Abstract
Skeletal muscle homoeostasis is maintained by a variety of cytoprotective mechanisms. Since ablation of the TauT (taurine transporter) gene results in susceptibility to exercise-induced muscle weakness in vivo, it has been suggested that TauT is essential for skeletal muscle function. However, the regulatory mechanisms of TauT expression remain to be elucidated. In the present study, we demonstrated that TauT was up-regulated during myogenesis in C2C12 cells. Treatment with bFGF (basic fibroblast growth factor), which inhibited muscle differentiation, abrogated myogenic induction of TauT. The promoter activities of TauT were up-regulated during muscle differentiation in C2C12 cells. Database analyses identified an MEF2 (myocyte enhancer binding factor 2) consensus sequence at −844 in the rat TauT gene. Truncation of the promoter region containing the MEF2 site significantly reduced the promoter activity, demonstrating the functional importance of the MEF2 site. Electrophoretic mobility-shift assays confirmed that MEF2 bound to the MEF2 consensus sequence and that DNA–protein complex levels were increased during differentiation. Promoter analyses using mutated promoter-reporter plasmids demonstrated that this site was functional. Importantly, transfection with a MyoD expression vector markedly enhanced TauT promoter activity in the (non-myogenic) 10T1/2 cells. Moreover, co-transfection with an MEF2 expression vector augmented MyoD-induced TauT promoter activity, suggesting that MEF2 is required for full activation of TauT expression. Finally, we examined the effects of taurine on myotube atrophy to clarify the biological significance of the up-regulation of TauT, and demonstrated that taurine attenuated muscle atrophy induced by dexamethasone. TauT expression is regulated under the control of the myogenic programme, and we propose that this is the mechanism for taurine-mediated resistance to muscle atrophy.
Keywords: muscle atrophy, myocyte enhancer binding factor 2 (MEF2), myogenesis, skeletal muscle, taurine transporter (TauT)
Abbreviations: bFGF, basic fibroblast growth factor; bHLH, basic helix–loop–helix; DEX, dexamethasone; DM, differentiation medium; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; FBS, foetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GM, growth medium; mdx, X-chromosome-linked muscular dystrophy; MEF2, myocyte enhancer binding factor-2; RT, reverse transcription; SRF, serum-response factor; TauT, taurine transporter; TonEBP, tonicity-response element-binding protein
INTRODUCTION
In the skeletal muscle, the development of cytoprotective systems accompanies cell differentiation. Several studies have demonstrated that the TauT (taurine transporter) was required for the maintenance of skeletal muscle functions. TauT is highly expressed in skeletal muscle [1] and plays a crucial role in taurine uptake in an Na+/Cl−-dependent manner [2]. Skeletal muscle stores 70% of the taurine in the whole body. Administration of taurine improves physical endurance in exercise [3] and modulates excitation–contraction coupling in skeletal muscle [4]. In addition, taurine exhibited antioxidant activity in skeletal muscles [5,6], indicating its cytoprotective activities. Importantly, ablation of the TauT gene resulted in muscle weakness with a remarkable decrease in the concentration of taurine in skeletal muscle [7]. Thus it is suggested that myogenic expression of TauT is a critical event for the maintenance of muscle homoeostasis, although it is still unclear whether the TauT-mediated uptake of taurine is required for the maintenance of skeletal myocytes.
The expression of TauT is regulated by a wide range of stimuli, such as osmotic stress and cytokines [8–10]. The promoter region of TauT gene has previously been identified [11]. The TauT promoter contains the binding sites of a number of transcription factors, including p53 and TonEBP (tonicity-response element-binding protein) [12,13]. TauT expression is positively and negatively regulated by these factors. It has been reported that p53 activation transcriptionally represses TauT expression in HEK-293 (human embryonic kidney cells) and NRK-52E renal cells [12], whereas TonEBP induces TauT mRNA in response to hyperosmolality in HepG2 hepatoma cells [13]. The regulatory mechanisms for TauT expression in skeletal muscle cells remain to be clarified, although skeletal muscle is the major organ of taurine storage.
In the present study, in order to elucidate the regulation of TauT expression in skeletal muscle cells, we investigated TauT mRNA and protein expression during muscle differentiation. We also analysed TauT promoter activity during myogenesis. Finally, to provide direct evidence for the importance of taurine in differentiated myocytes, we studied the effects of taurine on DEX (dexamethasone)-induced muscle atrophy in C2C12 myotubes. Our results indicate that the myogenic induction of TauT is a novel cytoprotective mechanism, regulated by the muscle differentiation programmes.
MATERIALS AND METHODS
Plasmid constructs
The plasmids pCGN-MyoD and pcDNA-MEF2C were kindly given by Dr K. Walsh (Whitaker Cardiovascular Institute, Boston University School of Medicine, Boston, MA, U.S.A.) and Dr E. N. Olson (Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX, U.S.A.) respectively.
The reporter plasmid containing the TauT promoter −862 to +46 nt region and its mutant were generated as follows: the −862 to +46 region was amplified by PCR using rat genomic DNA as the template. This amplicon was used as the template for PCR-mutagenesis to produce a DNA fragment that was mutated at the MEF2 site in the −862 to +46 PCR construct. The primer sequences were: forward, −862, 5′-GGGGTACCACACTCCCCATCTCCATTTAAAAATAGTCACATTTGCTGA-3′, and, −862 MEF2mut, 5′-GGGGTACCACACTCCCCATCTCCATGGCCGCTAGCTCACATTTGCTGA-3′, and reverse, +46, 5′-AAAAGCTTCTAGATGGCACGGGAGTTCA-3′ (the MEF2 consensus region is marked in bold, and mutated nucleotides of the MEF2 site are underlined). The wild-type and mutant DNA fragments were digested with KpnI/HindIII and inserted into the multicloning site of pGL3-Basic (Promega), and were designated as pTauT/−862-luc and pTauT/−862mut-luc respectively. The MEF2 site-mutated PCR product was digested with NheI to produce the MEF2-binding-site-deleted construct and inserted into pGL3-Basic, designated pTauT/−834-luc. The truncated plasmids, pTauT/−269-luc and pTauT/−99-luc, were similarly constructed as described previously [13]. The plasmid constructs were used for promoter assays after their sequences were confirmed.
Cell culture
C2C12 cells were maintained as myoblasts in GM (growth medium) containing DMEM (Dulbecco's modified Eagle's medium) supplemented with 20% (v/v) FBS (foetal bovine serum). To induce differentiation, the cells were cultured at a density of 1.9×105 cells/well in six-well dishes and were transferred to a serum-free ITS (insulin/transferrin/selenium) medium, i.e. DM (differentiation medium) consisting of DMEM supplemented with 5 μg/ml insulin, 5 μg/ml transferrin and 5 ng/ml sodium selenite.
C3H10T1/2 fibroblasts (RIKEN Cell Bank, Tsukuba, Japan) were grown in DMEM supplemented with 10% (v/v) FBS. For differentiation, C3H10T1/2 cells were transfected with the plasmid expressing MyoD (pCGN-MyoD) and then transferred to DMEM with 2% (v/v) horse serum.
In the experiments concerning the effects of taurine on muscle atrophy, C2C12 cells were cultured at a density of 1.9×105 cells/well in six-well dishes and differentiated into myotubes by maintaining cells in the DM for 3 days. Myotubes were exposed to 100 μM DEX in the presence or absence of taurine for 1 day. The phase-contrast microscope photographs of myotubes were taken randomly by one of us (T.I.), who also measured the width of more than 50 myotubes and was blinded to the cell culture conditions.
Northern-blot analysis
Northern-blot analysis was performed as described previously [13]. In brief, total RNA was prepared from C2C12 cells by using QIAzol (Qiagen) according to the manufacturer's instructions. Total RNA (10 μg) was separated by electrophoresis in a 1.5% agarose gel containing 2.2 M formamide and was transferred on to a Nylon membrane (Hybond N+; Amersham Biosciences). The membranes were pre-hybridized in QuickHyb hybridization solution (Stratagene) at 68 °C for 1 h, followed by hybridization with labelled probes at 68 °C for 1 h. The membranes were washed with the buffer containing 2×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate) and 0.1% (w/v) SDS, and then autoradiographed. The intensities of the TauT mRNA bands were measured using an image analyser (Fuji Film, Japan) and normalized to those for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA.
Western-blot analysis
Western-blot analyses were performed as described previously [13]. Cell lysates were prepared by collecting the C2C12 cell cultures in SDS/PAGE solution. Total proteins were separated by SDS/12.5% PAGE and were transferred on to a PVDF membrane (Millipore). After blocking with 5% (w/v) skimmed milk powder, the membrane was incubated with anti-TauT antibody (Alpha Diagnostic International). Horseradish-peroxidase-conjugated antibody (Santa Cruz Biotechnology) was used as the secondary antibody. The membrane was reprobed with anti-GAPDH antibody (Chemicon) as an internal control. An enhanced chemiluminescence system (Nacalai Tesque) was used for the detection. The band intensities were quantified by densitometry.
Promoter assays
Plasmids were transfected with the FuGENE™ 6 transfection reagent (Roche Applied Science) according to the manufacturer's recommendations. C2C12 cells were cultured in 12-well plates and were transfected with 0.3 μg of luciferase plasmid (pTauT/−862-luc, pTauT/−862mut-luc, or pTauT/−834-luc, pTauT/−269-luc, and pTauT/−99-luc) and 0.2 μg of the control Renilla plasmid (phRL-TK-luc; Promega). Cells were maintained under proliferation conditions for 20 h, transferred to DM for 2 days and then harvested.
Non-myogenic 10T1/2 cells were cultured in 12-well plates and were transfected with 0.18 μg of the luciferase reporter plasmids, 0.12 μg of the control Renilla plasmid, 0.10 μg of the pCGN-MyoD and 0.21 μg of pcDNA-MEF2 expression plasmid per well. Cells were maintained under proliferation conditions for 20 h, transferred to DM for 2 days and then harvested.
Luciferase activity was measured by using Dual-Luciferase Reporter Assay system (Promega).
EMSA (electrophoretic mobility-shift assay)
EMSAs were performed as described previously [14] with minor modification. Nuclear extracts were prepared from C2C12 cells according to the previous report. Briefly, C2C12 cells were washed twice with ice-cold PBS, lysed in buffer A [20 mM Hepes (pH 7.8), 20% (v/v) glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA (pH 8.0), 1 mM DTT (dithiothreitol), 0.1% (v/v) Nonidet P40 and protease inhibitor mixture (Sigma–Aldrich)] and incubated on ice for 10 min, followed by centrifugation at 2000 g for 5 min. The pellet was resuspended in buffer B [20 mM Hepes (pH 7.6), 20% (v/v) glycerol, 500 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA (pH 8.0), 1 mM DTT, 0.1% (v/v) Nonidet P40 and protease inhibitor mixture] and then homogenized using a 26-guage needle-tipped syringe. After incubation on ice for 30 min, the homogenates were centrifuged at 15000 g for 15 min, and the supernatants were used as nuclear extracts. The protein concentration was determined using the BCA (bicinchoninic acid) assay (Pierce Biotechnology) according to the manufacturer's protocol.
Binding reactions were performed by using 5 μg of nuclear proteins, combined with 100 fmol of TauT/−848 to −822 probe labelled with [γ-32P]dATP (PerkinElmer). Primer sequences were TauT/−860MEF2, 5′-CCATTTAAAAATAGTCACATTTGCTGA-3′, and TauT/−860 MEF2 mutation, 5′-CCATGGCCGCTAGCTCACATTTGCTGA-3′. The MEF2 consensus region is marked in bold. Mutated nucleotides of the MEF2 site are underlined. The DNA probe and nuclear extracts were incubated under the binding assay conditions [7.5 mM Hepes (pH 7.8), 0.9 mM EDTA, 3.6 mM MgCl2, 4.7 mM DTT, 10% (v/v) glycerol and 10 μg/ml poly(dI-dC)·(dI-dC)] at room temperature (20 °C) for 30 min. In the supershift experiments, nuclear extracts were incubated with 0.4 μg of anti-MEF2 antibody (Santa Cruz Biotechnology). After incubation, samples were resolved on 4% polyacrylamide 0.75×TBE (1×TBE is 45 mM Tris/borate/1 mM EDTA) gels. Gels were rinsed in fixing solution [ethanoic (acetic) acid/methanol/water: 1:1:8, by vol.] and exposed to X-ray film.
RT (reverse transcription)–PCR analysis
Total RNA was prepared from C2C12 cells using QIAzol, according to the manufacture's instructions. RT–PCR was performed as described previously [15] with minor modification. Briefly, 1 μg of total RNA was subjected to RT with Rever Tra Ace (Toyobo, Osaka, Japan), using oligo(dT) primer (Invitrogen), followed by PCR. The PCR primers used were atrogin-1: forward, 5′-CTGAATAGCATCCAGATCAGCAGG-3′, and reverse, 5′-TTGATAAAGTCTTGAGGGGAAAGTG-3′, and GAPDH: forward, 5′-CATCACCATCTTCCAGGAGCG-3′, and reverse, 5′-GAGGGGCCATCCACAGTCTTC-3′. The PCR for atrogin-1 was performed as follows: 10 min at 94 °C, and then 30 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C. The GAPDH mRNA levels were also estimated by RT–PCR, as an internal control, using the following PCR conditions: 5 min at 94 °C, and then 24 cycles of 30 s at 94 °C, 30 s at 60 °C and 30 s at 72 °C. RT–PCR products were subjected to 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV illumination.
Statistics
Statistical significance was determined using Student's t test. Results were expressed as the means±S.E.M., and P<0.05 was considered to be statistically significant.
RESULTS
TauT was up-regulated during skeletal muscle differentiation in C2C12 cells
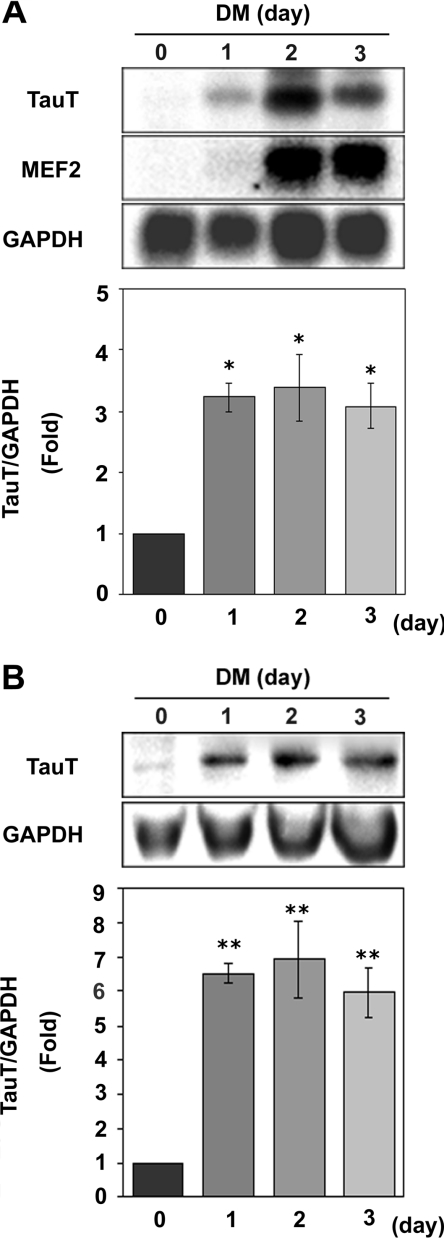
We analysed TauT expression during muscle differentiation in C2C12 cells, to address the molecular mechanisms of the expression of TauT in skeletal muscles. As shown in Figure 1(A), Northern-blot analyses revealed that TauT mRNA levels increased after the cells were transferred from GM to DM, indicating that TauT mRNA is up-regulated during myogenesis. Consistent with the increase in mRNA, TauT protein was also up-regulated during differentiation in C2C12 cells, as shown by Western blotting (Figure 1B).
Figure 1. Expression of TauT mRNA and protein during C2C12 differentiation.
C2C12 cells were maintained in GM (day 0). To induce the muscle differentiation, cells were transferred to DM and cultured for 1–3 days. (A) TauT, MEF2 and GAPDH mRNA expression was analysed by Northern blotting. The relative TauT band intensities were assessed as the ratio to the intensity for GAPDH. Results are means±S.E.M., n=3. (B) TauT expression was examined by Western-blot analysis with anti-TauT antibody. The relative TauT band intensities were assessed as the ratio to the intensity for GAPDH. Results are means±S.E.M., n=3. *, P<0.05 compared with myoblasts (day 0); **, P<0.01 compared with myoblasts (day 0).
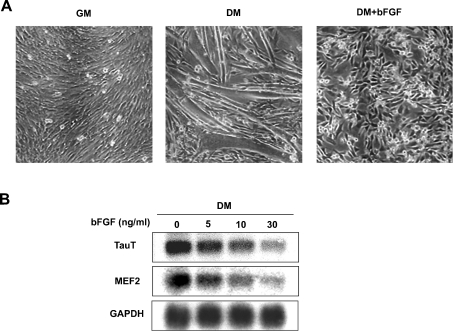
Next, we examined the effects of bFGF (basic fibroblast growth factor) on TauT expression, because bFGF is known to inhibit terminal differentiation of C2C12 cells [16,17]. In the presence of 10 ng/ml bFGF, C2C12 cells failed to differentiate into myotubes in DM (Figure 2A). Concomitant with the inhibitory effects of bFGF on muscle differentiation, up-regulation of TauT transcription was abrogated by bFGF in a dose-dependent manner (Figure 2B). Myogenic induction of MEF2 was also inhibited by bFGF, indicating that TauT expression is associated with myogenesis.
Figure 2. Inhibitory effects of bFGF on TauT expression.
(A) Morphological change of C2C12 cells induced by bFGF. C2C12 cells, maintained in GM, were transferred to DM and cultured in the presence or absence of bFGF (10 ng/ml) for 2 days. Representative phase-contrast images of C2C12 cells are shown. (B) Inhibitory effects of bFGF on TauT expression. C2C12 cells were cultured in DM in the presence of the indicated concentrations of bFGF for 2 days. The expression of TauT and MEF2 and GAPDH mRNAs were analysed by Northern blotting. The experiments were repeated three times with similar results.
Expression of TauT was transcriptionally enhanced during muscle differentiation
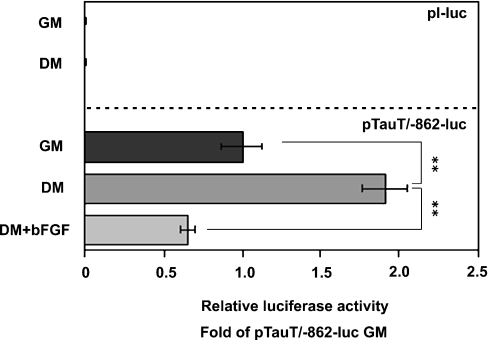
To address the mechanisms for the up-regulation of TauT mRNA during myogenesis, transcriptional activity was examined using a reporter gene assay. C2C12 cells were transfected with pTauT/−862-luc and cultured in either GM or DM for 2 days. Luciferase assays were performed to estimate the activity of TauT promoter. The −862/+46 TauT promoter was activated 2-fold in differentiating myocytes in DM, compared with C2C12 myoblasts in GM (Figure 3). In addition, treatment with bFGF abolished the enhancement of the promoter activity, indicating that TauT expression is transcriptionally regulated during myogenic differentiation.
Figure 3. Promoter activity of the TauT gene was enhanced during differentiation.
C2C12 cells were transfected with pTauT/−862-luc or pl-luc (basic vector) and pRL-TK. Cells were cultured in GM for 18 h, followed by the cultivation in GM or DM, in the presence or absence of bFGF (10 ng/ml) for 2 days. Promoter activity of the TauT gene was measured. The activity was normalized to the luciferase activity of pRL-TK. Assays were performed in triplicate. Results are means±S.E.M. The experiments were repeated three times with similar results. **, P<0.01.
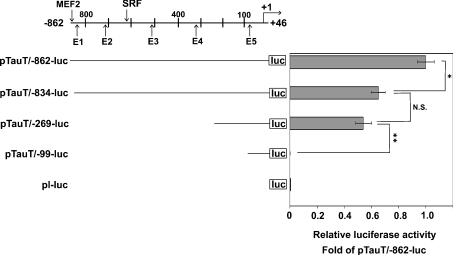
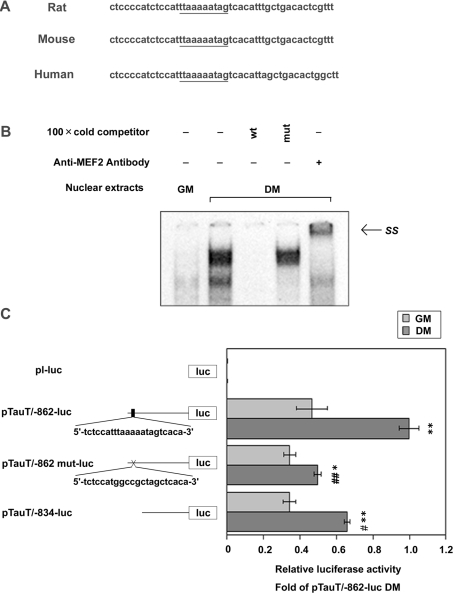
To examine whether TauT expression was directly regulated by muscle-specific transcription factors and/or myogenesis-related factors, we searched the TauT promoter for the consensus binding sequences of these factors. Several putative binding sites were identified, including one MEF2 site, one SRF (serum-response factor) site, and five E-boxes in the TauT promoter between its transcription initiation site and −979 (Figure 4). To clarify the biological significance of these binding motifs, three promoter plasmids containing deletion mutations of the TauT promoter (pTauT/−834-luc, pTauT/−269-luc and pTauT−99-luc) were generated, and promoter activity was measured under differentiation conditions. As shown in Figure 4, the deletion of the promoter region from −862 to −835, which contains the MEF2 site, significantly reduced the promoter activity. In contrast, although the deletion of the promoter region between −834 and −270, which contains four E-boxes (E1, E2, E3 and E4) and the SRF site, had a tendency to decrease the promoter activity, it was not significant. Despite the homology search failing to find any consensus binding sites for muscle-specific or myogenesis-related transcription factors, deletion of the fragment from −269 to −100 significantly impaired the promoter activity. This suggested that the −269 to −100 region might be involved in the promoter activity, possibly as the basic transcription element. The truncated plasmid pTauT/−99-luc contains an E-box, E5, but showed almost the same activity as the control luciferase vector (pl-luc). These findings suggest that MEF2 may be a key regulator in the myogenic induction of TauT, though other transcription factors probably participate in its regulation.
Figure 4. Promoter analyses of the rat TauT gene using a deletion mutant.
C2C12 cells were transfected with the TauT promoter-reporter plasmid (pTauT/−862-luc, pTauT/−834-luc, pTauT/−269-luc or pTauT−99-luc) and pRL-TK. After 18 h of cultivation in GM, cells were cultured in DM for 2 days. The promoter activity of the TauT gene was measured and normalized to the luciferase activity of pRL-TK. pl-luc basic vector was used as a promoterless control. Results are means±S.E.M., n=3. The putative binding sites of MEF2, the E-box for bHLH transcription factors, and SRF in the −862/+46 TauT promoter region are shown. The experiments were repeated three times with similar results. *, P<0.05; **, P<0.01.
To confirm the importance of the MEF2 consensus sequence at −844/−835 of the rat TauT gene, we compared the corresponding regions among rat, mouse and human TauT genes (Figure 5A). The MEF2 consensus sequence was completely conserved among these species. Moreover, the sequences surrounding the MEF2 site are also well conserved, indicative of the functional significance of the MEF2 sequence.
Figure 5. TauT mRNA expression is transcriptionally regulated by MEF2C during skeletal muscle differentiation in C2C12 cells.
(A) Sequence alignment of the −858/−813 rat TauT promoter and the equivalent regions in the human and mouse TauT genes. The MEF2 consensus region is underlined. (B) MEF2 binds directly to the MEF2 consensus sequence (−844/−835) of the TauT promoter. C2C12 myoblasts were differentiated into myotubes by culturing in DM for 2 days. Nuclear extracts were prepared from C2C12 myoblasts or myotubes. The probe containing the MEF2 consensus sequence was labelled and incubated with nuclear extracts. The formation of DNA–protein complexes was analysed by EMSAs in the presence or absence of 100-fold excess of unlabelled probes with (mut) or without (wt) mutation at MEF2 site. The supershift assay was carried out using anti-MEF2 antibody. (C) The MEF2-binding site at −844 to −835 functionally regulates TauT transcriptional activation in myotubes. C2C12 cells were transfected with promoter–reporter constructs (pTauT/−862-luc or pTauT/−862 mut-luc or pTauT/−834-luc or pl-luc) and pRL-TK and cultured in GM for 18 h. Then cells were cultured in GM or DM for 2 days and then harvested. The basic vector, pl-luc, was used as a promoterless control. The promoter activity of the TauT gene was measured and normalized against the luciferase activity of pRL-TK. Assays were performed in triplicate. Results are means±S.E.M., n=3. The experiments were repeated three times with similar results. *, P<0.05 and **, P<0.01 compared with GM; #, P<0.05 and ##, P<0.01 compared with DM pTauT/−862-luc. SS, supershifted band.
EMSAs were performed using labelled oligonucleotides of the TauT −848/−822 promoter region as a probe to examine the binding activity of this MEF2 consensus sequence (Figure 5B). Incubation of the probe with the myotube nuclear extracts resulted in the increased formation of DNA–protein complexes, compared with myoblasts. The complex formation was abrogated by unlabelled competitor −848/−822, but not by mutated MEF2 competitor. Furthermore, this complex was supershifted using anti-MEF2C antibody, indicating that MEF2C binds directly to the −844 to −835 region of the TauT promoter.
Next, the importance of the MEF2 consensus sequence for TauT transcriptional activity was estimated by using the plasmids pTauT/−834-luc and pTauT/−862mut-luc. Both the deletion and mutation constructs exhibited decreased luciferase activities in myotubes as compared with the wild-type construct, suggesting that the MEF2-binding site at −844 to −835 functionally regulates TauT transcriptional activation in myotubes (Figure 5C). It should be noted that the activities of pTauT/−862mut-luc and pTauT/−834-luc were enhanced in myotubes as compared with myoblasts, suggesting that other transcription factors may be involved in myogenic induction of TauT mRNA.
Gene expression of TauT is regulated by muscle-specific transcription factors
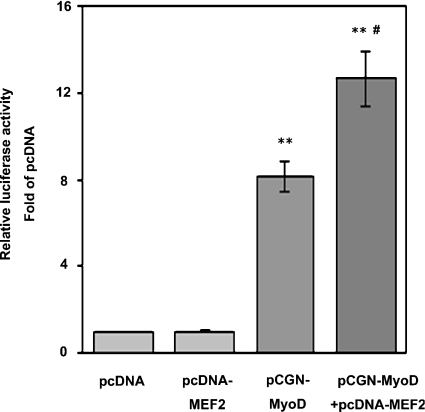
To confirm causality between TauT expression and myogenic differentiation, we examined the promoter activity of TauT gene in 10T1/2 fibroblasts expressing MyoD. After transfection with the expression vector encoding MyoD, 10T1/2 cells in DM differentiated into myotubes (Figure 6). The activity of pTauT/−862-luc was higher in 10T1/2 cells transfected with MyoD than in those with the control vector, providing the critical evidence that TauT was transcriptionally up-regulated during skeletal muscle differentiation. Importantly, co-expression of MEF2 with MyoD in 10T1/2 cells resulted in a marked increase in −862/+46 TauT promoter activity, whereas cells overexpressing MEF2 alone showed the basal level of promoter activity. Collectively, it is suggested that TauT is transcriptionally regulated during myogenesis and that MEF2 is required for full activation of the TauT promoter.
Figure 6. MEF2C enhanced TauT promoter activity in a muscle-differentiation-dependent manner.
Non-myogenic 10T1/2 fibroblasts were co-transfected with pTauT/−862-luc and pRL-TK, together with an expression plasmid for MyoD or MEF2C. Cells were cultured in GM for 18 h, and were then transferred to DM for 2 days before being assayed. Promoter activity was measured and normalized to the luciferase activity of pRL-TK. Results are means±S.E.M., n=3. The experiments were repeated three times with similar results.**, P<0.01 compared with pcDNA; #, P<0.05 compared with pCGN-MyoD.
Taurine attenuates muscle atrophy induced by DEX
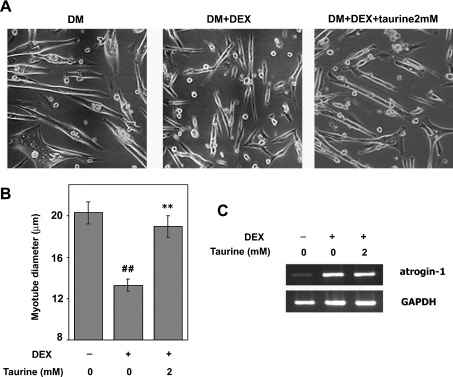
To clarify the biological significance of TauT expression during myogenic differentiation, the effects of taurine on DEX-induced muscle atrophy were examined in C2C12 myocytes. C2C12 cells were differentiated and treated with 100 μM DEX in the presence or absence of taurine. As shown in Figure 7(A), treatment with DEX resulted in obvious atrophic phenotypes, which were ameliorated by taurine treatment. To quantitatively evaluate the cytoprotective effects of taurine treatment, myotube diameters were measured (Figure 7B). A reduction in diameter was observed in myotubes treated with DEX, whereas taurine ameliorated the DEX-mediated reduction in muscle diameter. The anti-atrophic effects of taurine seemed to be distinct from hypertrophy, because treatment with taurine alone did not induce enlargement of muscle cells (results not shown).
Figure 7. Taurine exhibits cytoprotective effects on DEX-induced muscle atrophy.
(A) Representative microscopic photographs of C2C12 myotubes showing DEX-induced muscle atrophy. C2C12 cells were differentiated for 3 days, and then treated with vehicle (left) or 100 μM DEX in the absence (middle) or presence (right) of 2 mM taurine for 24 h. (B) Quantitative analyses of the anti-atrophic effects of taurine. Cells were cultured in the presence or absence of DEX and/or taurine for 24 h. Cell width was measured in two fields (more than 50 myotubes/field). Results are means±S.E.M. The experiments were repeated three times with similar results. (C) The expression of atrogin-1 was analysed by RT–PCR. C2C12 myotubes were treated with or without DEX and/or taurine for 24 h. Total RNA was prepared, and the expression of atrogin-1 mRNA was analysed by RT–PCR. GAPDH mRNA was used as an internal control. The experiments were repeated three times with similar results.
It has been reported that DEX induces muscle atrophy by up-regulating atrogin-1/MAFbx, a muscle-specific ubiquitin ligase [18]. Therefore we analysed the effects of taurine on DEX-mediated induction of atrogin-1 by RT-PCR (Figure 7C). C2C12 cells showed a remarkable increase in atrogin-1 mRNA in the presence of 100 μM DEX, as reported previously in [19]. However, expression of atrogin-1 was not affected by treatment with taurine.
DISCUSSION
In the present study, we demonstrated that TauT was up-regulated in C2C12 cells during myogenesis. The promoter analyses revealed that the TauT promoter contained an MEF2-binding site that was required for full TauT promoter activity. Transfection with the MyoD-expressing vector resulted in the activation of the TauT promoter in 10T1/2 cells and this activity was enhanced further when MEF2C was overexpressed. Finally, incubation of C2 myotubes with taurine ameliorated the DEX-induced muscle atrophy, indicating the importance of myogenic induction of TauT in cytoprotection.
Terminal differentiation is closely associated with the development of the cell survival system in skeletal muscles. Myoblasts exit from the cell cycle during muscle differentiation and this confers resistance to apoptosis [20–22]. It has also been shown that insulin-like growth factor, a paracrine factor, plays important roles both in muscle differentiation and in cell survival [23–25]. We propose that TauT expression is a novel cytoprotective mechanism that is under the control of the myogenic programme.
First, we demonstrated that TauT mRNA was up-regulated during myogenesis in C2C12 cells. In C2C12 cells, muscle differentiation was induced by changing the culture medium from medium containing FBS to low-growth-factor medium. However, it is unlikely that the up-regulation of TauT results merely from the depletion of FBS, because bFGF inhibited the muscle differentiation concomitant with the inhibition of TauT up-regulation under low-growth-factor conditions. In addition, TauT promoter activity in 10T1/2 cells (grown in DM) is enhanced by transfection with a MyoD-expressing vector, but not with an empty vector. Collectively, the results strongly suggested that myogenic expression of TauT is closely linked with myogenesis.
In the promoter region of the TauT gene, we identified an MEF2-binding site. Deletion or mutation of the MEF2-binding site impaired the activation of TauT promoter, indicating that the MEF2 site is functional. Although co-transfection with a plasmid vector expressing MEF2C enhanced the MyoD-induced activation of the TauT promoter in 10T1/2 cells, the overexpression of MEF2C alone did not affect its activity. These results suggest that MEF2C up-regulates TauT expression synergistically with other myogenic transcription factors. Indeed, it is well known that MEF2 forms a complex with bHLH (basic helix–loop–helix) transcription factors [26,27].
Ablation of the TauT gene results in the exercise-induced muscle weakness seen in TauT-null mice [7]. Therefore we examined the direct effects of taurine on the maintenance of muscle homoeostasis, using a DEX-induced muscle atrophy model in vitro. Interestingly, in the presence of taurine, myotubes showed resistance to DEX-induced atrophy, suggesting that the uptake of taurine autonomously promotes muscle viability. It has been reported previously that DEX up-regulates atrogin-1, a muscle-specific ubiquitin ligase, resulting in muscle atrophy [18]. Therefore we examined the effects of taurine on DEX-mediated induction of atrogin-1 [28]. RT–PCR analyses revealed that atrogin-1 was up-regulated by DEX treatment, but that its upregulation was not affected by taurine. This indicates that taurine does not regulate the signalling molecules upstream of atrogin-1. Consistent with this hypothesis, taurine did not inhibit the DNA-binding activity of the FOXO (forkhead box O) transcription factor, which is responsible for DEX-mediated induction of atrogin-1 (results not shown).
In the present study, we have demonstrated that overexpression of MyoD activated the TauT promoter in 10T1/2 cells. MyoD plays important roles, not only in muscle differentiation, but also in muscle regeneration. Consistent with our results, a recent report demonstrated that MyoD−/− muscle, which shows low-regeneration activity, contains a low level of taurine. In addition, skeletal muscle from mdx (X-chromosome-linked muscular dystrophy) mice, which shows high levels of muscular regeneration activity, contains high concentrations of taurine [29]. These results suggest that the TauT/taurine system may be closely associated with muscle regeneration. This suggestion is supported by the recent finding that taurine ameliorated the exercise-induced loss of muscle strength and Cl− conductance that are signs of muscle degeneration in mdx mice [30].
In summary, TauT is up-regulated during muscle differentiation, providing a molecular basis for taurine-mediated resistance to muscle atrophy. The taurine/TauT system may be a therapeutic target for protecting myotubes against muscle wasting.
Acknowledgments
We thank Ms Yasuko Murao (Graduate School of Pharmaceutical Sciences, Osaka University) for her secretarial work. This study is partially supported by a grant from Taisho Pharmaceutical Co. Ltd and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
References
- 1.Ramamoorthy S., Leibach F. H., Mahesh V. B., Han H., Yang-Feng T., Blakely R. D., Ganapathy V. Functional characterization and chromosomal localization of a cloned taurine transporter from human placenta. Biochem. J. 1994;300:893–900. doi: 10.1042/bj3000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchida S., Kwon H. M., Yamauchi A., Preston A. S., Marumo F., Handler J. S. Molecular cloning of the cDNA for an MDCK cell Na+- and Cl−-dependent taurine transporter that is regulated by hypertonicity. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8230–8234. doi: 10.1073/pnas.89.17.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yatabe Y., Miyakawa S., Miyazaki T., Matsuzaki Y., Ochiai N. Effects of taurine administration in rat skeletal muscles on exercise. J. Orthop. Sci. 2003;8:415–419. doi: 10.1007/s10776-002-0636-1. [DOI] [PubMed] [Google Scholar]
- 4.De Luca A., Pierno S., Camerino D. C. Effect of taurine depletion on excitation–contraction coupling and Cl− conductance of rat skeletal muscle. Eur. J. Pharmacol. 1996;296:215–222. doi: 10.1016/0014-2999(95)00702-4. [DOI] [PubMed] [Google Scholar]
- 5.Fang Y. Z., Yang S., Wu G. Free radicals, antioxidants, and nutrition. Nutrition. 2002;18:872–879. doi: 10.1016/s0899-9007(02)00916-4. [DOI] [PubMed] [Google Scholar]
- 6.Huxtable R. J. Physiological actions of taurine. Physiol. Rev. 1992;72:101–163. doi: 10.1152/physrev.1992.72.1.101. [DOI] [PubMed] [Google Scholar]
- 7.Warskulat U., Flogel U., Jacoby C., Hartwig H. G., Thewissen M., Merx M. W., Molojavyi A., Heller-Stilb B., Schrader J., Haussinger D. Taurine transporter knockout depletes muscle taurine levels and results in severe skeletal muscle impairment but leaves cardiac function uncompromised. FASEB J. 2004;18:577–579. doi: 10.1096/fj.03-0496fje. [DOI] [PubMed] [Google Scholar]
- 8.Lambert I. H. Regulation of the cellular content of the organic osmolyte taurine in mammalian cells. Neurochem. Res. 2004;29:27–63. doi: 10.1023/b:nere.0000010433.08577.96. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y. S., Ohtsuki S., Takanaga H., Tomi M., Hosoya K., Terasaki T. Regulation of taurine transport at the blood–brain barrier by tumor necrosis factor-α, taurine and hypertonicity. J. Neurochem. 2002;83:1188–1195. doi: 10.1046/j.1471-4159.2002.01223.x. [DOI] [PubMed] [Google Scholar]
- 10.Mochizuki T., Satsu H., Shimizu M. Tumor necrosis factor α stimulates taurine uptake and transporter gene expression in human intestinal Caco-2 cells. FEBS Lett. 2002;517:92–96. doi: 10.1016/s0014-5793(02)02584-x. [DOI] [PubMed] [Google Scholar]
- 11.Han X., Budreau A. M., Chesney R. W. Cloning and characterization of the promoter region of the rat taurine transporter (TauT) gene. Adv. Exp. Med. Biol. 2000;483:97–108. doi: 10.1007/0-306-46838-7_9. [DOI] [PubMed] [Google Scholar]
- 12.Han X., Patters A. B., Chesney R. W. Transcriptional repression of taurine transporter gene (TauT) by p53 in renal cells. J. Biol. Chem. 2002;277:39266–39273. doi: 10.1074/jbc.M205939200. [DOI] [PubMed] [Google Scholar]
- 13.Ito T., Fujio Y., Hirata M., Takatani T., Matsuda T., Muraoka S., Takahashi K., Azuma J. Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem. J. 2004;382:177–182. doi: 10.1042/BJ20031838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujio Y., Kunisada K., Hirota H., Yamauchi-Takihara K., Kishimoto T. Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J. Clin. Invest. 1997;99:2898–2905. doi: 10.1172/JCI119484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clegg C. H., Linkhart T. A., Olwin B. B., Hauschka S. D. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 1987;105:949–956. doi: 10.1083/jcb.105.2.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lathrop B., Thomas K., Glaser L. Control of myogenic differentiation by fibroblast growth factor is mediated by position in the G1 phase of the cell cycle. J. Cell Biol. 1985;101:2194–2198. doi: 10.1083/jcb.101.6.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 19.Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., Glass D. J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Walsh K. Resistance to apoptosis conferred by Cdk inhibitors during myocyte differentiation. Science. 1996;273:359–361. doi: 10.1126/science.273.5273.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Guo K., Wills K. N., Walsh K. Rb functions to inhibit apoptosis during myocyte differentiation. Cancer Res. 1997;57:351–354. [PubMed] [Google Scholar]
- 22.Fujio Y., Guo K., Mano T., Mitsuuchi Y., Testa J. R., Walsh K. Cell cycle withdrawal promotes myogenic induction of Akt, a positive modulator of myocyte survival. Mol. Cell. Biol. 1999;19:5073–5082. doi: 10.1128/mcb.19.7.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawlor M. A., Rotwein P. Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol. Cell. Biol. 2000;20:8983–8995. doi: 10.1128/mcb.20.23.8983-8995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart C. E., Rotwein P. Insulin-like growth factor-II is an autocrine survival factor for differentiating myoblasts. J. Biol. Chem. 1996;271:11330–11338. doi: 10.1074/jbc.271.19.11330. [DOI] [PubMed] [Google Scholar]
- 25.Florini J. R., Magri K. A., Ewton D. Z., James P. L., Grindstaff K., Rotwein P. S. “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 1991;266:15917–15923. [PubMed] [Google Scholar]
- 26.Kaushal S., Schneider J. W., Nadal-Ginard B., Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 27.Molkentin J. D., Black B. L., Martin J. F., Olson E. N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 28.Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIntosh L. M., Garrett K. L., Megeney L., Rudnicki M. A., Anderson J. E. Regeneration and myogenic cell proliferation correlate with taurine levels in dystrophin- and MyoD-deficient muscles. Anat. Rec. 1998;252:311–324. doi: 10.1002/(SICI)1097-0185(199810)252:2<311::AID-AR17>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 30.De Luca A., Pierno S., Liantonio A., Cetrone M., Camerino C., Fraysse B., Mirabella M., Servidei S., Ruegg U. T., Conte Camerino D. Enhanced dystrophic progression in mdx mice by exercise and beneficial effects of taurine and insulin-like growth factor-1. J. Pharmacol. Exp. Ther. 2003;304:453–463. doi: 10.1124/jpet.102.041343. [DOI] [PubMed] [Google Scholar]