Abstract
We have sequenced to high levels of accuracy 714-kb and 233-kb regions of the mouse and bovine X-inactivation centers (Xic), respectively, centered on the Xist gene. This has provided the basis for a fully annotated comparative analysis of the mouse Xic with the 2.3-Mb orthologous region in human and has allowed a three-way species comparison of the core central region, including the Xist gene. These comparisons have revealed conserved genes, both coding and noncoding, conserved CpG islands and, more surprisingly, conserved pseudogenes. The distribution of repeated elements, especially LINE repeats, in the mouse Xic region when compared to the rest of the genome does not support the hypothesis of a role for these repeat elements in the spreading of X inactivation. Interestingly, an asymmetric distribution of LINE elements on the two DNA strands was observed in the three species, not only within introns but also in intergenic regions. This feature is suggestive of important transcriptional activity within these intergenic regions. In silico prediction followed by experimental analysis has allowed four new genes, Cnbp2, Ftx, Jpx, and Ppnx, to be identified and novel, widespread, complex, and apparently noncoding transcriptional activity to be characterized in a region 5′ of Xist that was recently shown to attract histone modification early after the onset of X inactivation.
[The sequence data described in this paper have been submitted to the EMBL data library under accession nos. AJ421478, AJ421479, AJ421480, and AJ421481. Online supplemental data are available at http://pbil.univ-lyon1.fr/datasets/Xic2002/data.html and www.genome.org.]
In mammals, dosage compensation of X-linked genes is achieved by the transcriptional silencing of one of the two X chromosomes in the female cell during early development, a process known as X inactivation. Initiation of X inactivation involves recognition of the number of X chromosomes present in the cell, ensuring that the single X chromosome remains active in the diploid male cell and that only a single X chromosome is inactivated in the female diploid cell. This process, which is known as counting, is thought to involve an evaluation of Xic number against ploidy. Initiation also includes a recognition process linked to the choice of the X chromosome to be inactivated. Initiation of X inactivation and other early events are regulated by a master control region, the Xic (X-inactivation center). The XIC/Xic is a unique region of the X chromosome situated in Xq13 in man and in the syntenic mouse region that is necessary for the counting, the choice, and the subsequent nucleation of silent chromatin on the presumptive inactive X. Silencing spreads bidirectionally from the Xic into linked sequences, which need not be of X-chromosome origin (Lee and Jaenisch 1997). The study of chromosomal rearrangements in human culminated in the identification of a 680-kb to 1.2-Mb candidate region that shows full XIC function (Rastan 1983; Rastan and Brown 1990; Brown et al. 1991a). Subsequent efforts to delimit the XIC/Xic have concentrated on the use of transgenesis in the mouse to determine the candidate region that is sufficient for Xic function. Using the stringent criteria that single-copy transgenes must show full Xic function, these experiments have not as yet defined the minimum size of the Xic necessary for ectopic function. Although a single copy of a 35-kb cosmid transgene recapitulates some aspects of Xic function (Herzing et al. 1997), other studies of transgene copy-number dependence have suggested that even a 450-kb region may not contain all the elements necessary for autonomous, ectopic Xic activity (Heard et al. 1999).
As defined cytologically, Xic has been shown to contain at least five genes (Heard et al. 1997; Avner and Heard 2001). One of these, the Xist (X-inactive specific transcript) gene, which is the only gene known to be specifically transcribed from the inactive X chromosome in female somatic cells, codes for a 17-kb spliced, polyadenylated noncoding RNA (Borsani et al. 1991; Brockdorff et al. 1991; Brown et al. 1991b). Xist is necessary and sufficient for the initiation and spread of X inactivation but not for counting (Penny et al. 1996; Marahrens et al. 1997; Wutz and Jaenisch 2000).
At least some of the sequences required for counting must lie immediately 3′ to Xist as a 65-kb deletion of a region extending 3′ downstream of Xist exon 6 disrupts the counting process (Clerc and Avner 1998). This region contains a 17-mer minisatellite (Simmler et al. 1996), the Tsx gene (Cunningham et al. 1998), the DXPas34 locus, a CpG-rich minisatellite showing a highly characteristic pattern of hypermethylation on the active X chromosome (Courtier et al. 1995; Prissette et al 2001) and an associated CpG island that is the presumptive major initiation site for the Xist antisense transcript Tsix (Lee et al. 1999; Mise et al. 1999). Tsix may have a repressive role on Xist, with which it overlaps (Debrand et al. 1999; Lee and Lu 1999; Morey et al. 2001; Stavropoulos et al. 2001), and has also been implicated in imprinted X inactivation in murine extraembryonic tissues (Lee 2000; Sado et al. 2001). A spliced form of Tsix, initiating upstream of DXPas34, has been described (Sado et al. 2001) and it is likely that antisense transcriptional activity is widespread throughout the region (Debrand et al. 1999). The region between Xist and DXPas34, which includes the major Tsix initiation site, regulates Xist transcript accumulation and its retention at the site of transcription (Morey et al. 2001), probably through the activity of the Tsix antisense (Stavropoulos et al. 2001).
Another regulatory element within Xic is the X-controlling element, or Xce (Cattanach and Williams 1972). Xce, which lies 3′ to Xist and the counting region (Simmler et al. 1993; M. Prissette, unpubl. data), influences the choice of which X chromosome is to be inactivated (Heard et al. 1997). Xce is, however, only one element regulating choice, and other regions lying both within the Xist gene and 3′ to the Xist gene also influence chromosome choice (Avner and Heard 2001).
Several years ago, we obtained high-quality sequence of a 94-kb region encompassing the mouse Xist gene (Simmler et al. 1996) and the region lying 3′ to Xist, which was subsequently shown to be involved in the counting process and in choice (Clerc and Avner 1998; Morey et al. 2001). More recently, the Xist gene and a small region immediately up- and downstream have been sequenced in the vole (Nesterova et al. 2001) and compared with the homologous human and mouse sequences. The overall picture obtained from functional analysis of the mouse Xic is of a complex integrated locus in which functionally important elements may be located over several hundred kilobases both upstream and downstream of Xist itself. In this context, we decided to undertake a fully annotated comparative analysis of the human and mouse XIC/Xic based on the sequencing to high standards of a 714-kb region including the murine Xist gene, which was compared to the 2.3 Mb of available finished sequence for the orthologous region in human. Sequencing of a 233-kb core region of the bovine Xic around Xist to similarly high standards allowed a three-way species comparison of the core central region, including the Xist gene.
RESULTS
Global Description of the Region
We sequenced a region of 714 kb of the mouse X chromosome, centered on the Xist gene, and a region of 233 kb from the bovine Xist locus. The human sequence orthologous to the mouse locus was identified by a BLAST search and extracted from the Human Genome Project Working Draft Assembly (October 7, 2000, freeze; http://genome.ucsc.edu/; Lander et al. 2001). We searched for genes in the three species by using a combination of different approaches: ab initio exon prediction (GENSCAN, FGENESH), homology-based gene prediction (BLASTX, GENEWISE), ESTs, or cDNAs (BLASTN, SIM4; see Methods). GENSCAN or FGENESH exon predictions were considered as belonging to true genes only if they were confirmed by other evidence (cDNA sequencing or positive RT-PCR between adjacent exons or finding of ESTs covering adjacent exons). Mouse, human, and bovine sequences were compared with each other using SIM to identify conserved blocks corresponding to potential functional elements (coding or noncoding).
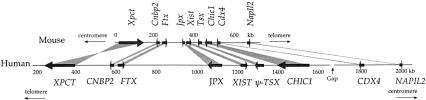
We identified 11 genes in the mouse Xic region. Seven of these genes were previously known: Xpct, Xist, Tsx, Tsix, Chic1 (formerly, Brx), Cdx4, and NapIl2 (formerly, Bpx). We characterized 4 new genes: Cnbp2, Ftx, Jpx, and Ppnx. Four of the 11 genes, Xist, Tsix, Ftx, and Jpx, are untranslated RNA genes (i.e., do not code for proteins). All the genes identified in mouse are conserved in human, except Ppnx and Tsix (see Fig. 1). In human, however, Tsx has become a pseudogene. The human XIC locus is considerably expanded compared to its mouse ortholog. It covers ∼2300 kb (the exact size is not known because there is a gap in the human assembly between the CHIC1 and CDX4 genes), that is, about three times larger than in the mouse (Figs. 1,2,3). Despite this major change in locus size, the order and orientation of genes is perfectly conserved in human and mouse, except for Xpct, which is at the same location but in the inverse orientation (Fig. 1).
Figure 1.
Comparative map of Xic region in mouse and human.
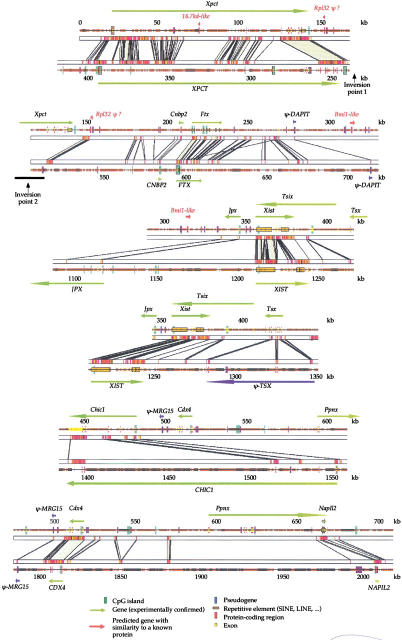
Figure 2.
Comparison of mouse and human genomic sequences in Xic region. Genomic sequences were first analyzed with RepeatMasker to identify and mask repeated elements and then aligned with SIM. Conserved blocks with a similarity score >30 are displayed (see Methods). An electronic version of this figure is available at http://pbil.univ-lyon1.fr/datasets/Xic2002/data.html. Because the alignment is very long and because of an inversion of the Xpct gene, it was not possible to display the whole human–mouse comparison in a single continuous line. The alignment, therefore, is displayed in six overlapping fragments. The overlaps are designed to allow the continuity of the fragments to be better appreciated.
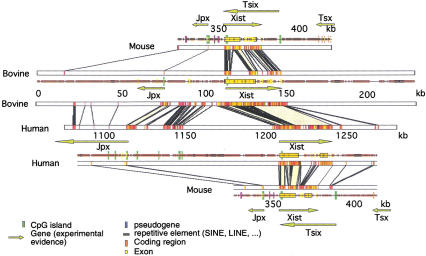
Figure 3.
Three-way comparison of mouse, human, and bovine genomic sequences around Xist. See Fig. 2 legend. An electronic version of this figure is available at http://pbil.univ-lyon1.fr/datasets/Xic2002/data.html.
The XIC/Xic region is relatively GC-poor in all three species (42% in mouse, 40% in human, and 39% in bovine) and repeat elements (essentially LINEs and SINEs) make up 38%, 57% and 58% of the mouse, human, and bovine loci, respectively (Table 1). In mouse, we identified 22 pseudogenes (i.e., sequences with similarity to known functional genes but which contain stop codons or frameshifts in the coding region). The density of pseudogenes in the mouse Xic region (31 pseudogenes/Mb) is relatively high compared to estimates from the human genome overall (6 per Mb; Goncalves et al. 2000). Protein-coding regions of confirmed genes, make up 0.84% of the Xic region in the mouse, 0.24% in the human XIC (compared with ∼1.8% for the whole human genome; Lander et al. 2001). Thus, globally, the region is relatively G+C-poor, repeat-rich, and poor in protein-coding genes.
Table 1.
Frequency of the major classes of interspersed repeated elements (in term of percent of the sequence length) in the autosomes, the X chromosome, in the Xic region taken as a whole and in introns of genes located in the Xic region
| Length (kb analyzed) | LINE (%) | SINE (%) | LTR (%) | DNA (%) | Total (%) | ||
|---|---|---|---|---|---|---|---|
| Mouse | Autosomesa | 223,057 | 10.5 | 9.4 | 6.9 | 0.5 | 27.4 |
| X chromosomea | 10,000 | 16.2 | 9.1 | 6.5 | 0.7 | 32.6 | |
| Xic region | 714 | 14.5 | 14.5 | 8.5 | 0.3 | 37.9 | |
| Xist introns | 9 | 0.0 | 10.2 | 0.0 | 0.0 | 10.2 | |
| Other intronsc | 190 | 17.2 | 13.2 | 3.1 | 0.4 | 33.9 | |
| Human | Autosomesb | 348,845 | 17.0 | 14.3 | 6.8 | 2.4 | 40.6 |
| X chromosomeb | 56,667 | 30.2 | 9.6 | 9.0 | 2.5 | 51.2 | |
| XIC region | 2300 | 38.9 | 8.4 | 8.6 | 1.3 | 57.2 | |
| XIST introns | 18 | 8.2 | 8.9 | 0.5 | 1.0 | 18.6 | |
| Other intronsc | 352 | 47.5 | 7.8 | 9.0 | 2.0 | 66.3 | |
| Bovine | XIC region | 233 | 46.5 | 9.3 | 1.3 | 0.8 | 57.9 |
| XIST introns | 17 | 17.8 | 5.8 | 0.6 | 0.0 | 24.2 |
Sequences extracted from GenBank release 124 (June 2001).
Data from Bailey et al. (2000).
Introns of protein-coding genes located in Xic region.
Gene Content
New Genes
Cnbp2: A Novel Gene Coding for a Zinc Finger Protein
This new gene, located ∼150 kb 5′ of the Xist gene, was predicted in the mouse by both GENSCAN and FGENESH and was confirmed by the identification in GenBank of a full-length cDNA (accession number AK015789). The predicted open reading frame (ORF) encodes a protein of 170 amino acids with strong similarity (75% identity) to cellular nucleic acid binding protein (CNBP). CNBP is a zinc-finger DNA-binding protein of unknown function, highly conserved in vertebrates, having two possible forms due to alternative splicing (De Dominicis et al. 2000). In human, CNBP maps to chromosome 3 (3q13.3-q24) and consists of five exons. The new predicted protein aligns perfectly with the shortest of the two forms of CNBP (170 amino acids long) and hence is likely to have similar biochemical activities. We therefore named our gene Cnbp2. The Cnbp2 gene contains a single exon and is conserved in human (Fig. 2), in which we found several matching ESTs. The fact that Cnbp2 is conserved and expressed both in human and mouse suggests that it is a real gene and not a pseudogene despite its intronless nature suggestive of a retroelement. This is also supported by the comparison of the human and mouse orthologs: The ratio of synonymous over nonsynonymous substitution rate of 3.6, (i.e., much greater than one, indicating that this gene is under selective pressure). We found no evidence of other closely related CNBP homologs in the human genome. An RT-PCR product was detected only in adult mouse testis RNA, suggesting that Cnbp2 is not widely transcribed.
Ftx: A Novel Conserved Noncoding Gene
We found in GenBank a mouse cDNA (AK020989) that spans five exons of a new gene, located ∼140 kb in 5′ of Xist and that we have named Ftx. We found several overlapping mouse ESTs (BB619145, BB628053, BB660326, BG920590, and BB305979) that identifed two additional exons and revealed at least four different transcripts resulting from alternative splicing. The seven exons identified all have perfect consensus splice sites. We identified a human cDNA (AK057701) spanning four exons in the corresponding region and sharing the same orientation. The 5′-part of the gene is well conserved and contains a CpG island in both species (Fig. 2). Both the mouse and the human cDNAs start in this conserved CpG island, which suggests that it might correspond to the promoter region. The intron–exon structure of the gene is poorly conserved: Only four of the seven mouse exons are located in regions that are conserved in human, and only two of them overlap with the exons identified in human. We found SINE elements both in human and mouse transcripts, and we detected no similarity with any known protein (after masking repeated elements). The longest mouse ORF contains only 87 codons and is not conserved in human, which suggests that Ftx encodes an untranslated RNA. RT-PCR studies, using primers designed from within exon 7, suggest that the Ftx gene is ubiquitously expressed (undifferentiated ES cells, liver, brain, kidney, and testis have been tested; data not shown).
Jpx: A Novel Conserved Noncoding Gene Subject to X Inactivation
A small region proximal to the Xist gene, which is well conserved between mouse, human, and bovine, was identified (Fig. 3). As this region also contains a conserved CpG island, the presence of a transcript was suspected. Several ESTs were found to match perfectly to this region in all three species, identifying a novel gene we have named Jpx. The murine Jpx gene, located in the mouse 10 kb upstream of Xist, is composed of three exons identified by three ESTs (accession numbers BE994494, AI391037, and BB564398), with alternative splice donor sites in the first exon generating 315-nucleotide- and 562-nucleotide-long transcripts (Fig. 4). A polyadenylation consensus motif (AATAAA) is present in the third exon, followed in several of the ESTs by a poly(A) tail 16 nt downstream, indicating that the third exon likely represents the true 3′ end of Jpx. The CpG island lies within the first exon, suggesting that exon 1 may represent the 5′ end of the gene. This first exon is highly conserved in all three species. In both human and bovine, transcription of this exon with correct splice signals is confirmed by several matching ESTs. Human and bovine ESTs (AV714079, AW484353, and BE485548) indicate the presence of three exons as observed in the mouse, sharing the same orientation. All putative exon–intron junctions have a consensus splice-site sequence. The second human exon is a homolog of mouse exon 1. However, the last two mouse exons are not conserved. Indeed, in the three species, the last two exons (except human exon 2) correspond to repeat elements: The mouse second and third exons show perfect homology with LTR/MaLR elements, whereas the bovine second and third exons correspond respectively to LINE and SINE repeats and the human third exon to a SINE repeat. It is highly unusual to find repeat sequences functioning as exons as in Jpx. In the mouse, neither of the two alternative transcripts contains an ORF large enough to encode a protein: the longest ORF is only 129 bp long (43 amino acids) and is not conserved in human or bovine. This suggests that Jpx, such as Ftx, Xist, and Tsix, encodes an untranslated RNA. The fact that this gene is expressed and clearly conserved in all three species suggests a functional role for Jpx.
Figure 4.
Cloning and characterization of Ppnx and Jpx genes. (A) Jpx gene. Schematic diagram showing the three exons (light grey boxes) of the murine Jpx gene, at positions 343745–343379 (exon 1), 340888–340796 (exon 2), 334465–334364 (exon 3), as defined by the three ESTs represented as dark grey boxes above the map. ESTs are identified by their GenBank accession number. The presence of an alternative splice at position 343626 nt in exon 1 generates either a 315- or 562-nt transcript. The presence of a poly(A) tail in EST BE994494 identifies the 3′ end of the gene. The first exon may correspond to the 5′ end of the gene, as a CpG island (diagonally hatched box) lies in the vicinity. The two last exons are LTR/MaLR-repeat elements (white boxes). (B) Northern analysis using a PCR probe corresponding to exon 1 of the Jpx gene after 5 d exposure. (C) Ppnx gene. Schematic diagram illustrating the six exons of the murine Ppnx gene. All exons and introns are represented to scale with the exception of introns 2 and 5 that are very large (21707 bp and >45389 bp, respectively). Each exon (grey) is identified by its coordinates: exon 1, 591535–591619; exon 2, 592146–592314; exon 3, 614021–614268; exon 4, 615349–615488; exon 5, 618116–>619619 (with three alternative splice donor sites at position 618262, 618719, and 618758), and exon 6, 665008–665306. The black boxes represent two pseudogenes predicted by GENSCAN; the small black arrow shows an antisense transcription detected in intron 2. The single-exon NapIl2 gene is shown diagonally hatched. Three independent clones A, B, and C obtained after screening a testis cDNA library are shown below the genomic structure. Clone A does not contain either a poly(A) consensus signal or a poly(A) track and has an in-frame stop codon present in its most 3′ part. The end of the 3′ UTR track is likely 300 bp downstream of the stop codon (unpubl.). Clones B and C possess a stop codon in exon 6 followed by a 3′ UTR of 177 bp, containing both a poly(A) signal and a poly(A) tail. Three of the RT-PCR products suggesting the existence of alternative splice donor sites in exon 5 are also represented (1, 2, and 3). (D) Northern analysis of Ppnx expression. A mouse multiple tissue Northern blot was hybridized with a probe corresponding to the third exon of Ppnx. The Ppnx hybridization signals were obtained after 8 d exposure.
RT-PCR studies suggest that the Jpx gene is ubiquitously expressed, with Northern analysis revealing several low-level transcripts of different size (Fig. 4). Although we have not definitively identified the site of initiation of these transcripts, the presence of a conserved CpG island lying within the first exon of Jpx and the lack of conservation of genetic elements lying proximal to the Jpx region among all three species, suggests that they originate in exon 1 rather than more proximally. We cannot, however, formally rule out that these Jpx transcripts are not the 3′ UTR sequences of an as-yet unidentified, ubiquitously expressed longer mRNA(s). It is unlikely however that these Jpx transcripts represent the 3′ end of a variant Tsix transcript because the expression profiles are clearly distinct.
The mouse Jpx gene was shown to be subject to X inactivation (see Methods) and transcribed only from the active X chromosome. These findings allow the 5′ end of the domain exclusively expressed from the inactive X, which includes Xist to be clearly delineated for the first time.
Ppnx: A Mouse-Specific Gene Expressed in Testis and ES Cells
Five of the six exons on the forward strand and lying in the vicinity of Nap1l2 that characterize the Ppnx gene were predicted by both GENSCAN and FGENESH. Each of these exons was shown by random RT-PCR to be strongly expressed in both undifferentiated ES cells and the adult testis. More detailed analysis revealed the presence of these transcripts in the testis from 14 days postcoitum onward, suggesting expression in germ cells having entered meiosis. Strand-specific RT-PCR showed that the exons belong to a single gene, with a centromere 5′–3′ telomere orientation. Subsequent Northern blot analysis of adult and embryonic tissues confirmed the presence of Ppnx expression in the adult testis (Fig. 4).
The Ppnx gene contains two particularly large introns (22 and >45 kb, respectively, for introns 1 and 5) and several alternative exons, giving rise to alternative splice forms (see Fig. 4). Two different ORFs of 1815 and 873 nt were identified from the three cDNA clones we obtained, depending on the start and stop codons and the poly(A) consensus signal or poly(A) track used (for more details, see Fig. 4). Virtual translation of these clones gave protein products of 604 and 290 amino acids, respectively. Both putative proteins contained a common secretion-motif associated with the first 20 amino acids. In silico searches for protein similarities using BLASTP showed that both proteins share 20%–30% identity with trypsin due to the presence of a serine protease-like domain.
Ppnx is not conserved between mouse and human, at least within the Xic/XIC regions (Fig. 1). The only human homolog of Ppnx that could be detected when we compared the different Ppnx splice forms against protein databases (using BLASTP) and EST or genomic databases (using TBLASTN) located to the human Y chromosome. This human-coding sequence is interrupted by two stop codons and does not match to any human EST, suggesting that it probably corresponds to a pseudogene.
Potential Genes
We identified three putative genes, both predicted by GENSCAN and FGENESH, that showed significant similarity to other known proteins: a BMI1-like gene, a 16.7 kd-like gene, and a RPL32-like gene (Fig. 2). None of these putative genes is conserved in human. BMI1-like and 16.7 kd-like match several ESTs with ∼90%–95% identity. However, the comparison of these ESTs with the assembly of the mouse genome (Ensembl database, v. 4.1.1; http://www.ensembl.org) revealed that they correspond to transcripts from closely related paralogous genes located in other regions of the X chromosome. In the absence of evidence of transcription activity, we are unable to conclude whether the BMI1-like and the 16.7 kd-like elements correspond to real genes. The RPL32-like element, on the other hand, is a retroelement that is 100% identical to the mouse ribosomal protein L32 (RPL32) mRNA but does not contain any introns and most likely corresponds to a recently inserted retropseudogene. This RPL32-like element is potentially functional, due to the fact that no frameshift or stop codon, which might stop it encoding a protein identical to RPL32, was noted.
Previously Known Genes
Xpct: Alternative Splicing in First Intron
The Xpct gene (X-linked PEST-containing transporter; Debrand et al. 1998) encodes a protein of 613 amino acids that belongs to a family of monocarboxylate transporters (Lafreniere et al. 1994; Price et al. 1998). The gene contains 6 exons that are perfectly conserved in both species. Xpct spans 117 kb in mouse and 147 kb in human. The first intron is very long (104 and 135 kb in mouse and human, respectively), and contains remarkably large conserved regions (Fig. 2). In mouse, we found one EST (accession number AW106665) that matches exons 2 and 3 and an alternative exon (with consensus splice signals) located in a conserved region of the first intron. This suggests that Xpct might undergo alternative splicing or alternative promoter use in both species.
Xist: Evolution of Its Gene Structure in Mammals
Although the Xist gene that is conserved in mouse, human, and bovine (Fig. 3) has been studied by many different groups, its precise exon–intron structure is still not fully established, notably at the 3′ end. We identified Xist exons in human and mouse by comparison with ESTs and with Xist cDNA sequences published in GenBank. Debate on the human XIST gene has concerned the seventh and eighth exons, these being found only in rare splice variants (Brown et al. 1992). Although Hong and colleagues (2000) failed to identify human exon 7, we found three ESTs from human hepatocellular carcinoma cells that clearly confirm the existence of XIST exons 7 and 8 (Fig. 5). In agreement with Hong et al. (1999), we found that the mouse cDNA published by Brockdorff et al. (1992) corresponds to seven exons rather than the six exons originally suggested. Based on sequence conservation and RT-PCR analysis, it has also been proposed that mouse Xist contained an additional 3′ exon, homologous to human exon 8 (Simmler et al. 1996; Sheardown et al. 1997). This eighth exon has been described in voles (rodent; Nesterova et al. 2001). We have confirmed the existence of mouse exon 8 by comparison with two ESTs from mammary gland (Fig. 5). As Hong and colleagues (1999, 2000) failed to identify this eighth exon, it may only be used, however, in rare splice variants. We conclude therefore on the presence of eight exons and of clear evidence of alternative splicing and poly(A) tail use at the 3′ end of the Xist gene in both species. Although in mouse and human the longest Xist mRNA forms span 17.9 and 19.3 kb, respectively, the splice variants of the alternative 3′-end exons (m8, h7, and h8; for nomenclature, see Fig. 5 legend) result in RNAs that are at least 5 kb shorter.
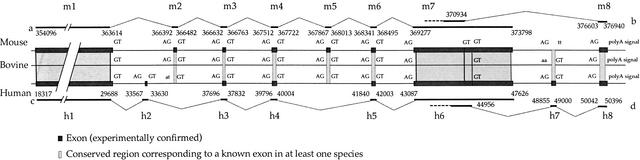
Figure 5.
Comparison of Xist gene intron–exon structure in mouse, bovine, and human. Introns and the first exon are not drawn to scale. Conserved regions corresponding to known exons (in at least one species) are indicated. Consensus splice signals that align with boundaries of known exons are in capitals. In mouse and human, 8 exons have been identified (Brown et al. 1992; Sheardown et al. 1997). Note, however, that some exons in mouse have no counterpart in the human and vice versa. To avoid ambiguities in exon numbering, we used the prefix “h” to identify human exons, and “m” to identify mouse exons. Mouse exons m1, m3, m4, m6, m7, and m8 are conserved and correspond to human exons h1, h3, h4, h5, h6, and h8, respectively. In both species, we identified an alternative donor splice site in exon m7–h6, although it is not located exactly at the same position in human and mouse. With the exception of this splice site, splice signals (donor and acceptor) of these six exons, as well as the polyadenylation signal in the last exon, are perfectly conserved in mouse, man, and bovine (which suggests that they also correspond to true exons in the latter species). Human exon h2 is located in a region that is not conserved in mouse or bovine. Mouse exon m2 is conserved in the three species, with correct splice signals, except in human where the splice acceptor is mutated (AT instead of AG), which suggests that this exon is no longer functional in human. Mouse exon m5 is conserved in the three species with correct splice signals and might correspond to an unidentified alternative exon in humans. Human exon h7 is located in a region that is conserved in the three species, but consensus splice signals are not found in mouse and bovine. a, mouse Xist cDNA (GenBank accession number L04961; Brockdorff et al. 1992); b, mouse ESTs (BE626785, BE632200, and R74734); c, mouse exon m7a reported by Hong et al. (1999); d, human XIST cDNA (M97168; Brown et al. 1992); e, human exon h6a reported by Hong et al. (2000); f, human ESTs (AV699347, AV700119, and AV700677). Exon positions in mouse (bp): m1, 354096…363614; m2, 366392…366482; m3, 366632…366763; m4, 367512…367722; m5, 367867…368013; m6, 368341…368495; m7a, 369277…376940; m7b, 369277…371600; and m8, 376603…376940. In human, exon positions are indicated relative to the XIST genomic sequence (U80460): h1, 18317…29688; h2, 33567…33630; h3, 37696…37832; h4, 39796…40004; h5, 41840…42003; h6a, 43087…50396; h6b, 43087…44956; h7, 48855…49000; and h8, 50042…50396.
We aligned the XIST/Xist genes in mouse, human, and bovine and compared the location of exons identified in mouse and human (Fig. 5). Seven out of the eight mouse exons are perfectly conserved in the three species. Six of these seven have been confirmed experimentally in human; one (exon m5) may correspond to an unidentified splice variant. The intron–exon structure, however, is not totally conserved. The human XIST gene includes two exons (h2 and h7) that are not present in mouse and bovine, and the exon m2 found in the mouse and bovine genes is probably not functional in human.
At the sequence level, Xist exons show on average 66% and 62% identity, respectively, between mouse and human, and mouse and bovine. This figure is close to the average conservation in 5′- and 3′-untranslated regions of human and mouse orthologous protein-coding genes (Makalowski and Boguski 1998). With the exception of several conserved blocks, Xist introns are weakly conserved and cannot be aligned between the mouse and the two other mammals. By comparison with the rest of the Xic locus, the Xist gene is relatively devoid of LINE elements, not only in exons (LINE elements make up 0%, 0.4%, and 1.9% of Xist exons in mouse, human, and bovine, respectivley), but also in introns (see Table 1). Note that, unlike Xist, introns of protein-coding genes are generally rich in LINE repeats (Table 1). These observations suggest that the insertion of LINE elements within the Xist gene has been strongly counterselected (see below).
Tsix: No Evidence of Conservation in Other Mammals
The Tsix gene, identified in mouse, encodes an untranslated RNA antisense to Xist. Initially, Tsix was described as a 40-kb RNA encoded by a single exon, initiating close to a CpG island located 15 kb 3′ of the previously described end of Xist (i.e., 12 kb 3′ of the Xist m8 exon; Fig. 3; Lee et al. 1999). However, Sado and colleagues (2001) found that Tsix is, at least in part, subject to processing. They identified four exons, along with a major promoter upstream of Tsix exon 2 and a minor initiation site located upstream of Tsix exon 1 (i.e., ∼28 kb 3′ of the Xist m8 exon). Lee and colleagues (1999) have identified three conserved blocks between human and mouse, two of which are located close to the start of the Tsix transcription unit; this led them to suggest that the Tsix gene is conserved in human. The first conserved block corresponds to Xist exons m8 and h7 and h8. The two other blocks are located within Tsix intron 3, downstream of the major promoter. It is noteworthy that neither of these two latter blocks are conserved in bovine (Fig. 3). Tsix exon 4 overlaps with the 5′ end of Xist exon 1. Although the 5′ part of Tsix exon 4 that is common to Xist exon 1 is conserved in the three species, the 3′ part is not conserved. Therefore, in contrast to Xist, which is well conserved in the three species, neither Tsix exons nor promoter regions are conserved in mammals. Thus, if Tsix does exist in human and bovine, it is clear that its primary sequence is not subject to a strong selective pressure.
Tsx: A Human Pseudogene
Tsx is a testis-specific gene of unknown function (Simmler et al. 1996). The mouse Tsx gene, which spans 10 kb, contains 7 exons and encodes a protein of 144 amino acids without homology to any known gene. The orthologous region in human spans 44 kb. Conserved blocks that correspond to exons 1, 3, 4, 5, and 6 are clearly identified (Fig. 2). Exon 2 is not conserved (although we found conserved blocks within introns 1 and 2) close to the exon-2 boundaries. Human exons 4 and 6 contain stop codons, exons 1 and 3 contain frameshifts, and intron 5 contains mutated splice-donor signals, all of which indicates that Tsx has become a pseudogene in human. We found no ESTs matching these conserved blocks in human, and no other regions with significant similarity to Tsx in the complete human genome sequence, which suggests that the inactivation of Tsx gene in the primate lineage has not been compensated for by another homologous gene located elsewhere in the genome. Note that the comparison of mouse and rat Tsx cDNAs has shown that this gene has evolved very fast in rodents, which suggests that it is under weak selective constraints (Simmler 1996).
Unexpectedly, a region with similarity to mouse Tsx exon 7 was found in human, translocated on to the opposite DNA strand. Interestingly, the region corresponds to XIST exon 8. The similarity is weak but spans the acceptor splice signal, the polyadenylation site, and a block within the last intron. This observation suggests a possible evolutionary relationship between the genes.
Chic1: A Revised Structure
Chic1 which belongs to the family of CHIC genes (cysteine-rich hydrophobic proteins) is specifically expressed in the brain (Simmler et al. 1997). The 653 nt originally identified Chic1 coding sequence was suspected to be incomplete as no in-frame stop codon was detected upstream of the first ATG of the cDNA and because an 8-kb transcript using a specific Chic1 probe was detected by Northern blot. However, a comparison of the mouse and human genomic sequences 5′ of this ATG failed to reveal conserved sequence with significant coding potential. Moreover, a comparison of Chic1 with others members of the family, all of which code for proteins of approximately the same size and which start with the same conserved motif, suggests that the 653-nt sequence almost certainly represents the complete coding region of Chic1. The 6-kb region distal to exon 6 of the Chic1 gene is highly conserved between human and mouse (Fig. 2) and sequence from this region match several ESTs, suggesting that the region is transcribed. However, the absence of an extended ORF suggests that the region is not translated. To test the hypothesis that this 6-kb conserved region corresponds to the 3′ UTR of the Chic1 gene, strand-specific RT-PCR was carried out on female brain RNA, using primers positioned at several points within the 6-kb region. Our results show the presence of a continuous transcription unit with the same orientation as Chic1, extending from the last exon of Chic1 defined by Simmler and colleagues (data not shown), compatible with it being a 3′ UTR. The complete Chic1 gene, including the six exons and the very long 3′ UTR are perfectly conserved in human and mouse, even though the gene is highly expanded in human compared to mouse. The CHIC1 gene spans 40 kb in mouse and 163 kb in human (Fig. 2).
Cdx4
Cdx4 encodes a protein of 282 amino acids and belongs to the caudal family of homeobox genes (Gamer and Wright 1993). The gene spans ∼8 kb in mouse and human and contains three exons that are perfectly conserved (Fig. 2).
Nap1l2: An Unusual Location in the Intron of a Gene Sharing the Same Orientation
Nap1l2 (nucleosome assembly protein 1–like 2) is a single-exon gene, encoding a protein of 460 amino acids conserved in human and mouse (Fig. 2), which is specifically expressed in neurons (Rougeulle and Avner 1996; Rogner et al. 2000). Surprisingly, we found the Nap1l2 gene to be entirely included in the last intron of Ppnx, a newly identified mouse gene (see above). Several other examples of genes located within introns have already been reported, but, to our knowledge, they are all oppositely orientated on the antisense strand, whereas Nap1l2 and Ppnx share the same orientation. Ppnx and Nap1l2, however, have different expression patterns. Although Ppnx is found only in the adult testis, Nap1l2 is exclusively expressed in nervous tissue (Rougeulle and Avner 1996). We suppose that the absence of transcription interferences between these genes is due to their different expression pattern and the fact that Nap1l2 is composed of only one exon and thus contains no splicing site that could interfere with the splicing of Ppnx.
Patterns of Conservation
Conserved Genes
Overall, conserved blocks between the mouse and human Xic/XIC represent 76 kb (10.6% of the mouse locus; 3.3% of the human locus). Of these 76 kb, 62 kb are located within known genes (introns and exons of Xpct, Cnbp2, Ftx, Jpx, Xist, Tsx, Chic1, Cdx4, and Nap1l2). Thus, 35 out of the 48 identified exons in the Xic mouse sequence are located within conserved blocks. The 13 nonconserved exons correspond to the 6 exons of the Ppnx gene (absent in human), 2 of the 7 Tsx exons (a pseudogene in human), 3 out of the 7 Ftx exons, and 2 out of the 3 Jpx exons. Conserved regions span 28% of the total gene length within the mouse Xic, consistent with previous observations (Jareborg et al. 1999), whereas only 3.4% of intergenic regions are conserved. This is considerably lower than a recently published estimate (15.8%), which was based on the analysis of intergenic regions of 100 pairs of human and mouse orthologs (Shabalina et al. 2001). This discrepancy is probably due to the fact that the data set analysed by Shabalina and colleagues (2001) was biased toward gene-rich regions. In gene-rich regions, many of the conserved blocks in intergenic regions correspond to regulatory elements, often located in close vicinity to genes. Given the low density of conserved blocks within intergenic regions of the XIC/Xic locus, we believe it is unlikely that many other conserved genes have escaped detection.
Conserved Retropseudogenes
We identified in human and mouse a conserved pseudogene related to the MRG15 gene (morf-related gene 15 protein; Fig. 2; Bertram et al. 1999). In both species, this pseudogene (called MRG15-psi) contains stop codons and frameshifts and lacks introns (unlike its functional counterparts), which indicate that it corresponds to a retropseudogene. MRG15-psi is located at the same position (downstream of Cdx4) and has the same orientation in both human and mouse. In both species, the retropseudogene corresponds to the 3′ end of the MRG15 mRNA (i.e., it is truncated in the 5′ part of the protein-coding region, probably as a consequence of incomplete retrotranscription). It is highly unlikely that two MRG15 retroelements have inserted independently at the same location during the evolution of the primate and rodent lineages as the MRG15-psi gene is not a member of a very large family of retropseudogenes. In all we identified only nine other MRG15-related pseudogenes in the complete human genome. This suggests that MRG15-psi has been a pseudogene since its insertion at this locus prior to the divergence of primates and rodents.
We identified another retroelement conserved in mouse and human, located at the same position and in the same orientation in both species, between the Xpct- and the Bmi1-like genes (Fig. 2), which is related to the DAPIT gene (accession number AJ271158; H.H. Paivarinne, unpubl.). In both species, this element is intronless, which suggests that it has been derived from the intron-containing DAPIT gene by retrotranscription. Again, this element does not belong to a large family. We found only three other DAPIT-related sequences in the complete human genome. Thus, this DAPIT-related retroelement was most probably inserted in this locus before the divergence between rodents and primates. The mouse retroelement contains two stop codons, which indicates that it has become a pseudogene. The human retroelement contains a complete ORF. However, we found no matching ESTs and our phylogenetic analysis suggests a total absence of selective pressure on the coding sequence: The ratio nonsynonymous over synonymous substitution rates in the primate DAPIT-related retroelement is 1.5, close to the ratio of 1 expected for a pseudogene.
LINE Elements
Nonrandom Distribution of LINE Elements on the Two DNA Strands in Introns and Intergenic Regions
Smit (1999) noticed that L1 elements inserted within human introns were twice as frequent on the antisense strand compared to the sense strand (relative to the orientation of the gene). He proposed that this bias was due to the presence of a transcriptional termination site within L1 elements that interferes with transcription when inserted into an intron on the sense strand. Insertion of L1 elements is counterselected because it induces premature termination of transcription. Consistent with this model, SINEs and DNA transposons that have no or only weak transcriptional termination sites do not show strand bias (Smit 1999). The analysis of intron-containing genes from the XIC/Xic region confirms this trend not only in human, but also in mouse and bovine (Table 2). Overall, in human (for which we identified seven intron-containing genes), we observed a 2.1-fold excess of LINEs on the antisense strand of introns (197 vs. 93 L1 copies), whereas in mouse (9 genes) the excess is 5.3-fold (117 vs. 22) and in bovine (2 genes) 7.8-fold (31 vs. 4). A prediction of the model proposed by Smit (1999) is that in a locus that is transcribed from both strands, all LINE elements insertions should be counterselected. The very low density of LINE elements in the 3′ part of the mouse Tsix gene that overlaps with Xist is in agreement with this model (see above and Tables 1, 2). Note that both in human and in bovine, the density of LINE elements is also relatively low in this region (Table 2).
Table 2.
Assymetric distribution of LINEs on the two DNA strands
| Region | Species | Length (kb) | Gene orientation | LINE+ | LINE− | χ2 | P |
|---|---|---|---|---|---|---|---|
| Xpct gene | human | 147 | − | 47 | 20 | 10.88 | <1% |
| mouse | 117 | + | 11 | 51 | 25.81 | <1% | |
| intergene | human | 186 | intergene | 31 | 63 | 10.89 | <1% |
| Xpct-Cnbp2 | mouse | 69 | 6 | 11 | 1.47 | ns | |
| intergene | human | 26 | intergene | 1 | 4 | 1.80 | ns |
| Cnbp2-Ftx | mouse | 7 | 2 | 0 | 2.00 | ns | |
| Ftx gene | human | 14 | + | 2 | 6 | 2.00 | ns |
| mouse | 16 | + | 1 | 3 | 1.00 | ns | |
| intergene | bovine | 62 | intergene | 63 | 20 | 22.28 | <1% |
| Ftx-Jpx | human | 425 | 70 | 192 | 56.81 | <1% | |
| mouse | 114 | 9 | 13 | 0.73 | ns | ||
| Jpx gene | bovine | 16 | − | 22 | 4 | 12.46 | <1% |
| human | 68 | − | 49 | 30 | 4.57 | <5% | |
| mouse | 9 | − | 2 | 0 | 2.00 | ns | |
| intergene | bovine | 38 | intergene | 5 | 13 | 3.56 | ns |
| Jpx-Xist | human | 95 | 32 | 58 | 7.51 | <1% | |
| mouse | 10 | 0 | 2 | 2.00 | ns | ||
| Xist gene | bovine | 35 | + | 0 | 7 | 7.00 | <1% |
| human | 32 | + | 0 | 8 | 8.00 | <1% | |
| mouse | 23 | + | 0 | 0 | nd | ||
| intergene | bovine | 82 | intergene | 49 | 31 | 4.05 | <5% |
| Xist-Tsx | human | 38 | 7 | 3 | 1.60 | ns | |
| (Tsix gene) | mouse | 35 | − | 14 | 3 | 7.12 | <1% |
| Tsx gene | human | 44 | − (pseudo) | 26 | 4 | 16.13 | <1% |
| mouse | 10 | − | 2 | 1 | 0.33 | ns | |
| intergene | human | 70 | intergene | 31 | 7 | 15.16 | <1% |
| Tsx-Chicl | mouse | 25 | 1 | 1 | 0.00 | ns | |
| Chicl gene | human | 157 | − | 61 | 36 | 6.44 | <5% |
| mouse | 32 | − | 8 | 5 | 0.69 | ns | |
| intergene | human | gap | intergene | nd | nd | nd | |
| Chicl-Cdx4 | mouse | 27 | 1 | 18 | 15.21 | <1% | |
| Cdx4-gene | human | 7 | − | 0 | 1 | 1.00 | ns |
| mouse | 8 | − | 1 | 0 | 1.00 | ns | |
| intergene Cdx4-NapIl2 | human | 193 | intergene | 47 | 84 | 10.45 | <1% |
| intergene | mouse | 77 | intergene | 10 | 42 | 19.69 | <1% |
| Cdx4-Ppnx | |||||||
| Ppnx gene | mouse | 74 | + | 1 | 36 | 33.11 | <1% |
LINE+, number of LINEs on the direct strand; LINE−, number of LINEs on the complementary strand. P; probability, ns; not significant, nd; not done.
Interestingly, all the intergenic regions, including the Xist–Tsx intergenic region show an excess of L1 elements on one strand compared to the other. The orientation of the bias is conserved in human and mouse, although it is not always statistically significant in both species (Table 2). If the model proposed by Smit is correct, this would suggest that most of these intergenic regions are transcribed and with the same orientation of transcription in both species. Because the pattern of conservation between the human and mouse intergenic regions does not suggest the presence of many additional unidentified genes, these intergenic regions may contain nonconventional genes like Tsix, which are not well conserved in sequence and do not code for a protein, or other transcribed sequences that may play a regulatory role through their transcriptional activity.
It is also possible that the nonrandom distribution of LINE elements on the two DNA strands in intergenic regions is linked to factors other than transcription (e.g., replication). We are not, however, aware of any mechanism that could bias the orientation of LINEs during their integration, although this orientation could reflect some constraint on chromatin structure.
LINE Elements Distribution in the Mouse: No Support for Lyon's Model?
Lyon (1998, 2000) has proposed that LINE elements may be responsible for spreading the inactivation signal along the X chromosome. Consistent with such a model, it has been observed in human that the density of L1 elements on the X chromosome is about twice as high as that of the autosomes (26.5% vs. 13.4%) (Smit 1999; Bailey et al. 2000). It is known that overall LINE density is negatively correlated with the G+C content of isochores in which they are located (Duret et al. 1995) and that the G+C content of the human X chromosome is lower than the average G+C content of autosomes. However, the excess of L1 elements on the X is not due to its low G+C content: The X chromosome is 1.5- to 2-fold enriched for L1 elements over autosomal regions even when isochores of comparable G+C content are compared (Smit 1999). Furthermore, Bailey and colleagues (2000) have shown that the chromosomal bands encompassing the human XIC locus (Xq13, Xq21) are even more enriched for L1 elements (45% and 39%), essentially due to an excess of young L1 elements. Our analysis of the sequence of the human XIC region has confirmed the high density of LINE elements (39%; Table 1). The bovine Xic region is also particularly rich in LINE elements (46%). However, this feature is not observed in mouse. The L1 density of the mouse Xic region (14.5%) is about three times lower than in human or bovine and is close to the overall density measured for the mouse genome (Table 1). Moreover, the L1 density of the Xic region is relatively low compared to the average L1 density of mouse genomic sequences of comparable G+C content (25% of L1 for sequences of 40%–42% G+C content; Smit 1999). Thus, in contrast with human or bovine, there is no excess of LINE elements associated with the mouse Xic region.
We have also analyzed the mouse genomic sequences available in GenBank to compare the density of transposable elements throughout the X chromosome with that of the autosomes. Although LINE elements are slightly more frequent on the X chromosome than on autosomes, the excess is limited compared to that observed in human (Table 1). Indeed, none of the major classes of interspersed repeats show a strong difference in density on the mouse X chromosome (and notably the Xic region) compared to the autosomes (Table 1). Our analysis therefore provides no support for Lyon's recent model of inactivation spreading.
DISCUSSION
Our sequencing of 714 kb of the mouse and 233 kb of the bovine Xic centered on the Xist gene has provided the basis for a fully annotated comparative analysis of the complete mouse Xic with the 2.3-Mb orthologous region in human and has allowed a three-way species comparison of the core central region, including the Xist gene. Comparative sequence analysis is an efficient way to identify functional elements within genomic sequences as selectively constrained regions generally evolve slower and hence are more evolutionary conserved than functionless sequences that rapidly diverge because of genetic drift. However, this approach requires that the compared sequences are sufficiently distantly related to allow constrained from nonconstrained sequences to be distinguished. Rodents and primates diverged ∼100 million years ago (Myrs) and many studies have shown the efficiency of human–mouse comparison to identify functional elements (Ansari-Lari et al. 1998; Jang et al. 1999; Jareborg et al. 1999; Mallon et al. 2000; Pennacchio and Rubin 2001). Notably, it has been established that 95% of protein-coding exons are conserved between human and mouse (Batzoglou et al. 2000). It is important to emphasize that the comparative approach we have used is also efficient in identifying untranslated genes such as Xist, Ftx, or Jpx, which are not identified by programs designed to allow ab initio identification of protein-coding gene. The power of such comparative approaches for similarly identifying functionally conserved noncoding regions, whether they be small (<10 nt), such as the binding sites for protein factors, or larger regions such as the locus control region, has been clearly established by analysis of the mammalian β-globin clusters, as well as other regions (Duret et al. 1993; Gumucio et al. 1996; Duret and Bucher 1997; Kondrashov 1999; Bouck et al. 2000; Mohrs et al. 2001). The assertion that all conserved regions are functional (Shabalina et al. 2001), however, is likely to be exaggerated. Our finding of conserved pseudogenes in the XIC/Xic locus in human and mouse indeed suggests that functionless sequences may have retained significant similarities since the divergence of primates and rodents, although it is very difficult to exclude that such pseudogenes are not acting in one way or another as controlling elements for neighboring genes (see below). In this context, functional analysis of the MRG15-psi and DAPIT-related retroelements is of obvious interest. The use of a three-way species comparison such as we have used for the core Xic region, involving species separated by ∼240–300 Myrs (the evolutionary distance between rodents and artiodactyls is as great as that between rodents and primates), increases the analytical power of such a comparative approach and contributes to avoiding the identification of similarities occurring purely by chance. In this context, it is important to note that mutation rates vary along the genome (Matassi et al. 1999; Perry and Ashworth 1999), and hence some regions may show greater conservation simply because they are less subject to mutations. We chose bovine as the third species because both rodents and primates are relatively distantly related to artiodactyls (∼80–100 Myrs), because the role of Xist in X inactivation in cows has been studied (De La Fuente et al. 1999), and because of the ready availability of bovine BAC genomic libraries for sequencing.
To further increase the specificity of such comparative approaches, it is necessary to consider even more distantly related species, such as birds or fish. We were constrained, however, by the functional specificity of our system since the XIC/Xic has no functional homolog in nonmammalian vertebrates. Indeed, the very mechanism of dosage compensation by Xist-induced X inactivation is specific to mammals, even if other species have alternative systems of dosage compensation which may present similarities at certain levels (Avner and Heard 2001; Park and Kuroda 2001)
The usefulness of adding a third species to the comparative sequence analysis is well illustrated by the short region conserved between human and mouse corresponding to the first exon of Jpx (lying 5′ of Xist). In the context of the repeat elements composing the second and third exons, Jpx would probably have escaped observation if we had not also detected conservation in the bovine (Fig. 3). The conservation of exon 1 taken together with the expression of the Jpx gene strongly suggests a functional role for Jpx. Taken together with the nonconservation of the repeats themselves between species, this may suggest that evolutionary constraints are acting on the promoter region itself rather than on the whole gene. However, the close-on 4% frequency of transposable element (TE) integration within human genes (Nekrutenko and Li 2001) makes it unlikely that the conserved involvement of repeats, albeit of differing classes, as in exons 2 and 3 of this gene, is totally fortuitous. Indeed, it could suggest a major role for the repeat elements in Jpx function. Such hypotheses are reinforced both by the key role of repeats in some other forms of genes silencing and the intriguing position of Jpx immediately upstream of Xist itself.
A second example of the utility of adding in a third species comes from the analysis of the conservation pattern of Tsix. Lee and colleagues (1999) identified two conserved blocks between human and mouse, close to the promoter region of Tsix, which led them to conclude that Tsix was conserved in human. However, these blocks are short and are conserved neither between mouse and bovine, nor between human and bovine. Thus, despite the mutational evidence from the mouse clearly establishing a role for Tsix antisense transcript and the associated DXPas34 locus in Xist regulation (Maxfield Boumil and Lee 2001) and in imprinted X-inactivation and -chromosome choice, it appears that the underlying sequence is not selectively constrained and is conserved in human and mouse simply by chance. Surprisingly, it turns out that the blocks of sequence conserved between human and mouse do not correspond to Tsix exons (Sado et al. 2001). Although the recent publication of Migeon and colleagues (2001) is suggestive of transcriptional activity antisense to XIST within the human XIC this activity presents few of the characteristics originally assigned to the murine Tsix antisense transcript. Our results raise specific questions as to the role of Tsix in species other than mouse and more generally as to the degree to which specific aspects and parameters of X inactivation may vary between mammalian species, even when the overall function such as that of the Xic or X inactivation is globally conserved (Looijenga et al. 1999). Differences between rodent and human X inactivation on one hand and marsupial X inactivation on the other include both the preferential inactivation of the paternal X chromosome and the tissue and species-specific nature of inactivation. Indeed, Xist has yet to be identified in marsupials. Differences between primates are likely less radical but several publications attest to differences in X inactivation occurring in human and murine extra-embryonic tissues and to potential differences in the regulation of the human and mouse XIST/Xist genes. It is interesting in this context to note that differences in the expression status of imprinted genes in mouse and human have been identified, and in the case of IGF2R/Igf2r this may be related to the nonexpression of the AIR antisense transcript in human (Oudejans et al. 2001).
Our work, although clearly showing the value of adding a third mammalian species (distantly related to the first two) in increasing the specificity of the comparative approach, does not clarify whether the choice of the species itself was optimal. Indeed the choice of bovine as a third species led to considerable difficulties in both the selection of bacterial artificial chromosomes (BACs) and sequencing due to the extremely high density of repetitive elements in the bovine Xic region. It would be useful to determine whether the genome of other mammalian model organisms, such as rabbit or dog, are less rich in repetitive elements, and thus better candidates for such sequencing.
Conserved elements identified include CpG islands and, more surprisingly, conserved pseudogenes. We identified 19 CpG islands (CGIs) in the mouse sequence, and 40 in the human one, that is, 2.7 and 1.7 CGIs per 100 kb, respectively, which is close to the density observed for the whole human genome (2.2 CGIs/100 kb; data from Lander et al. 2001). Five of these CGIs correspond to promoter regions of known gene (Xpct, Xist, Chic1, Cdx4, and Nap1l2), and three other to the putative promoter regions of Jpx and Ftx (NB: in mouse Ftx 5′ region there are two CpG islands very close to each other, that correspond to a single large CpG island in human). Thus, respectively, 42% (8/19) and 18% (7/40) of mouse and human CGIs correspond to known or putative promoter regions. This latter figure is consistent with a recent statistical analysis showing that 20% (373/1846) of human CGIs overlap with transcription start sites (Ponger et al. 2001). There are only nine CGIs that are conserved between human and mouse (Fig. 2). Interestingly, the eight CGIs mentioned previously are all conserved. Thus 89% (8/9) of conserved CGIs correspond to known promoter regions. The last conserved CGI, located downstream of Xpct (close to its 3′ end), might well correspond, moreover, to an unidentified promoter region. Although our gene sample is too small to be statistically reliable, these observations suggest that conserved CGIs frequently correspond to promoter regions. Comparative analysis of human and mouse CGIs, therefore, seems to be an efficient approach for identifying promoter regions.
The observation of conserved pseudogene orthologs raises the question of how such sequences can remain conserved since the time of divergence of rodents and primates and indeed what this might imply for their eventual functionality. Generally, retrotranscribed mRNAs are inactive from the moment of their insertion because they lack the promoter elements necessary for their expression. However, it can happen that a retroelement is expressed by chance after insertion downstream of an active promoter. This is unlikely to be the case for either the MRG15-psi or DAPIT-related retroelements we have studied in mouse or human as we found no matching EST. However, we obviously cannot exclude that they are expressed at a low level or in limited specific tissues or developmental stages that are not represented in the EST datasets. Under this scenario one might imagine that the DAPIT-related retroelement remained functional for a long period during primate and rodent evolution (which would explain their conservation) and only recently became a pseudogene in the rodent (and maybe also in primates, as suggested by the high ratio of nonsynonymous over synonymous substitution rates). However, in the case of MRG15-psi, it is clear that the coding region must have been truncated at the moment of its insertion. Thus a putative function (if any) cannot be related to protein-coding capacity. An RNA function or indeed a function in epigenetic control as, for example, a center of methylation, would not, however, necessarily require either conservation of protein-coding capacity nor indeed conservation of the entire genetic element. Functional tests will be necessary to further elucidate this.
Repeat elements and LINE (L1) elements, in particular, have had a major place in models seeking to explain various facets of the X-inactivation process and more recently imprinting (Greally 2002). Our computational analysis of the distribution of repeat elements in general and of LINE repeats, in particular, within the mouse Xic region, when compared to the rest of the genome did not however give support to the hypothesis of a role of these repeat elements in the spreading of X inactivation (Lyon 1998, 2000). Interestingly, though, our studies on L1 element distribution revealed that most intergenic regions within the mouse Xic, including the Xist–Tsx intergenic region, show an excess of L1 element on one strand compared to the other. If the model proposed by Smit (1999) is correct, this would suggest that most of these intergenic regions are transcribed. The orientation of strand bias is moreover conserved in human and mouse, reinforcing the notion of some underlying fundamental biological constraint. These intergenic regions could contain nonconventional genes like Tsix, Ftx, or Jpx, which are not well conserved in sequence and do not code for a protein but may play a regulatory role through their transcriptional activity. The intergenic region between Cnbp2 and Jpx contain many ESTs that might correspond to alternative transcripts of the Ftx and Jpx noncoding genes. ESTs corresponding to the other intergenic regions have yet to be found. The absence of ESTs, however, does not allow conclusions to be drawn systematically as to the absence of transcriptional activity associated with a genomic sequence, as ESTs are almost always derived from poly(A)+ cDNA libraries and generally correspond to the 3′ end of transcripts. The proven transcription activity at the Tsix locus does not, for instance, match that of any known EST.
Whether or not the increasing number of noncoding RNAs encoded within the Xic, other than Xist, play regulatory roles remains to be established. Such transcripts may turn out, even in mammals, to be part of a more widespread and widely used mechanism of epigenetic control. A region lying 50 kb upstream of the 5′ end of the murine Xist gene is one intergenic region where atypical transcriptional activity has recently attracted our attention during studies originally undertaken to validate GENSCAN predictions unsubstantiated by FGENESH. The region turned out to be characterized by multiple (as judged by their distinct expression profiles) apparently noncoding transcripts originating alternatively from both DNA strands. Strand-specific RT-PCR has indicated that these transcripts, unlike the Tsix and Xist transcripts, are not encoded over many kilobases of contiguous genomic DNA. Intriguingly this region was recently shown to correspond to part of a constitutive hot spot of Lys 9 methyl histone H3 activity that is associated with the onset of the X-inactivation process (Heard et al. 2001).
It is interesting to note that 3 of the 11 genes characterized in the murine Xic, including the newly characterized Ppnx and Cnbp2 genes, are exclusively or almost exclusively expressed in the testis. Although we have yet to confirm expression of the latter in male germ cells, the high proportion of Xic genes expressed in the testis is compatible with the X chromosome having a prominent role in male germ-cell development (Wang et al. 2001).
Our annotated sequence provides an excellent informative base for further genetic and molecular probing of the Xic. Comparison of our results for the Xic with chromosomal regions involved in other epigenetic phenomena, such as imprinting (Paulsen et al. 2001), may in turn allow other structural features to be identified and be subjected to functional analysis. Conversely, functional analysis of the Xic and further experimental validation of some of our computational observations is likely to improve both the understanding of the importance and role of some of the currently more poorly understood structural features we have identified and in the long term aid in identifying other regulatory motifs.
METHODS
Contig Construction and Sequencing
To sequence the murine Xic region, we established a contig of BAC clones from the 129/Sv mouse strain BAC library (Research Genetics, Inc.) by PCR screening using known primers. Overlapping BAC-end sequences were identified and the order verified by PCR. The entire mouse contig extending through from BAC 474E4 to 211B4 covers ∼1 Mb. A minimal tiling path set of five clones collectively spanning the region were selected for genomic sequencing. Although the telomeric and centromeric ends of clones 399K20 and 334L11 do not overlap, their ends are only 1178 pb apart and lie within the previously sequenced 94-kb Xist region.
Bovine BACs were identified by PCR screening of a bovine library maintained at the UR339, Unité de Génétique Biochimique et Cytogénétique, Institut National de la Recherche Agronomique. Primers corresponding to the 5′ region of the bovine Xist gene were designed to amplify a 650-bp product. End sequences of positive clones were used to rescreen the library, and three clones overlapping the 5′ end of the initial clone identified. Two BACs, covering a 233-kb interval from the bovine Xist region were retained for sequencing. The primers used are shown in supplemental data available at www.genome.org.
Sequencing of the clones was performed by ligating mechanically sheared 3-kb fragments of the BAC DNA into the pCDNA vector (Invitrogen), followed by random shotgun sequencing to 10-fold coverage. To increase sequence contiguity and facilitate assembly additional 10-kb fragment-size subclone libraries were prepared for mouse BACs 155J2 and bovine BAC 834C6. All plasmid clones were sequenced from both ends, using Licor sequencing technology. After assembly, sequence gaps and ambiguities were resolved using standard finishing techniques. Difficulties linked to the nature of the sequence meant that 436, 288, 384, 125, 198, 134, and 45 pairs of primers were used for finishing BACs 334L11, 399K20, 155J2, 561P13, 474E04, 356E2, and 834C6, respectively.
Sequence Analysis
The sequence of the human XIC region was extracted from the Human Genome Project Working Draft Assembly (October 7, 2000, freeze; http://genome.ucsc.edu/; Lander 2001). The human, mouse, and bovine sequence was analyzed for repeat sequences (LINEs, SINEs, etc.) using RepeatMasker (A.F. Smit, unpubl.; http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) and species-specific collection of repeat sequences from Repbase-Update (Jurka 2000). Exons of previously known genes were located with SIM4 (Florea et al. 1998). After masking of repeat sequences and already known exons, new genes were searched for using three complementary approaches. First, GENSCAN (Burge and Karlin 1997) and FGENESH (http://genomic.sanger.ac.uk/gf/gf.shtml) were used to identify potential exons. Exons predicted by GENSCAN were tested experimentally by RT-PCR. Secondly, genomic sequences were compared to protein databases with BLASTX (Altschul et al. 1997). Homologous proteins detected with BLASTX were then aligned to the genomic sequences with GENEWISE (Birney et al. 1996) to identify potential genes (http://www.sanger.ac.uk/Software/Wise2/). GENEWISE gene predictions were also performed by comparison of the genomic sequences to the PFAM database of protein domains. Finally, genomic sequences were compared with BLASTN (Altschul et al. 1997) to mRNA (EST or cDNA) sequences from GenBank (release 123). Detected mRNA sequences were then aligned to the genomic sequences with SIM4 to locate intron–exon junctions. Sequences with similarity to a functional genes but containing stop codons or frameshifts in the coding region were considered to be pseudogenes. Potential tRNA genes were searched for with tRNAscan-SE (Lowe and Eddy 1997).
Following Ponger et al. (2001), CpG islands are defined here as regions >500 bp with a G+C content >50% and a ratio CpGo/e (CpGo/e, number of observed CpG over number of expected CpG) of >0.6.
Pairwise local alignments of genomic sequences were performed with SIM (Huang and Miller 1991). SIM is a space-efficient implementation of the Smith and Waterman algorithm. It is much slower but also much more sensitive than heuristic methods such as BLAST or FASTA. We used SIM with the following scoring scheme: match = 1, mismatch = −1, gap opening penalty = 6, and gap extension penalty = 0.2. Only local alignments with a score >30 were retained (e.g., a score of 30 corresponds to a gap-free alignment with 100% identity >30 bp, or 80% identity >50 bp, or 65% identity >100 bp). Pairwise local alignments were visualized with LALNVIEW (Duret et al. 1996).
RT-PCR Analysis
Reverse transcription (RT) was performed on total RNA isolated with RNABle (Eurobio) and treated with RNAse-free DNAse I (Pharmacia; 10U/μg of RNA) for 30 min at 37°C to destroy genomic DNA. Genomic DNA contamination artifacts were controlled for in all RT reactions by including an RNA sample without reverse transcriptase. Random primed RT was performed on 10 μg of RNA by using SuperScriptII reverse transcriptase as recommended by the manufacturer (GIBCO-BRL) with random hexamers (Pharmacia) to prime first-strand cDNA synthesis in a 50-μL reaction volume for 1 h at 42°C. To determine the orientation of transcription, strand-specific RT was performed. For strand-specific RT, 20 μg of RNA was divided into five aliquots and reactions were performed in a 50-μL volume for 1 h at 50°C with specific primer (26 pM) with or without reverse transcriptase (RTase) as follows: (1) forward primer (plus strand), RTase present, to detect transcript with a telomere-centromere orientation; (2) forward primer, RTase absent; (3) reverse primer (minus strand), RTase present, to detect transcript with an opposite orientation; (4) reverse primer, RTase absent; and (5) no-primer control, RTase present. PCR amplification was performed on 2 μL of the RT reaction products and involved 40 cycles with forward and reverse primers under standard PCR conditions. To amplify DNA fragments >2 kb, the Expand Long Template PCR system (Boehringer) was used. Fifteen microliters of the 50-μL reaction mixture was loaded onto an ethidium bromide–stained agarose gel. Initial testing of gene predictions involved testing against RNAs from undifferentiated and differentiated male and female ES cells, adult somatic cells, and testis.
Inactivation Status
Female T16/Mai F1 mice undergo nonrandom X inactivation: Genes subject to X inactivation are expressed only from the T16H allele (Mus musculus domesticus) and not from the Mai allele (Mus musculus musculus). To determine the inactivation status of the Jpx gene reverse-transcribed mouse brain RNAs from the T16H, Mai, and T16H/Mai mouse strains (Rougeulle and Avner 1996) were PCR amplified using the Jpx 1Up and Jpx 1Lo primers (see below for sequences). RT-PCR products were purified using the Qiaquick PCR Purification kit (Qiagen) and digested with MspA1l (Promega) to reveal a restriction site known to be polymorphic between the Mai and T16H strains. Digestion products were analyzed by loading onto a 4% ethidium bromide-stained agarose gel (NuSieve Agarose, Tebu).
Northern Analysis
Poly(A)+mRNA Northern blots from Clontech were hybridized using the Express-Hyb solution (Clontech), as recommended by the manufacturer. Hybridizations exploited 32P random-primed double-stranded DNA probes (Megaprime DNA labeling kit, Amersham) obtained from PCR products amplified using the Jpx1Up/Jpx1Lo and Ppnx3Up/Ppnx3Lo primer pairs (Jpx1Up, 5′CGGCGTCCACATGTATACGTCC3′; Jpx1Lo, 5′TAGGAATGAGCCTCCCCAGCCT3′; Ppnx3Up, 5′AACCGTTATACCTGGACATTTC3′; Ppnx3Lo, 5′CATAACAGCTCTTGTATTGGCA 3′).
Isolation and Sequencing of Ppnx cDNA Clones
Three positive clones were isolated from an adult 129/Sv mouse testis cDNA library kindly provided by Colin Bishop and 2.5 × 105 lambda gt10 clones were plated out and colony lifts made with Hybond N+ membranes (Amersham). Screening was performed with the radioactive labeled 232-bp PCR fragment described in Northern analysis (see above) and hybridization carried out in 0.45 M sodium phosphate (pH 7.2), 1 mM EDTA, and 7% SDS at 65°C as modified from Church and Gilbert (1984), followed by washing in 2 × SSC, 0.1% SDS at 65°C. Single-pass sequencing of the three positive cDNA clones was carried out commercially.
WEB SITE REFERENCES
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker web site.
http://genome.ucsc.edu/; Human genome assembly.
http://genomic.sanger.ac.uk/gf/gf.shtml; FGENESH gene prediction software.
http://pbil.univ-lyon1.fr/datasets/Xix2002/data.html; Online supplementary material.
http://www.ensembl.org; Ensembl database, v. 4.1.1.; Mouse genome assembly.
http://www.sanger.ac.uk/Software/Wise2/; GENEWISE gene prediction software.
Acknowledgments
Marine Prissette was supported by a studentship from the Association pour la Recherche contre le Cancer (ARC), and Agnès Bourdet by a studentship from the Ministère de l'Enseignement Supérieur et de la Recherche. We thank Claire Rougeulle for communicating data concerning the Ftx gene and critical reading of the manuscript and to Jean Weissenbach for his continuing interest in the project. The project was partially financed by a grant from the A.R.C. to P.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pavner@pasteur.fr; FAX 0033145688656.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.152902.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, et al. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Avner P, Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc Natl Acad Sci. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES. Human and mouse gene structure: Comparative analysis and application to exon prediction. Genome Res. 2000;10:950–958. doi: 10.1101/gr.10.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, et al. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Thompson JD, Gibson TJ. PairWise and SearchWise: Finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 1996;24:2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler M-C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzni D, Lawrence C, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- Bouck JB, Metzker ML, Gibbs RA. Shotgun sample sequence comparisons between mouse and human genomes. Nat Genet. 2000;25:31–33. doi: 10.1038/75563. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, Ledbetter DH, Levy E, Craig IW, Willard HF. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991a;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafrenière RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991b;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence C, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Williams CE. Evidence of non-random X chromosome activity in the mouse. Genet Res. 1972;19:229–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Avner P. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet. 1998;19:249–253. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- Courtier B, Heard E, Avner P. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc Natl Acad Sci. 1995;92:3531–3535. doi: 10.1073/pnas.92.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham DB, Segretain D, Arnaud D, Rogner UC, Avner P. The mouse Tsx gene is expressed in Sertoli cells of the adult testis and transiently in premeiotic germ cells during puberty. Dev Biol. 1998;204:345–360. doi: 10.1006/dbio.1998.9004. [DOI] [PubMed] [Google Scholar]
- Debrand E, Heard E, Avner P. Cloning and localization of the murine Xpct gene: Evidence for complex rearrangements during the evolution of the region around the Xist gene. Genomics. 1998;48:296–303. doi: 10.1006/geno.1997.5173. [DOI] [PubMed] [Google Scholar]
- Debrand E, Chureau C, Arnaud D, Avner P, Heard E. Functional Analysis of the DXPas34 Locus, a 3′ regulator of Xist expression. Mol Cell Biol. 1999;19:8513–8525. doi: 10.1128/mcb.19.12.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Dominicis A, Lotti F, Pierandrei-Amaldi P, Cardinali B. cDNA cloning and developmental expression of cellular nucleic acid-binding protein (CNBP) gene in Xenopus laevis. Gene. 2000;241:35–43. doi: 10.1016/s0378-1119(99)00471-0. [DOI] [PubMed] [Google Scholar]
- De La Fuente A, Hahnel R, Basrur PK, King WA. X inactive-specific transcript (Xist) expression and X chromosome inactivation in the preattachment bovine embryo. Biol Reprod. 1999;60:769–775. doi: 10.1095/biolreprod60.3.769. [DOI] [PubMed] [Google Scholar]
- Duret L, Bucher P. Searching for regulatory elements in human noncoding sequences. Curr Opin Struct Biol. 1997;7:399–406. doi: 10.1016/s0959-440x(97)80058-9. [DOI] [PubMed] [Google Scholar]
- Duret L, Dorkeld F, Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: Potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993;21:2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J Mol Evol. 1995;40:308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- Duret L, Gasteiger E, Perriere G. LALNVIEW: A graphical viewer for pairwise sequence alignments. Comput Appl Biosci. 1996;12:507–510. doi: 10.1093/bioinformatics/12.6.507. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamer LW, Wright CV. Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech Dev. 1993;43:71–81. doi: 10.1016/0925-4773(93)90024-r. [DOI] [PubMed] [Google Scholar]
- Gonçalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome Res. 2000;10:672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci. 2002;99:327–332. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio DL, Shelton DA, Zhu W, Millinoff D, Gray T, Bock JH, Slightom JL, Goodman M. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the beta-like globin genes. Mol Phylogenet Evol. 1996;5:1–32. doi: 10.1006/mpev.1996.0004. [DOI] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Ann Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P. Yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol. 1999;19:3156–3166. doi: 10.1128/mcb.19.4.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the mark on the X chromosome during X-inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Herzing LBK, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- Hong YK, Ontiveros SD, Chen C, Strauss WM. A new structure for the murine Xist gene and its relationship to chromosome choice/counting during X-chromosome inactivation. Proc Natl Acad Sci. 1999;96:6829–6834. doi: 10.1073/pnas.96.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, Ontiveros SD, Strauss WM. A revision of the human XIST gene organization and structural comparison with mouse Xist. Mamm Genome. 2000;11:220–224. doi: 10.1007/s003350010040. [DOI] [PubMed] [Google Scholar]
- Huang X, Miller W. A time-efficient, linear-space local similarity. Algor Adv Appl Math. 1991;12:337–357. [Google Scholar]
- Jang W, Hua A, Spilson SV, Miller W, Roe BA, Meisler MH. Comparative sequence of human and mouse BAC clones from the mnd2 region of chromosome 2p13. Genome Res. 1999;9:53–61. [PMC free article] [PubMed] [Google Scholar]
- Jareborg N, Birney E, Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Comparative genomics and evolutionary biology. Curr Opin Genet Dev. 1999;9:624–629. doi: 10.1016/s0959-437x(99)00027-1. [DOI] [PubMed] [Google Scholar]
- Lafreniere RG, Carrel L, Willard HF. A novel transmembrane transporter encoded by the XPCT gene in Xq13.2. Hum Mol Genet. 1994;3:1133–1139. doi: 10.1093/hmg/3.7.1133. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X-inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to non-random X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Verkerk AJ, Van Putten WL, Oosterhuis JW. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet. 1999;64:1445–1452. doi: 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: A repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- ————— LINE-1 elements and X chromosome inactivation: A function for “junk” DNA? Proc Natl Acad Sci. 2000;97:6248–6249. doi: 10.1073/pnas.97.12.6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc Natl Acad Sci. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Platzer M, Bate R, Gloeckner G, Botcherby MR, Nordsiek G, Strivens MA, Kioschis P, Dangel A, Cunningham D, et al. Comparative genome sequence analysis of the Bpa/Str region in mouse and man. Genome Res. 2000;10:758–775. doi: 10.1101/gr.10.6.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes & Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Matassi G, Sharp PM, Gautier C. Chromosomal location effects on gene sequence evolution in mammals. Curr Biol. 1999;9:786–791. doi: 10.1016/s0960-9822(99)80361-3. [DOI] [PubMed] [Google Scholar]
- Maxfield Boumil R, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet. 2001;10:2225–2232. doi: 10.1093/hmg/10.20.2225. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Chowdhury AK, Dunston JA, McIntosh I. Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: Implications for X Inactivation. Am J Hum Genet. 2001;69:951–960. doi: 10.1086/324022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise N, Goto Y, Nakajima N, Takagi N. Molecular cloning of antisense transcripts of the mouse Xist gene. Biochem Biophys Res Commun. 1999;258:537–541. doi: 10.1006/bbrc.1999.0681. [DOI] [PubMed] [Google Scholar]
- Mohrs M, Blankespoor CM, Wang Z-E, Loots GG, Afzal V, Habeida H, Shinkai K, Rubin EM, Locksley RM. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- Morey C, Arnaud D, Avner P, Clerc P. Tsix-mediated repression of Xist accumulation is not sufficient for normal random X inactivation. Hum Mol Genet. 2001;10:1403–1411. doi: 10.1093/hmg/10.13.1403. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudejans CB, Westerman B, Wouters D, Gooyer S, Leegwater PA, van Wijk IJ, Sleutels F. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics. 2001;73:331–337. doi: 10.1006/geno.2001.6522. [DOI] [PubMed] [Google Scholar]
- Park Y, Kuroda MI. Epigenetic aspects of X-chromosome dosage compensation. Science. 2001;293:1083–1085. doi: 10.1126/science.1063073. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Takada S, Youngson NA, Benchaib M, Charlier C, Segers K, Georges M, Ferguson-Smith AC. Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res. 2001;11:2085–2094. doi: 10.1101/gr.206901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. The Xist gene is required in cis for X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Perry J, Ashworth A. Evolutionary rate of a gene affected by chromosomal position. Curr Biol. 1999;9:987–989. doi: 10.1016/s0960-9822(99)80430-8. [DOI] [PubMed] [Google Scholar]
- Ponger L, Duret L, Mouchiroud D. Determinants of CpG islands: Expression in early embryo and isochore structure. Genome Res. 2001;11:1854–1860. doi: 10.1101/gr.174501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NT, Jackson VN, Halestrap AP. Cloning and sequencing of four new mammalian monocarboxylate transporter (MCT) homologues confirms the existence of a transporter family with an ancient past. Biochem J. 1998;329:321–338. doi: 10.1042/bj3290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prissette M, El-Maarri O, Arnaud D, Walter J, Avner P. Methylation profiles of DXPas34 during the onset of X-inactivation. Hum Mol Genet. 2001;10:31–38. doi: 10.1093/hmg/10.1.31. [DOI] [PubMed] [Google Scholar]
- Rastan S. Non-random X chromosome inactivation in mouse X-autosome translocation embryos: Location of the inactivation centre. J Embryol Exp Morph. 1983;78:1–22. [PubMed] [Google Scholar]
- Rastan S, Brown SD. The search for the mouse X-chromosome inactivation centre. Genet Res. 1990;56:99–106. doi: 10.1017/s0016672300035163. [DOI] [PubMed] [Google Scholar]
- Rogner UC, Spyropoulos DD, Le Novere N, Changeux JP, Avner P. Control of neurulation by the nucleosome assembly protein-1-like 2. Nat Genet. 2000;25:431–435. doi: 10.1038/78124. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Avner P. Cloning and characterization of a murine brain specific gene Bpx and its human homologue lying within the Xic candidate region. Hum Mol Genet. 1996;5:41–49. doi: 10.1093/hmg/5.1.41. [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Kondrashov VA, Kondrashov AS. Selective constraint in intergenic regions of human and mouse genomes. Trends Genet. 2001;17:373–336. doi: 10.1016/s0168-9525(01)02344-7. [DOI] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AET, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- Simmler MC, Cattanach BM, Rasberry C, Rougeulle C, Avner P. Mapping the murine Xce locus with (CA)n repeats. Mamm Genome. 1993;4:523–530. doi: 10.1007/BF00364788. [DOI] [PubMed] [Google Scholar]
- Simmler MC, Cunningham DB, Clerc P, Vermat T, Caudron B, Cruaud C, Pawlak A, Szpirer C, Weissenbach J, Claverie JM, et al. A 94 kb genomic sequence 3′ to the murine Xist gene reveals an AT rich region containing a new testis specific gene Tsx. Hum Mol Genet. 1996;11:1713–1726. doi: 10.1093/hmg/5.11.1713. [DOI] [PubMed] [Google Scholar]
- Simmler MC, Heard E, Rougeulle C, Cruaud C, Weissenbach J, Avner P. Localisation and expression analysis of a novel conserved brain expressed transcript, Brx/BRX, lying within the Xic/XIC candidate region. Mamm Genome. 1997;8:760–766. doi: 10.1007/s003359900561. [DOI] [PubMed] [Google Scholar]
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X-inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]