hundred open reading frames that show sequence similarity to the identified enzymes (Figure 2). Thus enzymes of this family appear to be ubiquitous, existing in organisms ranging from bacteria to humans. There is a potentially wide variety of functions that the phosphatases of this family may possess, as the substrates so far include pyridoxal phosphate, UMP and p-nitrophenyl phosphate. All of these substrates are structurally similar in that they all contain an aromatic ring.
The original name of the family was the nitrophenyl phosphatase family, because initially only the yeast nitrophenyl phosphatases were known. Jang et al. [15] assumed that this was a PLPase family. Many open reading frames of the family are designated as sugar phophatases, probably because NAGD is within the N-acetylglucosamine operon, yet none of the characterized enzymes including NAGD are sugar phosphatases. Mis-annotation is common when the number of known enzymes is limited.
The nitrophenyl phosphatase family is a distinct family of the HAD superfamily [1]; BLAST searches of the known enzymes of the family retrieve one another and putative family members well before they retrieve other HAD superfamily members. Enzymes of the HAD superfamily are distinguished by three conserved motifs: motif I (DXDX[T/V][L/V]), motif II ([S/T]XX) and motif III (K-[G/S][D/S]X3[D/N]) [45], or more generally, an aspartate in motif I, a serine or threonine in motif II, and a lysine or arginine and one to three aspartates or glutamates in motif III [4]. While members of the nitrophenyl phosphatase family contain these motifs (Figure 6), there is considerably more sequence similarity amongst these family members (Figure 2). Much of this sequence similarity lies between motifs I and II. This region corresponds to the substrate specificity loop described by Lahiri et al. [5] and thus these additional conserved residues may dictate this family's substrate specificity. This family appears to recognize aromatic phosphates, and consistent with this substrate specificity, there is a conserved aromatic amino acid within an [F/W/Y][I/L/V] motif. There are also a number of conserved glycines and bulky aliphatics. More interestingly, there is a conserved [S/T]NN motif and a conserved serine residue within this region. Additionally, GKP is conserved within motif III instead of just a conserved lysine.
The [S/T]NN and [F/W/Y][I/L/V] motifs appear to be unique to the nitrophenyl phosphatase family. The following families of the HAD superfamily were analysed for these motifs: HADs, epoxide hydrolases, MDP-1 family members, Phospho-1 ethanolamine/choline phosphatases, P-type ATPases, histidinol phosphatases, phosphoglycolates and trehalose phosphatases. A BLAST search was performed on a known enzyme of each family, and five unique sequences were aligned for each. Upon visual observation, the [S/T]NN and [F/W/Y][I/L/V] motifs are absent in these families, thus verifying that these motifs can be used as predictive markers of the family.
When a hidden Markov model was built from the nitrophenyl phosphatase family, a mouse nitrophenyl phosphatase [15], not used to build the model, gave an E (expectation) value of 6×10−159, while enzymes from the other HAD families gave E values of approx. 1, comparable with the results for a completely unrelated DNA ligase. This hidden Markov model search verifies that the nitrophenyl phosphatases are considerably more conserved with one another than they are with other HAD superfamily members.
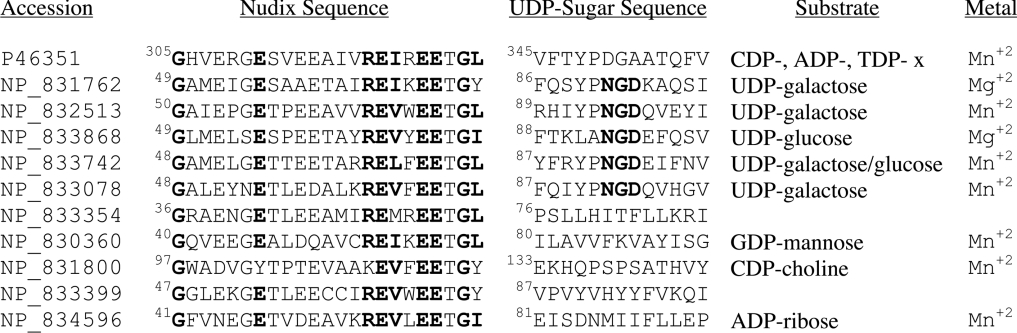
A standard BLAST search of non-redundant databases, using the last 163 amino acids of YZGD, was performed to uncover the related Nudix hydrolases. Of the top 20 hits, 12 are from B. anthracis, B. cereus or B. thuringiens (Figure 3), presumably due to close evolutionary relationships of the organisms. Fortuitously, the Nudix hydrolases of B. cereus have been characterized [16], and we anticipated that this study would help in determining the activity of YZGD. We did a BLAST search of the last 163 amino acids of YZGD, searching only the B. cereus ATCC14579 database, to identify as many unique paralogues of known function as possible. The first five hits are UDP-sugar hydrolases (Figure 7). Though YZGD does hydrolyse UDP-sugars, they are not the best Nudix substrates for YZGD (Table 2). The bioinformatics data corroborate this finding, as the UDP-sugar hydrolases contain an ‘NGD’ triad [16] that is missing in YZGD (Figure 7). However, the bioinformatics data were useful in that many of the B. cereus enzymes use Mn2+ instead of Mg2+ for their activation [16]. This observation was vital to our characterization of YZGD as YZGD also absolutely requires Mn2+. These bioinformatics results are consistent with YZGD being a unique Nudix hydrolase.
Figure 7. Comparison of YZGD and the Nudix hydrolases from B. cereus.
A BLAST search of the B. cereus ATCC14579 database using the last 163 amino acids of YZGD uncovered the Nudix hydrolases of B. cereus. This refined BLAST search was carried out because many of the top hits of the BLAST search of non-redundant databases turned up enzymes from Bacillus organisms, and the activities of most of the Nudix hydrolases from B. cereus ATCC14579 have been determined [16]. The top five hits are UDP-sugar hydrolases, but YZGD is not a UDP-sugar hydrolase. While both YZGD and the B. cereus enzymes contain the Nudix signature sequence, only the UDP-sugar hydrolases contain the ‘NGD’ UDP-sugar signature sequence. Note, however, that both YZGD and many of the Nudix hydrolases from B. cereus require Mn2+ instead of the more typical Mg2+ Nudix hydrolase activator.
Acknowledgments
This work was supported by a grant (J-497) from the Thomas and Kate Jeffress Trust Foundation, a Cottrell College Science Award (CC5180) from Research Corporation (Tucson, AZ, U.S.A.), and an AREA grant (GM066715-01) from the National Institutes of Health. We also acknowledge the expertise of Dr Shuba Gopal (Department of Biological Sciences, Rochester Institute of Technology, Rochester, NY, U.S.A.) in the use of the hidden Markov model analysis.
References
- 1.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Koonin E. V., Tatusov R. L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 1994;244:125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- 3.Bessman M. J., Frick D. N., O'Handley S. F. The MutT proteins or ‘Nudix’ hydrolases, a family of versatile, widely distributed, ‘housecleaning’ enzymes. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 4.Allen K. N., Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem. Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Lahiri S. D., Zhang G., Dai J., Dunaway-Mariano D., Allen K. N. Analysis of the substrate specificity loop of the HAD superfamily cap domain. Biochemistry. 2004;43:2812–2820. doi: 10.1021/bi0356810. [DOI] [PubMed] [Google Scholar]
- 6.Selengut J. D. MDP-1 is a new and distinct member of the haloacid dehalogenase family of aspartate-dependent phosphohydrolases. Biochemistry. 2001;40:12704–12711. doi: 10.1021/bi011405e. [DOI] [PubMed] [Google Scholar]
- 7.Roberts S. J., Stewart A. J., Sadler P. J., Farquharson C. Human PHOSPHO1 exhibits high specific phosphoethanolamine and phosphocholine phosphatase activities. Biochem. J. 2004;382:59–65. doi: 10.1042/BJ20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko Y., Toh E. A., Banno I., Oshima Y. Molecular characterization of a specific p-nitrophenyl phosphatase gene, PHO13, and its mapping by chromosome fragmentation in Saccharomyces cerevisiae. Mol. Gen. Genet. 1989;220:133–139. doi: 10.1007/BF00260867. [DOI] [PubMed] [Google Scholar]
- 9.Attias J., Bonnet J. L. A specific alkaline p-nitrophenyl phosphatase activity from baker's yeast. Biochim. Biophys. Acta. 1972;268:422–430. doi: 10.1016/0005-2744(72)90338-5. [DOI] [PubMed] [Google Scholar]
- 10.Attias J., Durand H. Further characterization of a specific p-nitrophenyl phosphatase from baker's yeast. Biochim. Biophys. Acta. 1973;321:561–568. doi: 10.1016/0005-2744(73)90199-x. [DOI] [PubMed] [Google Scholar]
- 11.Yang J., Dhamija S. S., Schweingruber M. E. Characterization of the specific p-nitrophenyl phosphatase gene and protein of Schizosaccharomyces pombe. Eur. J. Biochem. 1991;198:493–497. doi: 10.1111/j.1432-1033.1991.tb16041.x. [DOI] [PubMed] [Google Scholar]
- 12.Tuleva B., Vasileva T. E., Galabova D. A specific alkaline phosphatase from Saccharomyces cerevisiae with protein phosphatase activity. FEMS Microbiol. Lett. 1998;161:139–144. doi: 10.1111/j.1574-6968.1998.tb12940.x. [DOI] [PubMed] [Google Scholar]
- 13.Fonda M. L. Purification and characterization of vitamin B6-phosphate phosphatase from human erythrocytes. J. Biol. Chem. 1992;267:15978–15983. [PubMed] [Google Scholar]
- 14.Gao G., Fonda M. L. Identification of an essential cysteine residue in pyridoxal phosphatase from human erythrocytes. J. Biol. Chem. 1994;269:8234–8239. [PubMed] [Google Scholar]
- 15.Jang Y. M., Kim D. W., Kang T. C., Won M. H., Baek N. I., Moon B. J., Choi S. Y., Kwon O. S. Human pyridoxal phosphatase; molecular cloning, functional expression and tissue distribution. J. Biol. Chem. 2003;278:50040–50046. doi: 10.1074/jbc.M309619200. [DOI] [PubMed] [Google Scholar]
- 16.Xu W. L., Dunn C. A., Jones C. R., D'Souza G., Bessman M. J. The 26 Nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J. Biol. Chem. 2004;279:24861–24865. doi: 10.1074/jbc.M403272200. [DOI] [PubMed] [Google Scholar]
- 17. Reference deleted.
- 18.Lawhorn B. G., Gerdes S. Y., Begley T. P. A genetic screen for the identification of thiamin metabolic genes. J. Biol. Chem. 2004;279:43555–43559. doi: 10.1074/jbc.M404284200. [DOI] [PubMed] [Google Scholar]
- 19.Klaus S. M., Wegkamp A., Sybesma W., Hugenholtz J., Gregory J. F., III, Hanson A. D. A nudix enzyme removes pyrophosphate from dihydroneopterin triphosphate in the folate synthesis pathway of bacteria and plants. J. Biol. Chem. 2005;280:5274–5280. doi: 10.1074/jbc.M413759200. [DOI] [PubMed] [Google Scholar]
- 20.Abe M., Ito S. I., Kimoto M., Hayashi R., Nishimune T. Molecular studies on thiaminase I. Biochim. Biophys. Acta. 1987;909:213–221. doi: 10.1016/0167-4781(87)90080-7. [DOI] [PubMed] [Google Scholar]
- 21.Ames B. N., Dubin D. T. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 1960;235:769–775. [PubMed] [Google Scholar]
- 22.Fiske C. H., Subbarow Y. The colorimetric determination of phosphorous. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- 23.O'Handley S. F., Dunn C. A., Bessman M. J. Orf135 from Escherichia coli is a Nudix hydrolase specific for CTP, dCTP and 5-methyl-dCTP. J. Biol. Chem. 2001;276:5421–5426. doi: 10.1074/jbc.M004100200. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eddy S. R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 28.Costello C. A., Kelleher N. L., Abe M., McLafferty F. W., Begley T. P. Mechanistic studies on thiaminase I. Overexpression and identification of the active site nucleophile. J. Biol. Chem. 1996;271:3445–3452. doi: 10.1074/jbc.271.7.3445. [DOI] [PubMed] [Google Scholar]
- 29.Canales J., Pinto R. M., Costas M. J., Hernandez M. T., Miro A., Bernet D., Fernandez A., Cameselle J. C. Rat liver nucleoside diphosphosugar or diphosphoalcohol pyrophosphatases different from nucleotide pyrophosphatase or phosphodiesterase I: substrate specificities of Mg2+- and/or Mn2+-dependent hydrolases acting on ADP-ribose. Biochim. Biophys. Acta. 1995;1246:167–177. doi: 10.1016/0167-4838(94)00191-i. [DOI] [PubMed] [Google Scholar]
- 30.Sheikh S., O'Handley S. F., Dunn C. A., Bessman M. J. Identification and characterization of the Nudix hydrolase from the archaeon, Methanococcus jannaschii, as a highly specific ADP-ribose pyrophosphatase. J. Biol. Chem. 1998;273:20924–20928. doi: 10.1074/jbc.273.33.20924. [DOI] [PubMed] [Google Scholar]
- 31.Bessman M. J., Walsh J. D., Dunn C. A., Swaminathan J., Weldon J. E., Shen J. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A) J. Biol. Chem. 2001;276:37834–37838. doi: 10.1074/jbc.M107032200. [DOI] [PubMed] [Google Scholar]
- 32.O'Handley S. F., Frick D. N., Dunn C. A., Bessman M. J. Orf186 represents a new member of the Nudix hydrolases, active on adenosine(5′)triphospho(5′)adenosine, ADP-ribose, and NADH. J. Biol. Chem. 1998;273:3192–3197. doi: 10.1074/jbc.273.6.3192. [DOI] [PubMed] [Google Scholar]
- 33.Mildvan A. S., Xia Z., Azurmendi H. F., Saraswat V., Legler P. M., Massiah M. A., Gabelli S. B., Bianchet M. A., Kang L. W., Amzel L. M. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Frick D. N., Bessman M. J. Cloning, purification and properties of a novel NADH pyrophosphatase. Evidence for a nucleotide pyrophosphatase catalytic domain in MutT-like enzymes. J. Biol. Chem. 1995;270:1529–1534. doi: 10.1074/jbc.270.4.1529. [DOI] [PubMed] [Google Scholar]
- 35.Frick D. N., Townsend B. D., Bessman M. J. A novel GDP-mannose mannosyl hydrolase shares homology with the MutT family of enzymes. J. Biol. Chem. 1995;270:24086–24091. doi: 10.1074/jbc.270.41.24086. [DOI] [PubMed] [Google Scholar]
- 36.Lin J., Abeygunawardana C., Frick D. N., Bessman M. J., Mildvan A. S. The role of Glu 57 in the mechanism of the Escherichia coli MutT enzyme by mutagenesis and heteronuclear NMR. Biochemistry. 1996;35:6715–6726. doi: 10.1021/bi953071x. [DOI] [PubMed] [Google Scholar]
- 37.Maksel D., Gooley P. R., Swarbrick J. D., Guranowski A., Gange C., Blackburn G. M., Gayler K. R. Characterization of active-site residues in diadenosine tetraphosphate hydrolase from Lupinus angustifolius. Biochem. J. 2001;357:399–405. doi: 10.1042/0264-6021:3570399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao G., Fonda M. L. Kinetic analysis and chemical modification of vitamin B6 phosphatase from human erythrocytes. J. Biol. Chem. 1994;269:7163–7168. [PubMed] [Google Scholar]
- 39.Fonda M. L., Zhang Y. Kinetic mechanism and bivalent metal activation of human erythrocyte pyridoxal phosphatase. Arch. Biochem. Biophys. 1995;320:345–352. doi: 10.1016/0003-9861(95)90018-7. [DOI] [PubMed] [Google Scholar]
- 40.Merrill A. H., Jr, Henderson J. M. Diseases associated with defects in vitamin B6 metabolism or utilization. Annu. Rev. Nutr. 1987;7:137–156. doi: 10.1146/annurev.nu.07.070187.001033. [DOI] [PubMed] [Google Scholar]
- 41.Whyte M. P., Mahuren J. D., Vrabel L. A., Coburn S. P. Markedly increased circulating pyridoxal 5-phosphate levels in hypophosphatasia. Alkaline phosphatase acts in vitamin B6 metabolism. J. Clin. Invest. 1985;76:752–756. doi: 10.1172/JCI112031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T. K., Lumeng L., Veitch R. L. Regulation of pyridoxal phosphate metabolism in liver. Biochem. Biophys. Res. Commun. 1974;61:677–684. doi: 10.1016/0006-291x(74)91010-9. [DOI] [PubMed] [Google Scholar]
- 43.Yamada R. H., Tsuji T., Nose Y. Uptake and utilization of vitamin B6 and its phosphate esters by Escherichia coli. J. Nutr. Sci. Vitaminol. 1977;23:7–17. doi: 10.3177/jnsv.23.7. [DOI] [PubMed] [Google Scholar]
- 44.John R. A. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta. 1995;1248:81–96. doi: 10.1016/0167-4838(95)00025-p. [DOI] [PubMed] [Google Scholar]
- 45.Wang W., Kim R., Jancarik J., Yokota H., Kim S. H. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 Å resolution. Structure. 2001;9:65–71. doi: 10.1016/s0969-2126(00)00558-x. [DOI] [PubMed] [Google Scholar]