Abstract
A major theme of TBI (traumatic brain injury) pathology is the over-activation of multiple proteases. We have previously shown that calpain-1 and -2, and caspase-3 simultaneously produced αII-spectrin BDPs (breakdown products) following TBI. In the present study, we attempted to identify a comprehensive set of protease substrates (degradome) for calpains and caspase-3. We further hypothesized that the TBI differential proteome is likely to overlap significantly with the calpain- and caspase-3-degradomes. Using a novel HTPI (high throughput immunoblotting) approach and 1000 monoclonal antibodies (PowerBlot™), we compared rat hippocampal lysates from 4 treatment groups: (i) naïve, (ii) TBI (48 h after controlled cortical impact), (iii) in vitro calpain-2 digestion and (iv) in vitro caspase-3 digestion. In total, we identified 54 and 38 proteins that were vulnerable to calpain-2 and caspase-3 proteolysis respectively. In addition, the expression of 48 proteins was down-regulated following TBI, whereas that of only 9 was up-regulated. Among the proteins down-regulated in TBI, 42 of them overlapped with the calpain-2 and/or caspase-3 degradomes, suggesting that they might be proteolytic targets after TBI. We further confirmed several novel TBI-linked proteolytic substrates, including βII-spectrin, striatin, synaptotagmin-1, synaptojanin-1 and NSF (N-ethylmaleimide-sensitive fusion protein) by traditional immunoblotting. In summary, we demonstrated that HTPI is a novel and powerful method for studying proteolytic pathways in vivo and in vitro.
Keywords: calpain, caspase, degradome, high throughput immunoblotting (HTPI), proteomics, traumatic brain injury (TBI)
Abbreviations: BDP, breakdown product; CaMPK, calcium/calmodulin-dependent protein kinase; CASK, calcium/calmodulin-dependent serine protein kinase; CCI, controlled cortical impact; Cdc, cell division cycle; DTT, dithiothreitol; GST, glutathione S-transferase; HTPI, high throughput immunoblotting; MM, molecular mass; NSF, N-ethylmaleimide sensitive fusion protein; Psme3, proteasome activator subunit 3; SBDP, αII-spectrin BDP; SNARE, soluble NSF attachment protein receptor; SNAP, synaptosome-associated protein (numerical values 23 and 25 are kDa); TBI, traumatic brain injury; where the annotation A3 etc is given, A is template A etc, 3 is lane 3 etc, on HTPI gels
INTRODUCTION
TBI (traumatic brain injury) represents a major central nervous system disorder without any clinically proven therapy. However, significant progress has been made in understanding the biochemical mechanisms of injury. Indeed, protease over-activation is a major theme in traumatic and ischaemic brain injury. These cysteine proteases include calpain-1 and -2, caspase-3, cathepsin-B and -L [1], and metalloproteases e.g. MMP (matrix metallprotease)-2 and -9 [2–3], and the proteasome [4]. Of particular interest are calpains and caspase-3 [5]. Calpain is activated during both oncotic (necrotic) and apoptotic cell-death in neurons, whereas caspase-3 is strictly activated only in neuronal apoptosis. Evidence demonstrates that both necrotic and apoptotic cell death are present in traumatic or ischaemic brain injury. Our research has shown that calpain-produced and caspase-3-produced SBDPs (αII-spectrin breakdown products) are present following both traumatic and ischaemic brain injury. We have also shown that the same SBDPs can be found in cerebrospinal fluid following TBI in rats [6]. In addition, many other brain proteins have been independently identified as vulnerable to proteolytic attack after toxic neural insults, or to calpain and/or caspase-3 actions. These include CaMPK (calcium/calmodulin-dependent protein kinase)-IV and CaMPK-II [5,7,8].
Novel HTPI (high throughput immunoblotting) technology (PowerBlot™, BD Biosciences) [9–16] has recently been developed. This proteomic method employs a panel of 1000 monoclonal antibodies targeting human or rat and mouse proteins. A total of 5 large SDS/PAGE blots were produced. Subsquently each of these blots was separated into 40 lanes using a manifold system. Each lane was then probed with multiple monoclonal antibodies that target protein antigens with good separation data [MM (molecular mass) difference], thus achieving a high throughput analysis. Since HTPI is still a Western blot in principle, it is an excellent method for separating intact proteins and their potential BDPs. We argue that it is an excellent system with which to study proteolysis. We further hypothesize that HTPI can assist us in identifying the complete set of brain protein substrates (degradome) that undergo proteolytic degradation during and after TBI. We term this the ‘TBI degradome’, fashioned after the term ‘degradome’ coined by Lopez-Otin and Overall [17,22]. It is likely that following TBI, some proteins will be up- or down-regulated, rather than just being proteolytically modified. Thus to further identify those that are degradomic, we contrasted the TBI differential proteome in parallel with calpain-2- and caspase-3-mediated degradation patterns, as generated by in vitro digestion of a naïve hippocampal lysate with these two proteases. To our knowledge, this is the first report using an HTPI approach to explore proteolytic systems in both an in vitro and in vivo system. Our method might also prove useful in identifying protein substrates for novel proteases of unknown function.
EXPERIMENTAL
In vivo model of TBI
A CCI (controlled cortical impact) device was used to model TBI in rats as described [28]. Briefly, adult male (280–300 g) Sprague–Dawley rats (Harlan, Indianapolis, U.S.A.) were anaesthetized with 4% isofluorane in a carrier gas of O2/N2O, 1:1 (4 min duration) followed by maintenance anaesthesia with 2.5% isofluorane in the same carrier gas. Core body-temperature was monitored continuously by a rectal thermistor probe and maintained at 37±1 °C by placing an adjustable temperature controlled heating pad beneath the rats. Animals were supported in a stereotactic frame in a prone position and secured by ear and incisor bars. A midline cranial incision was made, the soft tissues revealed, and a unilateral (ipsilateral to the site of impact) craniotomy (7 mm diameter) was performed adjacent to the central suture, midway between bregma and lambda. The dura mater was kept intact over the cortex. Brain trauma was produced by impacting the right cortex (ipsilateral cortex) with a 5 mm diameter aluminum impactor tip (housed in a pneumatic cylinder) at a velocity of 3.5 m/s with a 1.6 mm (severe) compression and 150 ms dwell-time (compression duration). These injuries were associated with local cortical contusion and diffuse axonal damage. Velocity was controlled by adjusting the pressure (compressed N2) supplied to the pneumatic cylinder. Velocity and dwell-time were measured by a linear velocity displacement transducer (Lucas Shaevitz™ model 500 HR, Detroit, MI, U.S.A.) that produced an analogue signal which was recorded by a storage-trace oscilloscope (BK Precision, model 2522B, Placentia, CA, U.S.A.). Sham-injured control animals underwent identical surgical procedures but did not receive an impact injury. Pre- and post-injury management were in compliance with guidelines set forth by the University of Florida Institutional Animal Care and Use Committee and the NIH (National Institutes of Health) guidelines detailed in the Guide for the Care and Use of Laboratory Animals.
Hippocampal tissue collection and protein extraction
After CCI (48 h), animals were anaesthetized and immediately killed by decapitation. Brains were immediately removed, rinsed with ice-cold PBS and halved. For the injured hemispheres, the hippocampus was rapidly dissected out, rinsed in ice-cold PBS, snap-frozen in liquid nitrogen, and frozen at −80 °C until further use. For Western blot analysis, the brain samples were pulverized to a fine powder with a small mortar/pestle set over solid CO2. The pulverized hippocampal tissue powder was then lysed for 90 min at 4 °C with 50 mM Tris (pH 7.4), 5 mM EDTA, 1% (v/v) Triton X-100 and 1 mM DTT (dithiothreitol). Owing to the need for in vitro protease-mediated digestion, protease inhibitor cocktail was not used. Instead, extreme care was taken to keep samples as cold as possible and to work rapidly to reduce formation of post-mortem artefacts. The brain lysate was then centrifuged at 8000 g for 5 min at 4 °C, to clear and remove insoluble debris, snap-frozen and stored at −85 °C until further use.
Calpain-2 and caspase-3 digestion of naïve brain lysate and purified proteins
Hippocampal lysates were prepared as described above. For purified protein digestion, βII-spectrin and synaptotagmin were used. βII-Spectrin (as a subunit of the αII/βII-spectrin heterotetramer) was purified from rat brain as described previously [27]. Recombinant synaptotagmin-1 [as an N-terminal GST (glutathione S-transferase) fusion protein, approx. 90 kDa] was obtained from Abnova Corp (Taiwan).
In vitro protease digestion of the naïve rat hippocampal lysate (5 mg) or purified protein with purified proteases, human calpain-2 (Calbiochem; 1 μg/μl) and recombinant human caspase-3 (Chemicon; 1 unit/μl) (at a protein/protease ratio of 1:200 and 1:50 respectively), was performed in a buffer containing 100 mM Tris/HCl (pH 7.4) and 20 mM DTT. For calpain-2, 10 mM CaCl2 was also added, and the solution then incubated at room temperature for 30 min. For caspase-3, 2 mM EDTA was added instead of CaCl2 and was incubated at 37 °C for 4 h. The protease reaction was stopped by the addition of SDS/PAGE sample buffer containing 1% (w/v) SDS.
HTPI
Four sets of pooled samples (n=6; naïve rat hippocampus, TBI hippocampus, calpain-digested hippocampus, and caspase-3-digested hippocampus) were prepared and subjected to 5 sets of gel/blots (templates A–E). The electrophoresis and blots were performed at the BD Bioscience facility (Bringamham, KY, U.S.A.). Briefly, the gels were 13 cm×10 cm, SDS/4–15% gradient polyacrylamide and 0.5 mm thick (Bio-Rad Criterion IPG well comb). A gradient system was used so that a large size-range of proteins could be detected on one gel. The protein extract (200 μg) was loaded onto one large well spanning the entire width of the gel. This translates into approx. 10 μg of protein extract per lane on a standard 10-well mini-gel. The gel was run for 1.5 h at 150 V. Proteins separated in the gel were transferred to an Immobilon-P membrane (0.2 μm, Millipore) over 2 h at 200 mA. We used a wet electrophoretic transfer apparatus TE Series from Hoefer. After transfer, the membrane was dried and re-wet in methanol. The membrane was blocked for 1 h with blocking buffer (LI-COR Biosciences). Next, the membrane was clamped with a Western blotting manifold that isolates 40 channels across the membrane. In each channel, a complex antibody cocktail (4–6 antibodies) was added and allowed to hybridize for 1 h at 37 °C. Each blot has 39 usable lanes and one lane on the far right probed with antibodies against MM markers; l lane (number 40) in templates A, B, C and D was loaded with a cocktail composed of MM standards: p190 (190 kDa), adaptin β (106 kDa), STAT-3 (signal transducer and activator of transcription-3) (92 kDa), PTP1D (non-transmembrane protein-tyrosine phosphatase) (72 kDa), MEK-2 [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase] (46 kDa), RACK-1 (receptor for activated C-kinase) (36 kDa), GRB-2 (growth factor receptor-bound protein-2) (24 kDa) and Rap2 (21 kDa). Lanes 20 and 40 of template E blots were loaded with 2 standardization cocktails (1, 112, 83, 62, 55, 42, 28 and 15 kDa; 2, 190, 120, 101, 60, 50, 27 and 21 kDa). The blot was removed from the manifold, washed and hybridized for 30 min at 37 °C with secondary goat anti-mouse antibodies conjugated to an Alexa Fluor® 680 fluorchrome (Molecular Probes). The membrane was washed, dried and scanned at 700 nm for monoclonal antibody target detection using the Odyssey Infrared Imaging System (LI-COR Biosciences). Samples were run in triplicate and analysed using a 3×3 matrix comparison method.
Traditional immunoblotting
Tissue samples (20 μg) were subjected to electrophoresis, equal volumes of samples for SDS/PAGE were prepared in a 2-fold loading buffer [0.25 M Tris (pH 6.8), 0.2 M DTT, 8% SDS, 0.02% Bromophenol Blue and 20% glycerol in distilled H2O], and gels were run at 120 V for 2 h in a mini-gel unit (Invitrogen). Protein bands were transferred to PVDF membrane on a semi-dry Transblot unit (Bio-Rad) at 20 V for 2 h. After electrotransfer, blotting membranes were blocked for 1 h at ambient temperature in 5% non-fat milk in TBST [20 mM Tris/HCl (pH 7.4), 150 mM NaCl and 0.05% (w/v) Tween-20), then incubated with the primary monoclonal antibody in TBST/5% milk. Primary antibodies used were anti-αII-spectrin from Affiniti, anti-βII-spectrin, anti-CaMPK-II, anti-CaMPK-IV, anti-striatin, anti-synaptojanin-1, anti-synaptotagmin and anti-NSF (N-ethylmaleimide-sensitive fusion protein) (all from BD Bioscience). The blot was then washed 3 times for 15 min with TBST and exposed to biotinylated secondary antibodies (Amersham) followed by a 30 min incubation with streptavidin-conjugated alkaline phosphatase (colorimetric method). Colorimetric development was performed with a one-step 5-bromo-4-chloro-3-indolyl phosphate-reagent (Sigma). The MMs of intact proteins and their potential BDPs were assessed by running along-side rainbow coloured MM standards (Amersham). Semi-quantitative evaluation of protein and BDP levels was performed via computer-assisted densitometric scanning (Epson XL3500 high resolution flatbed scanner) and image analysis using Image-J 1.5 software (NIH).
RESULTS
αII-Spectrin immunoblot – a positive control before HTPI
Our experimental design called for 4 treatment groups: naïve hippocampal lysate (naïve), TBI (CCI with 1.6 mm deformation distance) hippocampal lysate, and in vitro calpain-2 and caspase-3 digestion of naïve hippocampal lysates. We initially considered using unpooled individual samples for HTPI analysis, but running n=6 samples for the 4 conditions (naïve, calpain, caspase and TBI) would mean a total of 24 samples. The cost of running such expansive analyses would be formidable and extremely time consuming. We have, therefore, decided on an alternative strategy of pooling 6 samples from each group to enhance signal-to-noise ratio and to identify ‘putative’ hits. Subsequently, our strategy was to rely on follow-up ‘hit’-confirmation using individual brain lysate sample analysis with traditional immunoblots. The naïve hippocampal lysate was separately prepared from 6 individual rats and 1 mg of protein from each sample was pooled. Similarly, TBI samples were prepared from 6 individual injured rats and 1 mg of protein from each sample was pooled. As for calpain-2 and caspase-3 digestion, 3 mg of protein from pooled naïve samples was digested separately with calpain-2 or caspase-3 at protease/substrate protein ratios of 1:200 and 1:50 respectively. This process was repeated twice to obtain up to 6 mg of protein digest.
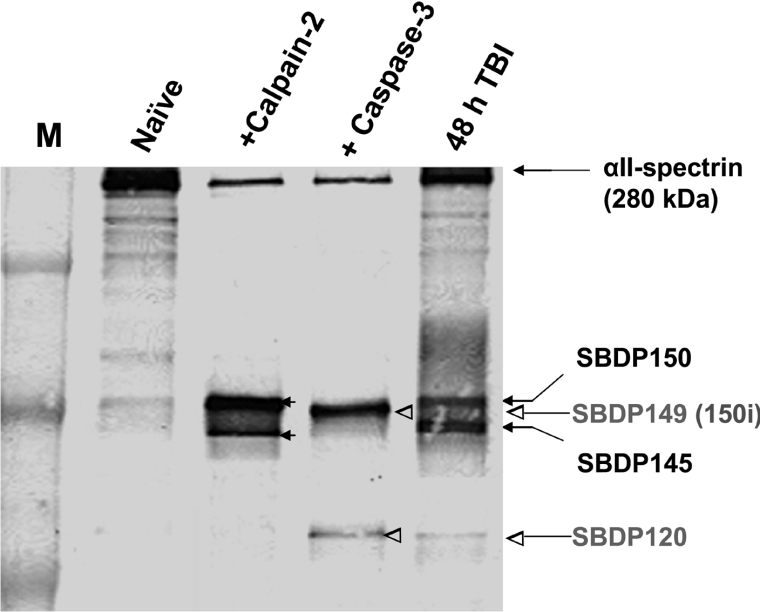
To ensure that the quality of the pooled samples was sound before subjecting them to the rigorous HTPI process, we subjected these pooled samples to traditional immunoblotting for αII-spectrin, a well established target of both calpains and caspases [5]. Naïve brains present only intact αII-spectrin of MM 280 kDa. Calpain-2 digestion decreased the levels of intact αII-spectrin and produced BDPs of 150 and 145 kDa (SBDP150 and SBDP145), whereas caspase-3 digestion also decreased the levels of intact αII-spectrin and yielded BDPs of 149 (SBDP149; also known as SBDP150i) and 120 kDa (SBDP120), as expected, based on our previous experience [5,18] (Figure 1). In the pooled TBI hippocampal samples, the decrease in αII-spectrin levels was not as notable, but a mixture of SBDP150/149, SBDP145 and SBDP120 was observed (Figure 1), as previously identified [19]. This confirmed that the 4 samples we prepared for this proteomic study have preserved the expected characteristic features.
Figure 1. αII-Spectrin immunoblot, a positive control before HTPI.
Pooled naïve rat hippocampal lysate, lysates digested with calpain-2 or caspase-3 in vitro, and rat TBI hippocampal lysate. Separate naïve and TBI hippocampi from 6 individual animals respectively were pooled. To confirm that the extent of proteolysis in the last 3 samples was comparable, we analysed 20 μg of protein from each of these samples by traditional SDS/6% PAGE, immunoblotting and probing with monoclonal anti-αII-spectrin antibody (Affiniti anti-fodrin). Intact protein (280 kDa) was observed under all conditions. Calpain-2 digestion produced major fragments SBDP150 (150 kDa) and SBDP145 (145 kDa) (solid arrows), whereas caspase-3 digestion produced SBP149 (SBDP150i, 149 kDa) and SBDP120 (120 kDa) (open arrow heads) [5]. In TBI samples, a mixture of SBDP150, SBDP149, SBDP145 and SBDP120 was observed. M, molecular mass marker.
Each pooled sample was run in triplicate on each of the 5 HTPI templates (A–E). Of the 1000 antibodies probed, 550 of them gave a recognized and quantifiable signal after development. In addition, triplicate naïve samples developed in template A blots showed relatively high-reproducibility in overall banding-profiles (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/394/bj3940715add.htm). However, some variation in the intensity of selected protein bands was observed, as expected with this type of analysis. Thus it is important that samples are always run and compared in triplicate. Other templates (B–D) also showed similar levels of same-sample-consistency (results not shown).
Calpain-2 and caspase-3 degradomes and the TBI differential proteome identified by HTPI
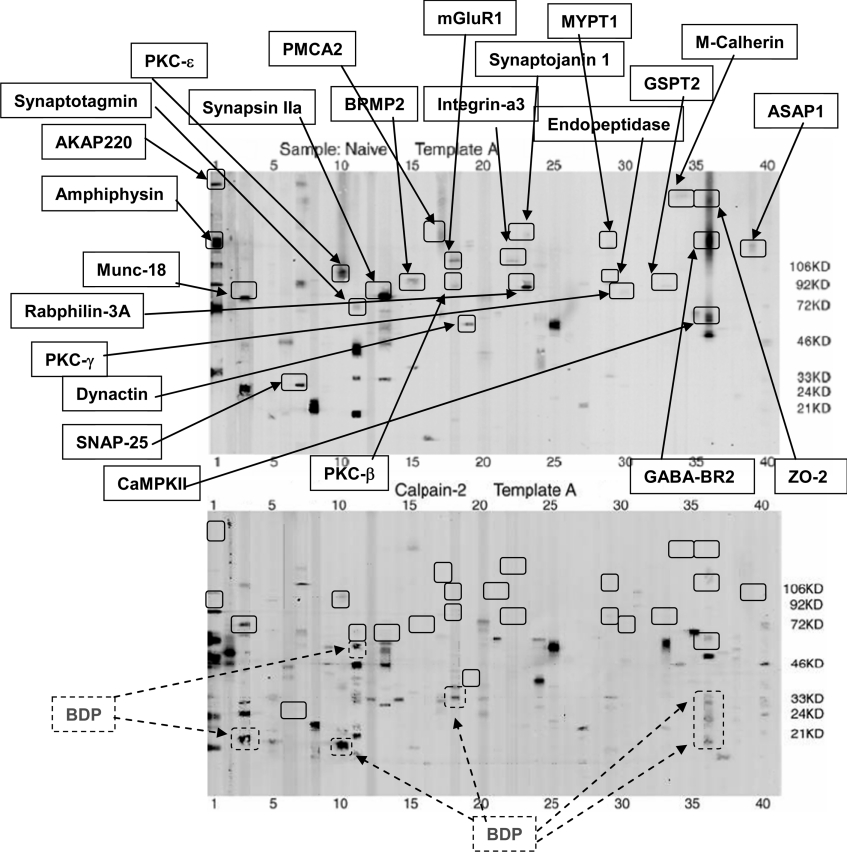
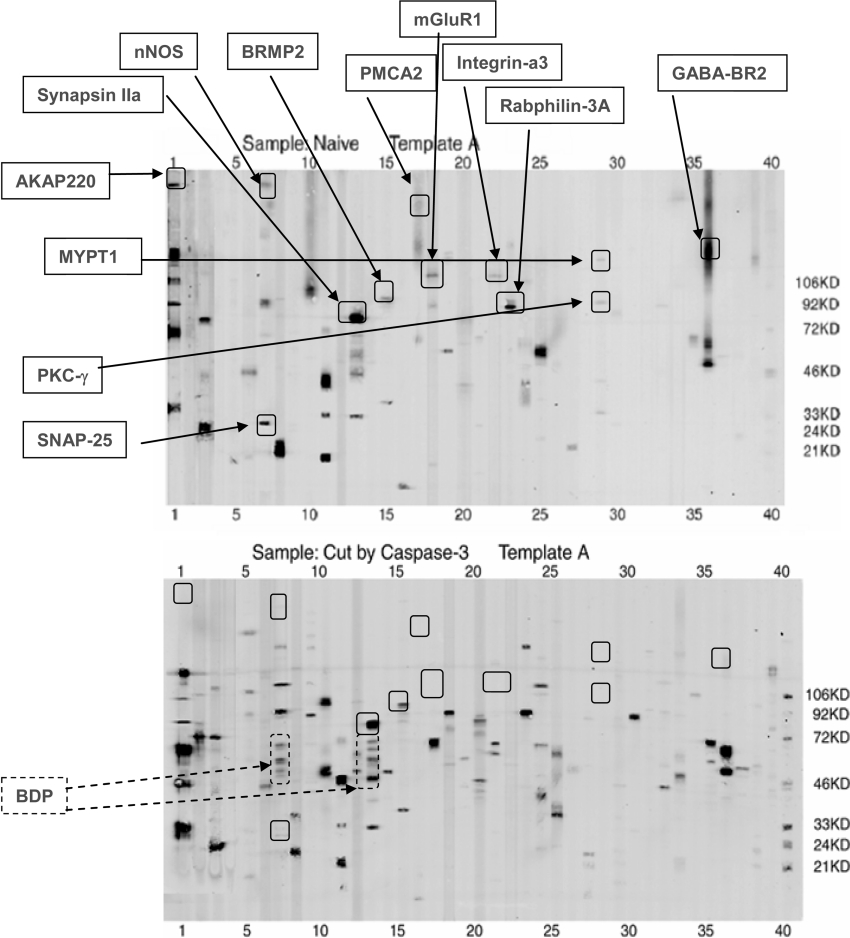
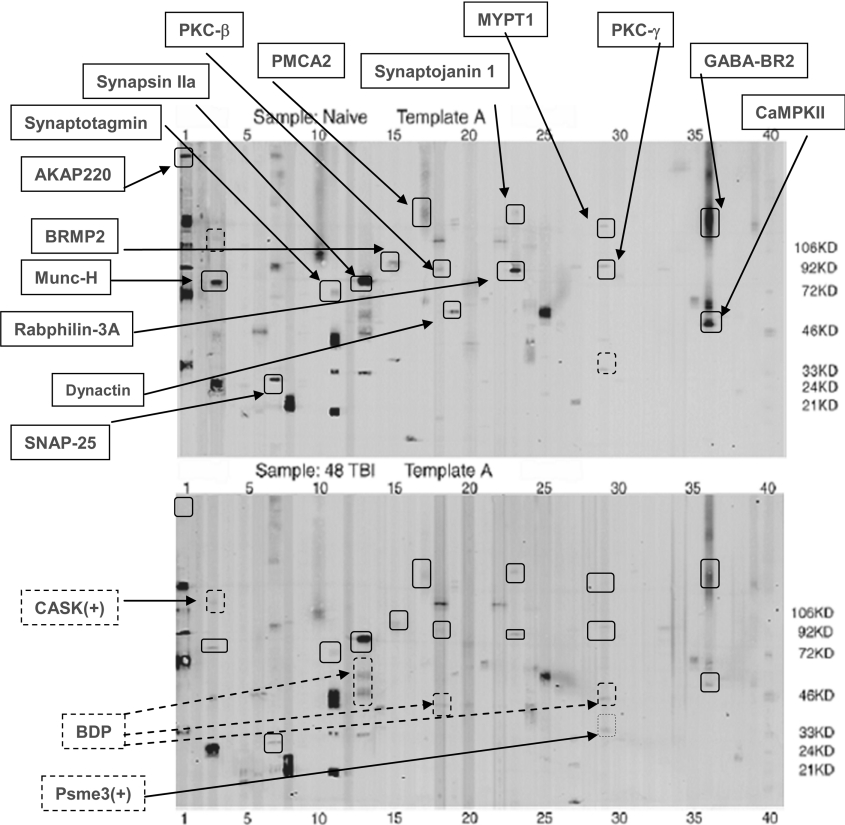
Since samples were run in triplicate, for each pairwise comparison (e.g. calpain-2 versus naïve), a 3×3 matrix comparison was made to cover all 9 combinations. Based on vigorous densitometric-computer-assisted and manual comparisons, we focused on parent protein-bands with significantly decreased intensity while also looking for the appearance of potential BDP bands after calpain-2/caspase-3 digestion (Figures 2 and 3) or after TBI (Figure 4). For example, representative template A blots for naïve and calpain-2 digestion are contrasted in Figure 3. We noted that 23 parent proteins had an average intensity that was reduced more than 2-fold after calpain digestion whereas 9 potential BDP bands (lanes 2, 10, 11, 13 and 36) were observed after calpain-2 digestion (Figure 2, lower panel). Similarly, 12 parent proteins in template A were significantly reduced in intensity after caspase-3 digestion as a result of proteolysis, with at least three identifiable BDPs observed in lanes 8 and 13 (Figure 3). Again, when template A for naïve hippocampus was compared with that of the TBI counterpart, 16 parent proteins were reduced in intensity, and therefore were either expression-down-regulated proteins or were potential proteolytic substrates (Figure 4). In addition, 2 potential BDPs can be readily observed (lanes 13, 18 and 29). In parallel, 2 proteins were found at up-regulated levels after TBI [CASK (calcium/calmodulin-dependent serine protein kinase), template A, lane 3, and Psme3 (proteasome activator subunit 3), A29] (Figure 4).
Figure 2. Example of the calpain-2 degradome (Template A).
A-templates from naïve hippocampus (upper panel) and calpain-2-digested hippocampal lysate (lower panel) were compared in triplicate (9 comparisons in total). One set of representative blots is shown. MM markers (lane 40) are indicated on the right. Protein bands with sufficient intensity were subsequently decoded and quantified using computer software as described in the Experimental section. We noted that for a number of proteins (solid box; upper panel), their average intensity decreased more than 2-fold after calpain digestion and several BDPs were also observed (BDP, dotted box; lower panel). ZO, zona occludin; for further definitions see Table 1, legend.
Figure 3. Example of the caspase-3 degradome (Template A).
A-templates from naïve hippocampus (upper panel) and caspase-3-digested hippocampal lysate (lower panel) were compared in triplicate. Only 1 set of representative blots is shown. MM markers (lane 40) are indicated on the right. Similarly to Figure 3, the expression of 9 parent proteins in template A (solid box, upper panel) was significantly decreased after caspase-3 digestion as a result of proteolysis, and several BDPs were observed (dotted box; lower panel). For definitions see Table 1, legend.
Figure 4. Example of the TBI differential proteome (Template A).
Template A for naïve hippocampus (upper panel) was compared with that for the TBI (1.6 mm deformation distance, 48 h) counterpart (lower panel). Comparisons were made in triplicate. A set of representative blots is shown. MM markers (lane 40) are indicated on the right. A total of 13 proteins in Template A were decreased in average intensity (down-regulated) after TBI (solid box; upper panel). In addition, several BDPs were readily observed (dotted box; lower panel). The only 2 proteins found to be up-regulated after TBI were CASK (A3) and Psme3 (A29) (dotted boxes). For definitions please see Table 1, legend.
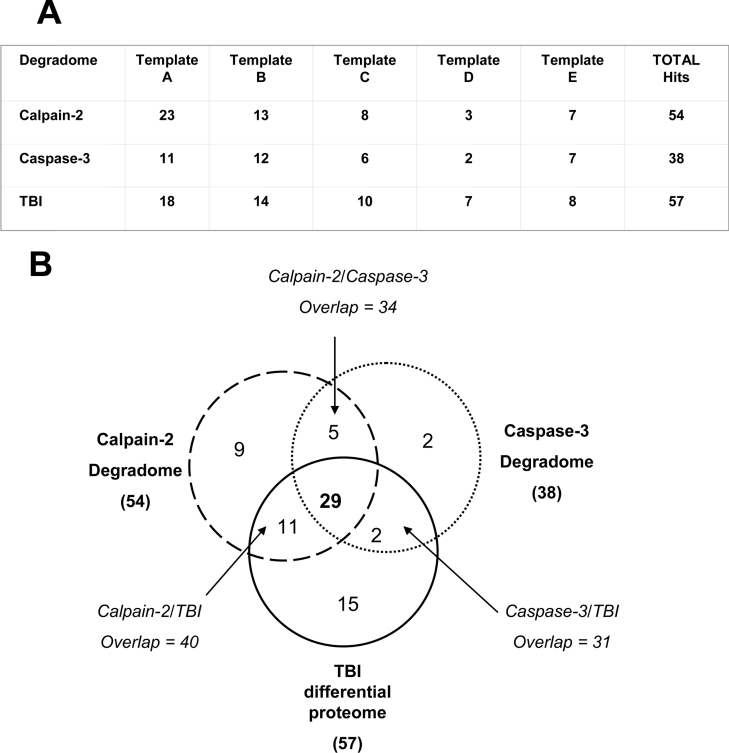
Template B comparisons also identified 13 to 12 potential proteolytic targets for calpain-2 and caspase-3 respectively and 14 differentially regulated proteins in TBI (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/394/bj3940715add.htm). In addition, templates C–E showed very similar proteomic patterns (results not shown). Table 1 summarizes the number of hits from each template when naïve hippocampal lysate was compared with calpain-2, caspase-3 and TBI treated samples respectively. In all, 54 and 38 proteins were putatively sensitive to calpain-2 and caspase-3 proteolysis respectively, whereas 48 proteins appeared to be down-regulated or degraded following TBI, and only 9 proteins were up-regulated [CASK, Psme3, α-actinin, ceruloplasmin, cdk2 (cyclin-dependent kinase2), NES1 (serine protease inhibitor kallikrein), TBP (TATA box-binding polypeptide), GS27 (Golgi SNARE 27) and Smg] (Figure 5A). Parent-protein signal reduction ranged from 2-fold to more than 10-fold. Based on 1000 antibodies used, the ‘hit’ rate for calpain digestion versus naïve lysate, and caspase-3 digestion versus naïve lysate, and TBI versus naïve lysate were 5.4%, 3.8% and 5.7% respectively. Furthermore, 40 of these proteins were common to the calpain-2 degradome and the TBI differential proteome, whereas 31 proteins were common to the caspase-3 degradome and the TBI differential proteome, as illustrated by the Venn diagram (Figure 5B). There were also significant overlaps (34 proteins) between the calpain-2 and caspase-3 degradomes. Lastly, 29 proteins were identified as putative degradomic targets under all three treatment conditions. However, it is important to note that the TBI differential proteome is not necessarily entirely degradative, but, in part, a result of changes in protein expression. Besides the 42 proteins in TBI samples that overlapped with the calpain/caspase-3 degradomes, we also found 6 additional proteins with decreased signal but with no calpain/caspase degradation counterparts [RIP2/RICK (receptor-interacting protein kinase2/Rip-like interacting caspase-like apoptosis-regulatory protein kinase), C26; syntaxin-6, C7; RONa, D27; PEX19 (peroxisome assembly factor 19), D29; SCAMP1 (secretory carrier-associated membrane protein1), D38 and SLK (Ste20-like kinase), E13] (Figure 5A, Table 1). Table 1 further details the identity of putative calpain-2 and caspase-3 substrates and TBI differential target proteins. As expected, some previously known calpain substrates were identified by HTPI, including CaMPK-II, (A36) [7], dynamin (B26) [19], PKC (protein kinase C)-α (B25) -β (A18) and -γ (A30) [5,18]. SNAP25 (synaptosome-associated protein 25)- (A7) was described as a calpain substrate, although it was not well studied [20]. Previously reported dual calpain/caspase-3 substrates identified by HTPI included the PMCA2 (plasma membrane calcium pump isoform2) (A17) [20,21], βII-spectrin (E8) [18] and CaMPK-IV (C12) [8] (Table 1). Of 48 TBI-down-regulated proteins, (Table 1), 9 proteins are associated with synaptic vesicle docking and trafficking: synaptojanin-1 (a synaptic inositol 1,4,5-trisphosphate-kinase), synaptotagmin-1 (a synaptic vesicle-exocytosis calcium sensor) and NSF, and synapsin-Ia and -II, SNAP-25, Munc-18 (non-neuronal syntaxin binding protein), α/β-SNAP, amphiphysin and rabphilin-3A. These data suggest that proteolysis might play a significant role in synaptic dysfunctions following TBI.
Table 1. Identity of the degradomic protein targets for calpain-2 and capase-3, and differentially regulated proteins after TBI.
Protein names or abbreviations are listed in the far left column. Their template and lane location are listed on the second column. Swiss Pro ID and predicted MMs are also listed. Where a protein is altered in levels in one of the three conditions (calpain-2, capase-3 or TBI), the fold-change of the parent protein is indicated in the 3 columns on the far right. Since the majority of proteins showed decreased levels in the treatment group versus control (naive), the fold-change always indicates decrease as the default. When a protein level is increased (for some proteins in TBI), the fold-change increase is indicated with a (+) sign and the fold-change is given inside brackets. Also, we have used high stringency inclusion criteria. Only proteins with a band decrease or increase of at least 1.5-fold in all 9 possible comparisons between 3 treatment replicas and 3 control replicas are shown. *, Fold changes for each treatment group are given against controls (naïve). AKAP220, A-kinase anchoring protein; Arp, actin-related protein; ASAP1, Arf GTPase-activating protein; BRMP2, amphiphysin; c-Cbl, casitas B-lineage lymphoma; CtBP1, C-terminal-binding protein 1; cdk2, cyclin-dependent kinase2; DRBP76, double-stranded RNA-binding nuclear protein76; GABA-B-R2, γ-aminobutyric acid-B-receptor2; GS27, Golgi SNARE 27; GSTP, GTP-binding protein appearing to be essential for the G1 to S phase transition of the cell cycle; hPRP17, human homologue of yeast Prp17; MEF2D, myocyte-enhancer-binding factor 2D; mGluR1, metabotropic glutamate receptor1; MEK, MAPK/ERK kinase; Munc-18, non-neuronal syntaxin binding protein; MYPT1, myosin phosphatase target protein1; Nck, Nck adaptor protein 1; Nek2, NIMA (never in mitosis A)-related kinase; NES1, serine protease inhibitor kallikrein; nNOS, neuronal nitric oxide synthase; NSP1, non-structural protein 1; PEX19, peroxisome assembly factor 19; PKC, protein kinase C; PMCA2, plasma membrane calcium pump isoform2; RIP2/RICK (receptor-interacting protein kinase2/Rip-like interacting caspase-like apoptosis-regulatory protein kinase; SCAMP1, secretory carrier-associated membrane protein1; SCAR-1, suppressor of cAMP receptor-1; SLK, Ste20-like kinase; Smg/GDS, stimulatory GDP/GTP exchange protein; TBP, TATA box-binding polypeptide; TFIIS, transcription factor IIS; TNIK, Traf2 and Nck-interacting kinase.
| Protein | Lane | Swiss Pro ID | MM (kDa) | Calpain-2* (fold-decrease) | Caspase-3* (fold-decrease) | TBI* [fold-decrease/(increase)] |
|---|---|---|---|---|---|---|
| Template A | ||||||
| AKAP220 | 1 | Q62924 | 220 | >10 | >10 | >10 |
| Amphiphysin | 1 | Q95163 | 125 | >10 | ||
| ASAP1 | 39 | Q9QWY8 | 130 | >10 | ||
| BRMP2 | 15 | O08539 | 96/89 | >10 | 7.31 | |
| CaMPK-II | 36 | P11275 | 52 | 5.23 | 5.67 | |
| CASK | 3 | Q62915 | 120 | (+5.8) | ||
| Dynactin | 19 | Q13561 | 50 | >10 | >10 | >10 |
| Endopeptidase | 29 | P42676 | 80 | >10 | ||
| GABA-BR2 | 36 | O088871 | 130 | >10 | >10 | >10 |
| GSTP2 | 33 | O88180 | 88 | 4.77 | ||
| Integrin-α3 | 22 | Q62470 | 135 | >10 | >10 | >10 |
| M-calherin | 34 | P10287 | 130 | >10 | ||
| mGluR1 | 19 | P23385 | 133 | >10 | ||
| Munc-18 | 3 | Q99PV2 | 68 | >10 | >10 | |
| MYPT1 | 29 | Q9DBR7 | 130 | 3.38 | 3.93 | 5.00 |
| nNOS | 7 | P29475 | 155 | >10 | ||
| PKC-β | 18 | P05771 | 80 | >10 | 7.31 | |
| PKC-ϵ | 10 | Q02156 | 90 | 6.50 | ||
| PKC-γ | 30 | P05129 | 80 | >10 | 3.30 | >10 |
| PMCA2 | 17 | P11506 | 133 | >10 | >10 | >10 |
| Psme3 | 29 | Q12920 | 36 | (+4.7) | ||
| Rabphilin-3A | 23 | P47709 | 75 | >10 | >10 | |
| SNAP-25 | 7 | P13795 | 25 | >10 | >10 | >10 |
| Synapsin-IIa | 13 | Q63537 | 74 | >10 | 1.70 | 10.10 |
| Synapsin-1 | 23 | Q62910 | 140 | >10 | >10 | |
| Synaptotagmin | 11 | P21579 | 65 | 2.91 | 1.50 | |
| Template B | ||||||
| α-Actinin | 9 | AAA51582 | 104 | (+8.41) | ||
| Adaptin | 29 | P17426 | 112 | >10 | 6.12 | |
| Bad | 30 | Q61337 | 23 | 10.80 | 3.23 | 3.00 |
| β-Catenin | 5 | Q02248 | 92 | >10 | >10 | >10 |
| Cathepsin L | 22 | P07711 | 43 | 7.18 | >10 | 7.71 |
| Ceruloplasmin | 37 | Q61147 | 150 | (>+10) | ||
| Dynamin | 26 | P21575 | >100 | 10.69 | >10 | 4.41 |
| Ki-67 | 8 | P46013 | 395 | >10 | >10 | |
| MEF2D | 3 | Q63943 | 70 | >10 | >10 | |
| Nck | 2 | P16333 | 47 | >10 | ||
| NSP1 | 24 | Q9Y2X4 | 72 | >10 | >10 | >10 |
| p150Glued | 26 | P28023 | 150 | >10 | >10 | >10 |
| p190 | 3 | Q13017 | 190 | >10 | >10 | >10 |
| PKC-α | 25 | P17252 | 82 | 3.67 | >10 | |
| SII/TFIIS | 32 | P23193 | 38 | >10 | >10 | >10 |
| Smac/Diablo | 6 | Q9NR28 | 22 | >10 | >10 | >10 |
| DRBP76 | 7 | Q13906 | 90 | >10 | ||
| Template C | ||||||
| Arp-3 | 18 | P32391 | 50 | >10 | >10 | |
| CaMPK-IV | 12 | Q16566 | 60 | 3.11 | 2.63 | 4.26 |
| c-Cbl | 9 | P22681 | 120 | >100 | ||
| cdk2 | 9 | P24941 | (>+10) | |||
| hPRP-17 | 15 | O60508 | 66 | 3.33 | 2.36 | |
| MEK2 | 5 | P51955 | 46 | >10 | ||
| NES1 | 38 | O43520 | 30 | (>+10) | ||
| p55-Cdc | 8 | Q9BW56 | 55 | >10 | ||
| Profilin | 28 | P07737 | 15 | >10 | >10 | |
| RIP2/RICK | 26 | AAH04553 | 61 | 2.59 | ||
| α/β-SNAP | 21 | P54920 | 35/36 | >10 | 13.79 | |
| Syntaxin 6 | 7 | Q63635 | 31 | 3.81 | ||
| Striatin | 22 | P70483 | 110 | 9.31 | 3.01 | 6.51 |
| TBP | 26 | P20226 | 37 | (+7.31) | ||
| TNIK | 38 | Q9UKE5 | 150 | 2.59 | ||
| Template D | ||||||
| Catenin/p120 | 32 | P30999 | 120 | >10 | >10 | >10 |
| GS27 | 10 | O14653 | 27 | (>+10) | ||
| p190-B | 28 | Q13017 | 195 | >10 | >10 | >10 |
| PEX19 | 29 | P40855 | 38 | 6.31 | ||
| RONa | 27 | Q04912 | 40 | >10 | ||
| SCAMP1 | 38 | O15126 | 36 | 7.40 | ||
| Synapsin-Ia | 38 | P09951 | 80 | >10 | 9.90 | |
| ATP synthase | 21 | P15999 | 55 | >10 | 5.84 | >10 |
| βII-Spectrin | 8 | Q01082 | 240 | >10 | >10 | 6.30 |
| β-Raf | 25 | P15056 | 95/72 | >10 | >10 | 2.71 |
| Clathirin heavy chain | 39 | P11442 | 120 | >10 | >10 | 7.43 |
| CtBP1 | 26 | Q91W16 | 48 | >10 | >10 | |
| NSF | 13 | P46459 | 82 | >10 | 3.30 | 2.59 |
| SCAR-1 | 4 | Q92588 | 80 | >10 | 7.50 | 3.23 |
| SLK | 13 | NP_055535 | 125 | 4.00 | ||
| Smg/GDS | 13 | CAA45067 | 57 | (>+10) |
Figure 5. Summary of the calpain-2 and caspase-3 degradomes and differential TBI proteome results from HTPI.
(A) The number of putative degradomic hits for each template based on calpain-2 versus naïve, caspase-3 versus naïve, and TBI versus naïve comparisons were tabulated. The total number of degradome hits is listed on the far right. (B) Venn diagram showing overlap of protein targets in the 3 degradomes (calpain, dashed line; caspase-3, dotted line; TBI, solid line). The total number of protein targets is in brackets. Overlaps and triple overlap numbers are indicated.
Cytoskeleton-associated proteins dynamin and dynactin, as well as the actin-binding protein, profilin, were also identified as TBI-proteolytic substrates. Adhesion molecules (M-calherin and integrin-α3) and adaptor proteins β-catenin and adaptin were also identified. Again, proteolysis of these proteins can lead to cytoskeletal degradation and compromise cell shape. Two neurotransmitter receptors, mGluR1 (metabotropic glutamate receptor 1) and GABA-B-R2 (γ-aminobutyric acid-B receptor 2), also appeared to be sensitive to proteolysis (Table 1).
Two cell cycle proteins [Ki-67 and p55-Cdc (cell division cycle)] were also identified in the TBI differential proteome. The TBI differential proteome also includes two apoptosis-associated proteins that have not previously been identified as sensitive to proteases, Bad protein (B36) that translocates to mitochondria, as well as the mitochondria-released Smac/Diablo (B6) that binds inhibitor of apoptosis proteins 1 and 2, thus facilitating the induction of apoptosis. How proteolysis influences the functions of some of these proteins remains to be elucidated.
Degradome and TBI differential proteome target validation
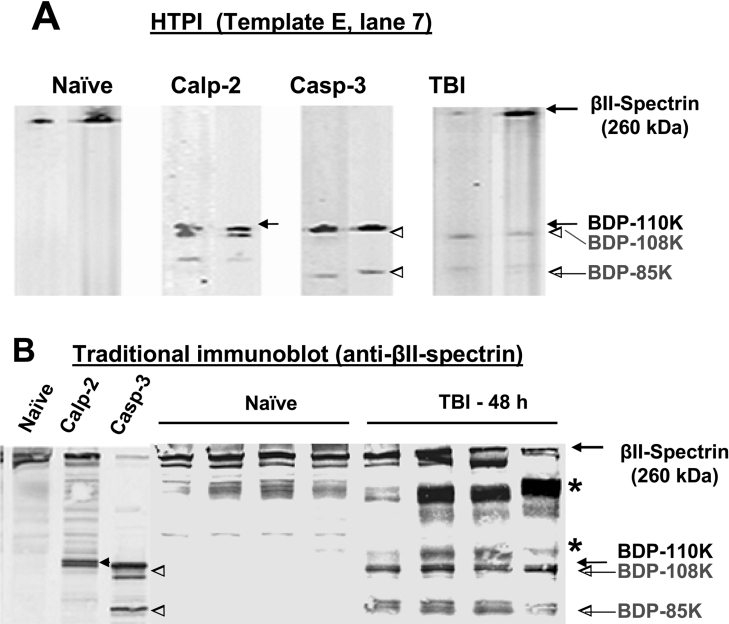
In order to assess the confidence of the degradomic target assignment based on HTPI, we first asked how the HTPI results compared with traditional immunoblotting for a specific protein. From E8, we observed that out of all triplicate runs, the βII-spectrin (240 kDa) level consistently diminished upon protease treatments, whereas putative BDPs of 110 kDa were identified upon calpain digestion, and BDPs of 108 kDa and 85 kDa were observed upon caspase-3 digestion. TBI also produced loss of intact βII-spectrin and fainter BDP bands of 110/108 and 85 kDa (see Figure 8A). In parallel to HTPI, we applied the pooled naïve hippocampal lysate and those subjected to calpain-2 and capase-3 digestion to traditional SDS/PAGE followed by immunoblotting with a monoclonal anti-βII-spectrin antibody (BD Biosciences) that was identical to that used in the HTPI. As illustrated in Figure 6(B), although naïve brains contained no BDPs, calpain-2 and capase-3 digestion produced BDPs of 110 kDa and of 108 and 85 kDa respectively, virtually identical to the patterns generated by HTPI (Figure 8A). In addition, we also assessed the integrity of βII-spectrin in 4 individual naïve and 4 individual TBI hippocampal samples. Consistent with the HTPI data shown in Figure 7(A), we again observed, in traditional immunoblots, the presence of BDPs of 110, 108 and 85 kDa in all 3 TBI hippocampal samples, but not in the naïve samples (Figure 8B).
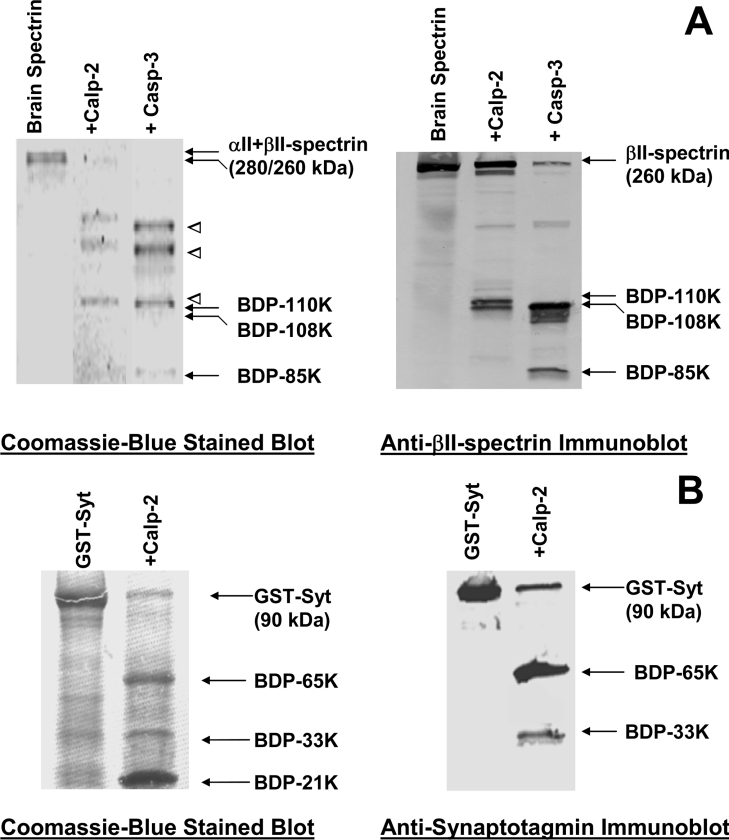
Figure 8. Examples of 2 purified proteins confirmed as proteolytic targets for calpain/caspase-3: βII-spectrin and synaptotagmin-1.
Purified βII-spectrin (as a subunit of rat brain αII/βII-spectrin) (A) and recombinant GST–synaptotagmin-1 (B) were subjected to calpain-2/caspase-3 digestion, and calpain-2 only digestion respectively. Both Coomassie Blue stained blotting-membrane (left panels) and immunoblotting analysis with anti-βII-spectrin and anti-synaptotagmin-1 antibodies (right panels) were performed. Intact proteins and major BDPs are labelled with arrows. Several major αII-spectrin BDPs were also identified (open triangles).
Figure 6. An HTPI approach allows rapid target confirmation.
(A) Extracted lanes (E7) from template E of the HTPI gel: intact βII-spectrin (240 kDa) expression level was shown to be significantly decreased by calpain-2 digestion, caspase-3 digestion and after TBI. A calpain-mediated BDP of 110 kDa (black label) and 2 caspase-mediated BDPs of 108 and 85 kDa respectively (grey labels) were tentatively identified. These 3 BDPs were also tentatively identified in the TBI samples. (B) Traditional SDS/PAGE and Western blotting were also performed using identical monoclonal anti-βII-spectrin antibodies. Samples analysed were naïve (pooled) versus calpain-2 and capase-3 digestion (left 3 lanes), as well as 4 separate naïve and TBI samples. Again, BDPs of 110 kDa (solid arrow) and of 108 and 85 kDa (open arrow heads) were observed. * Indicates rat heavy-chain IgG and fragments from contaminating blood that cross-react with the secondary anti-(mouse IgG) antibody detection system used.
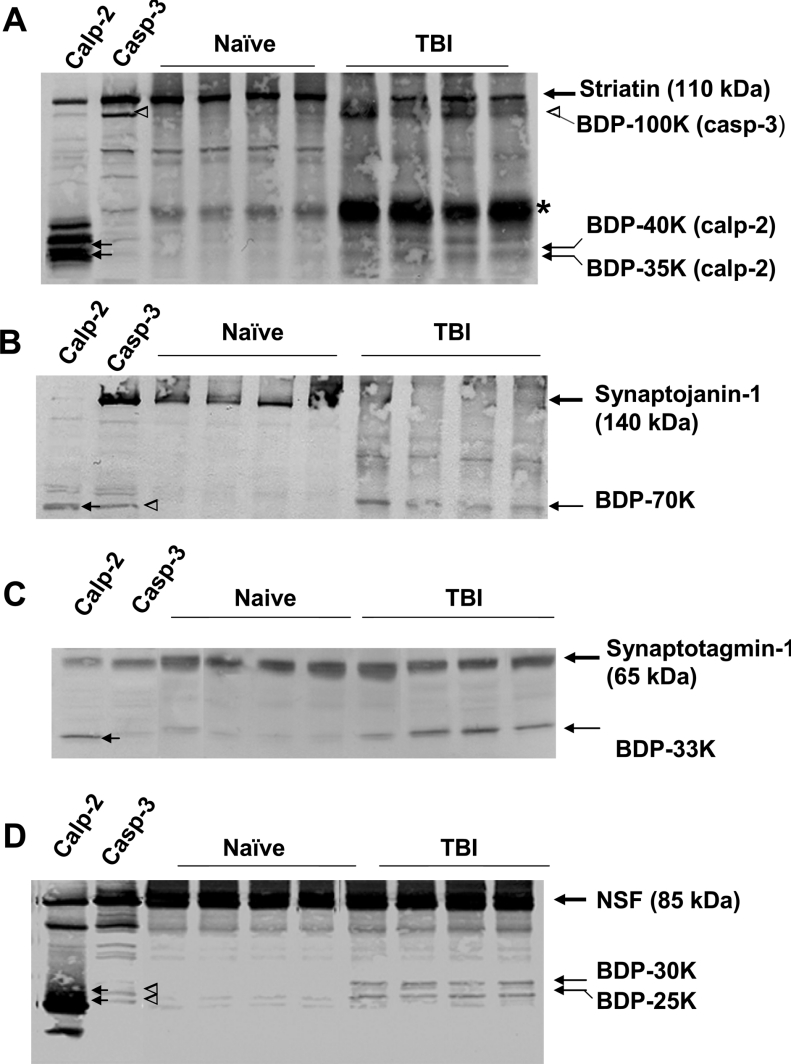
Figure 7. Examples of 4 proteins identified by HTPI as novel proteolytic targets.
A total of 4 proteins were identified as proteolytic targets for either calpain-2, capase-3 and/or in TBI: striatin (A), synaptojanin-1 (B), synaptotagmin-1 (C) and NSF (D). Traditional SDS/PAGE and Western blotting were also performed using monoclonal antibodies against striatin, synaptojanin-1, synaptotagmin (isoform I) and NSF. Samples analysed were calpain-2 and capase-3 digestions (2 left-hand lanes), as well as 4 separate naïve and TBI samples. In (A–D), intact proteins are shown with bold arrows (with MM in brackets). Calpain-2-mediated BDPs are shown with solid arrows. Caspase-3-mediated BDPs are shown with open arrow heads. MMs are as indicated. * In (A) indicates rat light-chain IgG from contaminating blood that cross-reacts with the secondary anti-(mouse IgG) antibody detection system used.
In this study, we identified over 30 novel protease substrates (see Table 1). To ascertain that these are truly proteolytic substrates, traditional immunoblots were again performed for 4 selected ‘novel’ degradomic targets: striatin (C22) and NSF (E13) as a calpain-2/caspase-3/TBI triple target; and synaptojanin-1 (A23) and synaptotagmin-1 (A11) as calpain/TBI double targets (see Table 1). Traditional immunoblotting results showed that striatin (110 kDa) was indeed sensitive to calpain-2 digestion, producing BDPs of 40 kDa and 35 kDa as predicted from HTPI. Caspase-3 digestion also produced a high MM fragment of 100 kDa (Figure 7A), which was not readily observed in HTPI. TBI samples also showed both a caspase-produced BDP of 100 kDa and calpain-produced BDPs of 40 and 35 kDa. Next, we confirmed that synaptojanin-1 (140 kDa) was highly sensitive to calpain digestion, producing a BDP of 70 kDa. Caspase-3 digestion also partially degraded synaptojanin-1 to a faint 70 kDa fragment, which was not observed in HTPI, probably due to sensitivity differences. Importantly, following TBI, intact synaptojanin-1 protein was almost completely degraded to the 70 kDa BDP (Figure 7B). Synaptotagmin-1 (65 kDa, A11) was degraded by calpain-2 to a BDP of 33 kDa but not by caspase-3 (Figure 7C). The BDP-33 kDa was also readily observed in all 4 TBI hippocampal samples (Figure 7C). Lastly, NSF was degraded by calpain and caspase-3 (to a lesser extent) to BDPs of 30 and 25 kDa (Figure 7D). We also established that both of these BDPs were readily observed in TBI samples but not in naïve samples (Figure 7D).
Finally, to directly prove that purified HTPI-identified target proteins are indeed vulnerable to calpain and/or caspase-3 digestion, we tested two proteins. We obtained purified βII-spectrin (as a subunit of the rat brain αII/βII-spectrin heterotetramer) and recombinant GST–synaptotagmin-1, and subjected them to calpain/caspase-3 digestion. Coomassie Blue staining of an SDS gel revealed that both αII- and βII-spectrin subunits (280 and 260 kDa respectively) were degraded by calpain and caspase-3, producing multiple fragments (Figure 8A). To ascertain that βII-spectrin was indeed a substrate for calpain and caspase-3, immunoblotting of the same samples probed with anti-βII-spectrin antibody was performed. βII-Spectrin breakdown patterns (BDP-110 kDa for calpain, BDP-108 kDa and -85 kDa for caspase-3) (Figure 8A) was virtually identical to those observed after HTPI of the hippocampal lysate digest (Figure 6). Similarly, recombinant GST–synaptotagmin-1 was vulnerable to calpain-2 proteolysis, producing 3 major fragments (65, 33 and 21 kDa) (Figure 8B). Again, to ensure that the fragments were derived from the intact synaptotagmin portion of the fusion protein, we performed immunoblotting with anti-synaptotagmin antibody and identified 2 immunoreactive polypeptides (65 and 33 kDa) (Figure 8B). Although the 65 kDa fragment represents the full-length synaptotagmin truncated from the GST portion (appearing as a 21 kDa fragment) at the artificial linker region, the 33 kDa synaptotagmin BDP was identical in size to that produced when the hippocampal lysate was digested with calpain (Figure 7).
DISCUSSION
In the present study, we combined two powerful and emerging areas in proteomics: degradomics [22] and high-throughput monoclonal antibody panel-based immunoblotting (HTPI) for protein identification [9,11]. Based on others' and our previous evidence showing dual attacks on neural proteins by calpains and caspase-3 under neural injury situations [5], we hypothesized that there would be significant overlap between the neuronal proteins vulnerable to proteolysis by direct calpain and caspase-3 digestion, and following TBI (i.e. calpain, caspase-3, and TBI-degradomes).
For our specific study, we compared and contrasted naïve with traumatically injured hippocampal lysates. Control craniotomies (sham operated) were performed, but not included in this study as this procedure also produced a mild form of brain trauma. As for the classical and ubiquitously distributed calpain-1 and -2, our evidence and others' has concluded that they share identical substrate sets [23]. Thus for simplicity, only calpain-2 was used for the in vitro digestion of hippocampal lysate. Similarly, various caspases are activated in brain-injury induced neuronal apoptosis (caspase-3, -8, -9 and -12) [23–26]. Among the execution caspases, the most relevant and well studied is caspase-3. Therefore it was selected for the present comparative degradomic study. The calpain-2 and caspase-3 degradomic data generated were then compared with TBI degradomic data (Table 1, Figure 5).
Using HTPI, we identified 43 proteins in the rat hippocampal proteome that were putatively degraded after TBI, whereas 54 and 38 proteins respectively were vulnerable to calpain-2 and caspase-3 proteolysis (Table 1). We further identified significant overlaps among the calpain-2 and caspase-3 degradomes, and TBI differential proteome, with 29 of these proteins common to the three degradomes (Figure 5). Within the calpain-2 and caspase-3 degradomes previously identified are calpain and capsase-3 substrates such as βII-spectrin, CaMPK-II and CaMPK-IV (Table 1). We identified and confirmed a number of previously unknown protease-sensitive target proteins in TBI, such as striatin, synaptotagmin-1, synaptojanin-1 and NSF, using traditional immunoblotting analysis of treated and untreated hippocampal lysates (Figure 7). We further confirmed that purified βII-spectrin and synaptotagmin-1 were in vitro substrates of calpain and/or caspase-3 (Figure 8).
Based on the significant overlaps among the calpain degradome, caspase-3 degradome and TBI differential proteome, it appears that these two proteases are operating in concert in TBI by attacking a subset of cellular proteins that are important to neuronal functions. For example, many novel TBI proteolytic targets are synaptic vesicle proteins (Table 1; Figure 7). It is tempting to suggest that proteolysis plays a significant role in the synaptic dysfunction following TBI. Many cytoskeleton proteins (dynamin, dynactin, and profilin and βII-spectrin) and cell adhesion proteins (adaptin and β-catenin) also appear to be at risk from proteolysis following TBI (Table 1). It should be noted that there are differences between the TBI differential proteome and the combined calpain/caspase-3 degradomes: although 42 TBI-down regulated proteins overlapped in calpain/caspase-3 degradomes, 6 other TBI-down-regulated proteins have no calpain/caspase counterparts. In addition, there are also 9 prominently up-regulated proteins in TBI lysates that could not be accounted for in calpain/capase-3 degradomes (Table 1, Figure 7).
To date, there are only a handful of published studies using HTPI/PowerBlot™ to address a biological problem [9–14]. Interestingly, only one previous paper used HTPI, in order to study post-translational modification protein conjugation to ISG15 (interferon stimulated gene15), a ubiquitin-like protein [12]. Thus to our knowledge, the present paper is the first report to use HTPI to study protein proteolysis. Although there are now several emerging proteomic technologies, including tryptic peptide analysis by tandem MS antibody microarray [29], the HTPI is the most ideally suited proteomic method to rapidly identify potential targets for a protease system. Most proteomic methods (MS/MS, antibody microarray) cannot readily distinguish intact proteins from their fragmented forms. However, HTPI, by contrast, is built on traditional immunoblotting technology. Thus intact proteins and their potential fragments were first resolved by one-dimensional SDS/PAGE before progression to electrotransfer and antibody probing. This method has been proven to be extremely powerful in identifying the occurrence of proteolysis, as well as distinct protein fragments (Figure 5B). We determined this to also be the case for HTPI (Figure 5A). Another powerful aspect of HTPI is the relative ease of protein identification and ‘hits’ confirmation. Since all the protein bands in the 5 templates are already annotated based on the applied monoclonal antibodies, putative protein identification is very rapid. Furthermore, since the exact antibodies used in the HTPI analysis are individually available, hit confirmation is rapid and robust (Figures 7 and 8). One potential drawback of using an antibody array approach is that antibody recognition of antigens might be species-specific. However, the 1000 antibody sets we employed were tested for species cross-reactivity (human/rat/mouse) and over 90% cross-react with protein antigen in human, rat and mouse. Of the 74 total hits in Table 1, all but 3 have confirmed rat-reactivity (95% cross-reactivity). The 3 exceptions are DRBP76 (double-stranded RNA-binding nuclear protein76), cathepsin L and p55-Cdc.
One of the potential limitations of the HTPI method is that it is not exhaustive. Currently the expansion of HTPI is limited by the availability of antibodies to specific protein antigens. However, in only a few years, the HTPI panel has already grown from 700 [9] to over 1000 monoclonal antibodies (present study). Another concern in using HTPI to identify proteolytic substrates in vivo is that this method will also detect proteins with significantly reduced expression levels rather than those that are degraded. However, we have compared the in vivo TBI differential proteome with in vitro protease degradomes (Figures 2–4), followed by traditional immunoblots which confirms the detection of BDPs (Figure 6). Therefore our approach adds another level of confidence to our interpretation of the degradomic data. Finally, any degradomic targets identified by HTPI (Figures 2–4) should be confirmed independently in follow-up studies, including cell-based studies where proteases of interest can be activated.
In summary, we have demonstrated that HTPI is a powerful and novel method for studying proteolytic pathways. In the present study, we have used the hippocampal proteome as an example to demonstrate the feasibility of using HTPI to study proteolytic targets in vivo and in vitro. This platform technology is applicable to the identification of potential targets for novel proteases with unknown functions. In addition, it is possible to identify specific protein hydrolysis/processing in a unique organ or cell system under physiological or pathological conditions.
Online data
Acknowledgments
The authors would like to acknowledge support from Department of Defense grants DAMD17-03-1-0066 and DAMD17-01-1-0765, and NIH grant R01 NS049175-01 A1. This paper has been reviewed by the Walter Reed Army Institute of Research and there is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. K. K. W. W. and R. L. H. hold equity in Banyan Biomarkers, Inc.
References
- 1.Yamashima T. Implication of cysteine proteases calpain, cathepsin and caspase in ischemic neuronal death of primates. Prog. Neurobiol. 2000;62:273–295. doi: 10.1016/s0301-0082(00)00006-x. [DOI] [PubMed] [Google Scholar]
- 2.Asah M., Asahi K., Jung J. C., del Zoppo G. J., Fini M. E., Lo E. H. Role for matrix metalloproteinase 9 after focal cerebral ischemia, effects of gene knockout and enzyme inhibition with BB-94. J. Cereb. Blood Flow Metab. 2000;20:1681–1689. doi: 10.1097/00004647-200012000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Clark A. W., Krekoski C. A., Bou S. S., Chapman K. R., Edwards D. R. Increased gelatinase A (MMP-2) and gelatinase B (MMP-9) activities in human brain after focal ischemia. Neurosci. Lett. 1997;238:53–56. doi: 10.1016/s0304-3940(97)00859-8. [DOI] [PubMed] [Google Scholar]
- 4.Phillips J. B., Williams A. J., Adams J., Elliott P. J., Tortella F. C. Proteasome inhibitor PS519 reduces infarction and attenuates leukocyte infiltration in a rat model of focal cerebral ischemia. Stroke. 2000;31:1686–1693. doi: 10.1161/01.str.31.7.1686. [DOI] [PubMed] [Google Scholar]
- 5.Wang K. K. W. Calpain and caspase, can you tell the difference. Trends Neurosci. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- 6.Pike B. R., Flint J., Johnson E., Glenn C. C., Dutta S., Wang K. K. W., Hayes R. L. Accumulation of calpain-cleaved non-erythroid βII-spectrin in cerebrospinal fluid after traumatic brain injury in rats. J. Neurochem. 2001;78:1297–1306. doi: 10.1046/j.1471-4159.2001.00510.x. [DOI] [PubMed] [Google Scholar]
- 7.Hajimohammadreza I., Nath R., Raser K. J., Nadimpalli R., Wang K. K. W. Neuronal nitric oxide synthase and calmodulin-dependent protein kinase IIα undergo neurotoxin-induced proteolysis. J. Neurochem. 1997;69:1006–1013. doi: 10.1046/j.1471-4159.1997.69031006.x. [DOI] [PubMed] [Google Scholar]
- 8.McGinnis K. M., Whitton M., Gnegy M. E., Wang K. K. W. Calmodulin-dependent protein kinase IV differentially cleaved by caspase-3 and calpain in SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 1998;273:1999–2000. doi: 10.1074/jbc.273.32.19993. [DOI] [PubMed] [Google Scholar]
- 9.Pasinetti G. M., Ho L. From cDNA microarrays to high-throughput proteomics. Implications in the search for preventive initiatives to slow the clinical progression of Alzheimer's disease dementia. Restor. Neurol. Neurosci. 2001;18:137–142. [PubMed] [Google Scholar]
- 10.Castedo M., Ferri K. F., Blanco J., Roumier T., Larochette N., Barretina J., Amendola A., Nardacci R., Metivier D., Este J. A., Piacentini M., Kroemer G. Human immunodeficiency virus 1 envelope glycoprotein complex-induced apoptosis involves mammalian target of rapamycin/FKBP12-rapamycin-associated protein-mediated p53 phosphorylation. J. Exp. Med. 2001;194:1097–1110. doi: 10.1084/jem.194.8.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melnick M., Chen H., Min-Zhou Y., Jaskoll T. The functional genomic response of developing embryonic submandibular glands to NF-κB inhibition. BMC Dev. Biol. 2001;1:15. doi: 10.1186/1471-213X-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malakhov M. P., Kim K. I., Malakhova O. A., Jacobs B. S., Borden E. C., Zhang D. E. High-throughput immunoblotting. Ubiquitiin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 2002;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 13.Yoo G. H., Piechocki M. P., Ensley J. F., Nguyen T., Oliver J., Meng H., Kewson D., Shibuya T. Y., Lonardo F., Tainsky M. A. Docetaxel induced gene expression patterns in head and neck squamous cell carcinoma using cDNA microarray and PowerBlot. Clin. Cancer Res. 2002;8:3910–3921. [PubMed] [Google Scholar]
- 14.Gifford S. M., Cale J. M., Tsoi S., Magness R. R., Bird I. M. Pregnancy-specific changes in uterine artery endothelial cell signaling in vivo are both programmed and retained in primary culture. Endocrinology. 2003;144:3639–3650. doi: 10.1210/en.2002-0006. [DOI] [PubMed] [Google Scholar]
- 15.Lorenz P., Ruschpler P., Koczan D., Stiehl P., Thiesen H. J. From transcriptome to proteome, differentially expressed proteins identified in synovial tissue of patients suffering from rheumatoid arthritis and osteoarthritis by an initial screen with a panel of 791 antibodies. Proteomics. 2003;3:991–1002. doi: 10.1002/pmic.200300412. [DOI] [PubMed] [Google Scholar]
- 16.Cicala C., Arthos J., Selig S. M., Dennis G., Jr, Hosack D. A., Van Ryk D., Spangler M. L., Steenbeke T. D., Khazanie P., Gupta N., et al. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favors virus replication. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9380–9385. doi: 10.1073/pnas.142287999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overall C. M., McQuibban G. A., Clark-Lewis I. Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning, a new tool for degradomics. Biol. Chem. 2002;383:1059–1066. doi: 10.1515/BC.2002.114. [DOI] [PubMed] [Google Scholar]
- 18.Wang K. K. W., Posmantur R., Nath R., McGinnis K., Whitton M., Talanian R. V., Glantz S., Morrow J. Simultaneous degradation of αII- and βII-spectrin by caspase 3 (CPP32) in apoptotic cells. J. Biol. Chem. 1998;273:22490–22497. doi: 10.1074/jbc.273.35.22490. [DOI] [PubMed] [Google Scholar]
- 19.Santella L., Kyozuka K., Hoving S., Munchbach M., Quadroni M., Dainese P., Zamparelli C., James P., Carafoli E. Breakdown of cytoskeletal proteins during meiosis of starfish oocytes and proteolysis induced by calpain. Exp. Cell Res. 2000;259:117–126. doi: 10.1006/excr.2000.4969. [DOI] [PubMed] [Google Scholar]
- 20.Wang K. K. W., Villalobo A., Roufogalis B. D. Calmodulin-binding proteins as calpain substrates. Biochem. J. 1989;262:693–706. doi: 10.1042/bj2620693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab B. L., Guerini D., Didszun C., Bano D., Ferrando-May E., Fava E., Tam J., Xu D., Xanthoudakis S., Nicholson D. W., Carafoli E., Nicotera P. Cleavage of plasma membrane calcium pumps by caspases, a link between apoptosis and necrosis. Cell Death Differ. 2002;9:818–831. doi: 10.1038/sj.cdd.4401042. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Otin C., Overall C. M. Protease degradomics, a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 23.Wang K. K. W., Yuen P. W. Calpain inhibition: an overview of its therapeutic potentials. Trends Pharmacol. Sci. 1994;15:412–419. doi: 10.1016/0165-6147(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 24.Posmantur R., McGinnis K., Nadimpalli R., Gilbertsen R., Wang K. K. W. Characterization of CPP32-like protease activity following apoptotic challenge in SH-SY5Y neuroblastoma cells. J. Neurochem. 1997;68:2328–2337. doi: 10.1046/j.1471-4159.1997.68062328.x. [DOI] [PubMed] [Google Scholar]
- 25.Larner S. F., McKinsey D. M., Torres M., Pike M., Hayes R. L., Wang K. K. W. Upregulation of caspase-12 after traumatic brain injury in rats. J. Neurochem. 2004;88:78–90. doi: 10.1046/j.1471-4159.2003.02141.x. [DOI] [PubMed] [Google Scholar]
- 26.Beer R., Franz G., Krajewski S., Pike B. R., Hayes R. L., Reed J. C., Wang K. K., Klimmer C., Schmutzhard E., Poewe W., Kampfl A. Temporal and spatial profile of caspase 8 expression and proteolysis after experimental traumatic brain injury. J. Neurochem. 2001;78:862–873. doi: 10.1046/j.1471-4159.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 27.Levilliers N., Peron-Renner M., Coffe G., Pudies J. Gelation and fodrin purification from rat brain extracts. Biochim. Biophys. Acta. 1986;882:113–126. doi: 10.1016/0304-4165(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 28.Pike B. R., Zhao X., Newcomb J. K., Posmantur R. M., Wang K. K. W., Hayes R. L. Regional calpain and caspase-3 proteolysis of α-spectrin after traumatic brain injury. NeuroReport. 1998;9:2437–2442. doi: 10.1097/00001756-199808030-00002. [DOI] [PubMed] [Google Scholar]
- 29.Wang K. K. W., Ottens A., Haskins W. E., Liu M. C., Denslow N., Chen S. S., Hayes R. L. Neuroproteomic studies of traumatic brain injury. Int. Rev. Neurobiol. 2004;61:215–240. doi: 10.1016/S0074-7742(04)61009-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.