Abstract
To facilitate high-throughput 3D imaging of brain gene expression, a new method called voxelation has been developed. Spatially registered voxels (cubes) are analyzed, resulting in multiple volumetric maps of gene expression analogous to the images reconstructed in biomedical imaging systems. Using microarrays, 40 voxel images for 9000 genes were acquired from brains of both normal mice and mice in which a pharmacological model of Parkinson's disease (PD) had been induced by methamphetamine. Quality-control analyses established the reproducibility of the voxelation procedure. The investigation revealed a common network of coregulated genes shared between the normal and PD brain, and allowed identification of putative control regions responsible for these networks. In addition, genes involved in cell/cell interactions were found to be prominently regulated in the PD brains. Finally, singular value decomposition (SVD), a mathematical method used to provide parsimonious explanations of complex data sets, identified gene vectors and their corresponding images that distinguished between normal and PD brain structures, most pertinently the striatum.
[All study results and supplementary data are available on the web at http://www.pharmacology.ucla.edu/smithlab/genome_multiplex and at http://www.genome.org. Microarray data are also available at GEO, http://www.ncbi.nlm.nih.gov/geo, under the series accession no. GSE30.]
The molecular basis for many neuropsychiatric disorders remains obscure (Owen et al. 2000). These diseases frequently have important genetic contributions, but it has been difficult to identify the relevant genes because of the complexities of human genetic analyses. Importantly, the neuroanatomical regions responsible for the deficits of the neuropsychiatric disorders are often uncertain. Vivid insights into the pathogenesis of these disorders could be obtained if it were possible to obtain an extensive sampling of gene expression patterns in three dimensions for both normal and diseased specimens. In unicellular systems, useful understanding of gene networks have been obtained from high-throughput gene expression methodologies, exemplified by microarrays (Brown and Botstein 1999), gene chips (Lipshutz et al. 1999), and SAGE (Velculescu et al. 1995). Nevertheless, these valuable techniques have yet to be systematically applied to understanding how the three-dimensional (3D) structure of multicellular organisms is constructed by their genomes. Classical technologies, such as in situ hybridization (ISH) or immunohistochemistry, give high resolution images of gene expression within the brain, but are low-throughput procedures, making it difficult to obtain a representative survey of the genome under a variety of experimental situations. It is possible to image metazoan gene expression in vivo, but currently, these technologies only permit, at most, the examination of a few genes at a time (Gambhir et al. 1999; Louie et al. 2000; Zacharias et al. 2000).
Here, a method called voxelation is used to investigate the gene expression changes that occur in the mouse brain as a result of a pharmacological model of Parkinson's disease (PD). The name of the method is derived from the term voxel, which is a cubic 3D image element. Conceptually, voxelation is simple, and uses the direct creation of voxels (cubes) spatially registered with the brain. RNA extracted from the voxels is analyzed using high-throughput techniques, allowing 3D gene expression patterns to be deduced. Voxelation thus results in multiple volumetric maps of gene expression in the brain, similar to the images reconstructed using biomedical imaging systems, such as PET and CT. The essence of the idea is the simplification of complex 3D anatomy into arrays of biochemical samples, facilitating a high-throughput analysis.
RESULTS
A Mouse Model of PD
The PD model was created by administration of toxic doses of methamphetamine (MA) to C57BL/6J mice. At the doses used, the MA model of PD has been reported to cause substantial loss (45%) of tyrosine hydroxylase-positive dopaminergic cells in the substantia nigra, as well as destruction of dopaminergic nerve terminals in the neostriatum (Sonsalla et al. 1996). A more widely used pharmacological model of PD, the MPTP model, results in similar neuropathological effects.
To confirm the induction of the PD phenotype by MA, the striata of treated mice and untreated controls were assayed for dopamine (DA), and its metabolites dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA; Fig. 1A–C). Statistically significant decreases in all three analytes were found in the striata of MA-treated mice. In addition, levels of tyrosine hydroxylase (TH), the rate-limiting enzyme for biosynthesis of dopamine, were assessed in the substantia nigra using real-time quantitative RT-PCR (QRT-PCR; Fig. 1D). Consistent with the reported loss of 45% of TH-positive dopaminergic neurons in the substantia nigra (Sonsalla et al. 1996), the real-time QRT-PCR studies found a statistically significant decrease of 66% in TH mRNA. The specificity of changes in DA and its metabolites was assessed by quantitating levels of a distinct neurotransmitter system. No significant changes were found in striatal levels of 5-hydroxytryptamine (5-HT, or serotonin) and its metabolite 5-hydroxyindoleacetic acid (5-HIAA; Fig. 1E,F).
Figure 1.
Neurotransmitter and metabolite changes in the MA-treated mice. For all histograms, except D, n = 4 controls, 4 MA-treated. The histograms show mean ± SEM, (**) p < 0.01, (*) p < 0.05. (A) Changes in striatal dopamine. One-tailed t-test, t = 2.53, df = 6, p = 0.02. (B) Changes in striatal dihydroxyphenylacetic acid (DOPAC). One-tailed t-test, t = 3.97, df = 6, p = 0.004. (C) Changes in striatal homovanillic acid (HVA). One-tailed t-test, t = 3.84, df = 6, p = 0.004. (D) Changes in substantia nigra tyrosine hydroxylase (TH). One-tailed t-test, t = 6.18, df = 2, p = 0.013. The peak levels of substantia nigra TH from the normal and MA brains, left and right, were estimated from real-time QRT-PCR analysis of voxels G3, G4, H3, and H4 (Fig. 4B). n = 2 controls (left and right), 2 MA-treated (left and right). (E) Changes in striatal 5-hydroxytryptamine (5-HT). Two-tailed t-test, t = 0.139, df = 6, p = 0.89. (F) Changes in striatal 5-hydroxyindoleacetic acid (5-HIAA). Two-tailed t-test, t = 1.44, df = 6, p = 0.20.
Gene Expression Relationships
Brains from control and MA-treated mice were divided into 40 voxels by slicing each brain into 10 coronal sections, and cutting each of the slices into four voxels, consisting of superior and inferior, left and right (Fig. 2). Each of the 40 voxels was then analyzed using a 9000-gene microarray. To explore the relationships between brain regions, we first examined the correlations between each of voxels in terms of gene expression levels. A subset of the entire 9000-gene data set was used for this analysis, consisting of those genes most strongly differentially expressed between the anterior half (20 voxels) and posterior half (20 voxels) of the normal brain. To identify these genes, we chose the outliers with p < 0.05 based on a T statistic, resulting in a total of 1189 genes. The rationale here was that the large differences may indicate interesting genes involved in brain development.
Figure 2.
Voxelation scheme. The slices are labeled A through J, from anterior to posterior. Slice A corresponds to section 8 (6.94 mm interaural, 3.14 mm bregma) of the Mouse Brain Library (Rosen et al. 2000; Williams 2000; http://www.nervenet.org/MBL/mbl.html), slice B to section 11 (5.78 mm interaural, 1.98 mm bregma), slice C to section 12 (5.50 mm interaural, 1.70 mm bregma), slice D to section 14 (4.39 mm interaural, 0.74 mm bregma), slice E to section 16 (3.80 mm interaural, 0.00 mm bregma), slice F to section 17 (2.98 mm interaural, −0.82 mm bregma), slice G to section 20 (1.68 mm interaural, −2.12 mm bregma), slice H to section 24 (0.00 mm interaural, −3.80 mm bregma), slice I to section 28 (−1.31 mm interaural, −5.09 mm bregma), slice J to section 32 (−2.44 mm interaural, −6.24 mm bregma). The anterior–posterior coordinates of the Mouse Brain Library (interaural and bregma relative distances) are as described (Franklin and Paxinos 1997). Odd-numbered voxels (e.g., A1, A3, B1) are from the right side of the brain, and even-numbered voxels (e.g., A2, A4, B2) are from the left.
The resulting voxel × voxel kinships are shown using spatial correlation matrices in Figure 3A. To assess the replicability of the voxelation strategy, the data were divided into separate spatial correlation matrices for the left and right halves of both the normal and MA brains. Because there are 20 voxels for each half of the brain, 20 × 20 cross-correlation matrices were obtained, which are shown as 20 × 20 voxel images, with the matrix elements intensity-color-coded to show the correlations (which can range from +1 to −1). Each of the relevant images includes a color bar to show correlation values. The rows and columns in the matrices are numbered from 1 to 20, which follow the order in which voxels were harvested from the brains. Hence, voxels 1, 2, 3, … from the left half correspond to voxels A2, A4, B2, …, whereas voxels 1, 2, 3, … from the right half correspond to voxels A1, A3, B1, … (see Fig. 2).
Figure 3.
Correlation matrices. (A) Spatial (voxel × voxel) correlation analyses using genes significantly differentially expressed between the anterior and posterior halves of the brain. The matrices for the normal and PD brain are shown with the data for the left and right halves separated out. The correlation between any pair of voxels as judged by gene expression levels is read by looking along the relevant row and column, and finding the intersection. The color of the corresponding element gives the correlation between that pair of voxels by reference to the scales (right). The diagonals are the autocorrelations of the gene expression patterns for each gene and are (and should be) equal to 1. All other correlations must be between 1 and −1. The numbering is such that voxels A2, A4, …, I2, I4 correspond to voxels 1, 2, …, 19, 20, respectively for the left halves of the brain, and voxels A1, A2, …, I1, I3 correspond to voxels 1, 2, …, 19, 20 for the right halves. (B) Expression (gene × gene) correlation analyses using the 6000 most strongly expressed genes when averaged across the 40 voxels. The correlation over voxels for any pair of genes is read by looking along the relevant row and column, and finding the intersection. The genes in the normal omnibus matrix (left and right halves combined) were parsimoniously ordered based on a similarity metric. The same gene order was used to construct the corresponding matrix for the MA brain, and the data were then separated into matrices for the left and right halves of the brains. (C) Monte Carlo analysis comparing similarity of gene expression matrices shown in B for left normal with right normal, and left MA with right MA. The distributions for each brain show the discrepancy between randomly selected pairs of permuted matrices quantitated using the Frobenius norm of the resulting difference matrix. The vertical lines show the differences between the actually observed left and right matrices for each brain. For both the normal and MA brains, the similarity of the gene expression matrices between the left and right halves was highly significant (p < 0.0001 for the normal brain; p = 0.005 for the MA brain). (D) Correlation analyses identify conserved networks of highly correlated gene expression clusters. The gene expression correlation matrices show the subset of genes common to both specimens that are anterior/posterior differentially expressed and display a spatial expression correlation coefficient of >0.75 with at least one other gene within the same brain. The genes in the normal omnibus matrix (left and right halves combined) were parsimoniously ordered using a similarity metric, and the same gene order was used to construct the corresponding matrix for the MA brain. The data were then separated into matrices for the left and right halves of the brains. Two mutually exclusive clusters of coregulated genes emerged: cluster 1 (genes 1–23) and cluster 2 (genes 24–55).
Because the brain shows a high degree of bilateral symmetry, we would expect that the left and right correlation matrices should be comparable. Figure 3A shows that the left and right matrices for both the normal and MA brains are, in fact, strikingly similar. This similarity extends both within experimental groups (e.g., left normal and right normal) and between the groups (e.g., left normal and left MA). A Monte Carlo analysis, conceptually similar to one described below, showed that the similarity between the four correlation matrices was highly significant (p < 0.0001 in all cases), implying excellent reproducibility of the voxelation strategy. Another feature might also be expected from the correlation matrices: the closer together two voxels are (i.e., nearer the diagonal), the more correlated they might be anticipated to be in terms of gene expression, whereas the further apart (i.e., toward the lower left and upper right parts of the matrices), the less correlated. This expected regional kinship can be readily confirmed from the matrices.
The correlation analyses discussed so far are spatial (i.e., voxel × voxel) and provide information on the correlation between voxels based on gene expression levels. However, it is also important to know the correlations between different genes—for example, if one gene shows a strongly regulated expression pattern across voxels, how does this correlate (positively or negatively) with the expression patterns of other genes? To investigate this, we used the 40 voxels as 40 realizations of gene expression, and took a cross correlation with respect to genes to reveal the degree to which the expression of one gene is correlated with that of others across the voxels (gene × gene matrices). For the purposes of this analysis, we used the 6000 most strongly expressed genes when averaged across the 40 voxels of the normal brain to construct gene expression correlation matrices for both control and MA-treated mice (Fig. 3B). The genes in the normal matrix were parsimoniously ordered based on a similarity metric, and the same gene order was used to construct the corresponding matrix for the MA brain. As an independent assessment of the replicability of voxelation, the data were then separated into matrices for the left and right halves of the brain for both control and MA-treated groups.
Strikingly, the matrices for the left and right brain halves were very similar within the control and MA groups (i.e., comparing left normal with right normal, and left MA with right MA), as judged using Monte Carlo statistics (p < 0.0001 for the normal brain; p = 0.005 for the MA brain; Fig. 3C), showing excellent reproducibility of the voxelation strategy. Although not as visually obvious, this statistically significant similarity also extended between groups (i.e., normal left vs. MA left, p = 0.045; normal right vs. MA right, p = 0.007), indicating conservation of gene expression relationships between the control and MA-treated brains, despite the shifts in gene expression that occur as a result of the MA treatment.
To gain further insights into gene expression in the normal and MA brains, a subset of the data was extracted from the anterior/posterior differentially expressed genes, consisting of those genes with a spatial expression correlation coefficient of >0.75 with at least one other gene in both normal and MA brains. This procedure should identify networks of coregulated genes conserved between the two brains. The results of the analysis are presented in Figure 3D as gene expression correlation matrices (gene × gene; see also Table 1). Genes in the normal matrix were parsimoniously ordered based on the similarity metric used for Figure 3B, and the same gene order was used to construct the corresponding matrix for the MA brain. For both brains, the data were again separated into matrices for the left and right halves. Similar to the overall data for the 6000 most strongly expressed subset of genes (Fig. 3B), there was highly significant left/right correspondence (i.e., left normal and right normal, left MA and right MA) for the coregulated gene subset in Figure 3D (Monte Carlo simulation, p < 0.0001), confirming replicability of the voxelation strategy. This conservation was also present in comparisons between experimental groups (i.e., left normal and left MA, right normal and right MA), implying that the coregulated networks of genes are independently maintained in both the normal and MA brains.
Table 1.
Coregulated Genes
| Gene # | Name/Unigene symbol | Accession no. | Function |
|---|---|---|---|
| 1 | Scyd1 | AA272232 | Small inducible cytokine subfamily D, 1 |
| 2 | Siat9 | AA185328 | Sialyltransferase 9 (CMP-NeuAc:lactosylceramide alpha-2,3-sialyltransferase) |
| 3 | EST | AA268133 | Novel |
| 4 | EST | AA051564 | Novel |
| 5 | Ptk2b | AA162543 | Protein tyrosine kinase 2 β |
| 6 | EST | AA000180 | Novel |
| 7 | EST | AA119581 | Novel |
| 8 | EST | AA178003 | Moderately similar to CGMP-dependent 3′,5′-cyclic phosphodiesterase (Rattus norvegicus) |
| 9 | EST | AA250443 | Novel |
| 10 | EST | AA049584 | Novel |
| 11 | lap | AA038556 | Intraolsternal A particles |
| 12 | Bcl11b | AA219828 | B-Cell lymphoa/leukaemia 11B |
| 13 | EST | AA163717 | Highly similar to hypothetical protein KIAA0029 (H. sapiens) |
| 14 | Hivep2 | AA277071 | Human immunodeficiency virus type I enhancer-binding protein 2 |
| 15 | Cspg5 | AA016861 | Chondroitin sulfate proteoglycan 5 |
| 16 | Cugbp2 | AA177976 | CUG triplet repeat, RNA-binding protein 2 |
| 17 | Capn12 | AA403949 | Calpain 12 |
| 18 | 6130400C22Rik | AA110791 | RIKEN cDNA 6130400C22 gene product |
| 19 | EST | AA271272 | Novel |
| 20 | 1200017E07Rik | AA458211 | RIKEN cDNA 1200017E07 gene product |
| 21 | 2810204M08Rik | AA031140 | AIKEN cDNA 2610204M08 gene product |
| 22 | EST | AA030405 | Novel |
| 23 | EST | AA154129 | Weakly similar to T43459 hypothetical protein DKFZp434F0721.1 (Homo sapiens) |
| 24 | EST | AI452297 | Novel |
| 25 | EST | AA008591 | Novel |
| 26 | EST | AA016374 | Highly similar to α-2-macroglobulin precursor (Raltus norvegicus) |
| 27 | EST | AA217217 | Novel |
| 28 | 3632451O06Rik | AA261708 | RIKEN cDNA 3632451O06 gene product |
| 29 | 5031422I09Rik | AI587794 | RIKEN cDNA 5031422I09 gene product |
| 30 | 2610024A01Rik | AA285896 | RIKEN cDNA 2610024A01 gene |
| 31 | 2010012F07Rik | AA277366 | RIKEN cDNA 2010012F07 gene product |
| 32 | Cbln1 | AA016422 | Cerebellin 1 precursor protein |
| 33 | Etohi6 | AA185218 | Ethanol induced 6 |
| 34 | EST | AA185648 | Novel |
| 35 | EST | AA168416 | Novel |
| 36 | 9430010M12Rik | AA270916 | RIKEN cDNA 9430010M12 gene product |
| 37 | Usp9x | AA178383 | Ubiquitin-specific protease 9, X chromosome |
| 38 | Rnf13 | AA189865 | Ring finger protein 13 |
| 39 | Nfl | AI385738 | Neurofilament, light polypeptide |
| 40 | Sepp1 | AA278440 | Selenoprotein P, plasma, 1 |
| 41 | Enpp2 | AA177363 | Ectonucleotide pyrophosphataea/phosphodiesterase 2 |
| 42 | Clptm1 | AA120831 | Cleft lip and palate associated transmembrane protein 1 |
| 43 | EST | AA544722 | Novel |
| 44 | 2700055K07Rik | AA498788 | RIKEN cDNA 2700055K07 gene product |
| 45 | Pla2g7 | AA073983 | Phospholipase A2 group VII (platelet-activating factor acetylhydrolase, plasma) |
| 46 | Abca2 | AA451270 | ATP-binding cassette, subfamily A (ABC1), member 2 |
| 47 | Crys2 | AA231358 | Crystallin, α B |
| 48 | Mbc2 | AA123008 | Membrane-bound C2-domain-containing protein |
| 49 | EST | AA049361 | Novel |
| 50 | Nfl | AA253725 | Neurofilament, light polypeptide |
| 51 | Sept4 | AA498275 | Septin 4 |
| 52 | Pkaca | AA051526 | Protein kinase, cAMP dependent, catalytic, α |
| 53 | Mbp | AA059540 | Myelin basic protein |
| 54 | EST | AA498757 | Novel |
| 55 | EST | W87059 | Novel |
Spatial Gene Expression Patterns
Interestingly, the sorted data in the correlation matrices of the coregulated subset shown in Figure 3D revealed two mutually exclusive clusters of genes. Cluster 1 (genes 1–23) was positively correlated within itself, and negatively correlated with cluster 2 (genes 24–55), and vice versa. The spatial map of gene expression variation across the voxels for the selected subset of genes in both the normal and MA brains is shown in Figure 4A. The figure shows that for both the normal and PD brains, cluster 1 is most strongly expressed in the anterior part of the brain, whereas cluster 2 is most strongly expressed in the posterior. The region in which cluster 2 is most strongly expressed corresponds to voxels 33–36 (I1–I4), and includes the cerebellum (Fig. 2), suggesting that the genes in cluster 2 may be particularly important in specifying this region of the brain. Figure 4A also shows that although the mutually dependent network of spatially coregulated gene clusters is maintained within each brain, the expression patterns are modified in the MA brain compared with the normal brain, both for cluster 1 and cluster 2.
Figure 4.
Spatial gene expression patterns for the subset of correlated genes. (A) Spatial expression patterns of the genes shown in Figure 3D for the normal and MA brains. The relative level of expression of any gene in any voxel is read by looking along the relevant row and column, finding the intersection, and referring to the scales. The voxel numbering in the columns of the matrix is such that voxels A1, A2, A3, …, I2, I3, I4 correspond to voxels 1, 2, 3, …, 37, 38, 39, 40, respectively. The genes are in the same order as for Figure 3D. The two clusters of genes are apparent, and although these have highly conserved patterns of expression within the normal and PD brains, these patterns are somewhat divergent between the two brains. (B) Level of TH expression as judged using real-time QRT-PCR. TH expression levels are found to peak in voxels corresponding to the olfactory bulb (A1–A4, voxels 1–4) and the substantia nigra (G3, G4, corresponding to voxels 27, 28; and H3, H4 corresponding to voxels 31, 32). However, the level of TH in the substantia nigra of the PD brain is substantially decreased compared with the normal brain (Fig. 1D). (C) The Nfl gene is present as two separate spots on the microarrays, corresponding to genes 39 and 50 in A. This provided an opportunity to assess within-array replicability, which was excellent (for the normal brain, r = 0.96, F[1,38] = 443.65, p < 0.0001; for MA brain, r = 0.90, F[1,38] = 153.80, p < 0.0001). (D) Expression pattern of the precerebellin-1 gene in the normal and PD brain. The line drawing shows the anatomy of the midlevel transverse section employed, which corresponds to section 8 of the Mouse Brain Library (interaural 5.40 mm, bregma −4.60 mm). (Olf) Olfactory lobes; (Str) striatum; (Hi) hippocampus; (Ce) cerebellum. Imaging software smoothed the expression patterns over the voxels. Gene expression levels can be deduced by reference to the pseudocolor scales.
Strikingly, when genes with a spatial expression correlation coefficient of >0.75 were extracted from the 6000 most strongly expressed genes rather than the anterior/posterior differentially expressed subset, a similar pattern of two gene clusters with anterior and posterior regional expression was uncovered (Supplementary Fig. 1; Supplementary Table 1 available at www.genome.org). This suggests that a fundamental property of gene expression in the brain is the distinction between anterior and posterior, including the cerebellum.
To confirm the anatomical registration of voxels from the normal and MA brains, RNA from the voxels was assayed for tyrosine hydroxylase (TH) abundance using real-time quantitative RT-PCR (QRT-PCR). TH is strongly expressed in the olfactory bulbs and the substantia nigra (Min et al. 1994), and was therefore expected to be expressed in voxels A1–A4 (voxels 1–4; olfactory bulbs) and the ventral voxels of slices G and H, that is, G3, G4 (voxels 27, 28), and H3, H4 (voxels 31, 32; substantia nigra). The real time QRT-PCR was performed using the same RNA samples used for the microarray studies. Figure 4B shows that the expected anatomical registration was confirmed, and that, in addition, there was a significant decrease of 66% in TH mRNA in the MA brains compared with the normal brains (Fig. 1D), consistent with previously published results (Sonsalla et al. 1996).
One of the genes within cluster 2 (Figs. 3D and 4A), the neurofilament light chain gene, Nfl (Yaworsky et al. 1997), was present on the microarrays as two independent spots (AI385738, gene 39; AA253725, gene 50), and this gave an opportunity to assess within-array reproducibility (Fig. 4C). There was a highly statistically significant correlation coefficient for the Nfl expression profiles independently obtained from the two spots on the microarrays, for both the normal and MA brains, confirming excellent within-array reproducibility.
Interestingly, one of the genes in cluster 2, the precerebellin-1 gene (gene 5), is known to be strongly expressed in the cerebellum (Kavety and Morgan 1998). A graphic representation of the spatial expression for precerebellin-1, in both the normal and PD brains, is shown in Figure 4D. The images have strong bilateral symmetry, again emphasizing the good replicability of the technology.
There were some interesting biological relationships within the coregulated clusters of genes. As mentioned above, the neurofilament light chain gene, Nfl, was present in cluster 2 of both the normal and MA brains. Two additional neurofilament genes, neurofilament medium polypeptide, Nfm (W64752; Myers et al. 1987), and α-internexin, Ina (AA218283; Chan and Chiu 1996), showed a high level of correlation within cluster 2 of the normal brain, but this relationship was not maintained within the MA brain. Consequently these genes are not shown in Figures 3D and 4A. The pattern of coregulation for the three neurofilament genes within the normal brain presumably reflects their related functions. The relaxation of this coregulation within the MA brain is interesting in light of the fact that aggregated neurofilament subunits are a major protein component of Lewy bodies (LBs), intracytoplasmic inclusion bodies that feature prominently in subcortical neurons of patients with Parkinson's disease (Trojanowski et al. 1998). A bioinformatics analysis found regulatory regions shared between the neurofilament genes, which is shown for two of them (Nfl and Ina) in Figure 5. Also found within cluster 2 are the genes for ring-finger protein 13, Rnf13 (AA189868, gene 38) and ubiquitin-specific protease 9, Usp9X (AA178383, gene 37), both of which are involved in ubiquitin-mediated protein degradation, with ring-finger proteins providing specificity to ubiquitin conjugation (Joazeiro and Weissman 2000).
Figure 5.
Putative regulatory elements shared between groups of correlated and anticorrelated genes. There were three groups of correlated (+) genes: (1) SIAT9, HIVEP2, SEPP1, SCYD1, NEFL, INA; (2) CUGBP2, MBP, CBLN1; (3) CBLN1, USP9X, PNUTL2; and one group of anticorrelated (−) genes: (1) MBP, NEFL, CBLN1, PNUTL2, USP9X. The groups are indicated by square brackets. Genes are indicated by UniGene symbol or name (http://www.ncbi.nlm.nih.gov/UniGene). Exons are indicated by short vertical lines and the direction of transcription by horizontal arrows. Homology searches were performed on human homologs of the relevant mouse genes. In all cases, the human gene has the same symbol as the mouse, except for ahuman NEFL = mouse Nfl, and bhuman PNUTL2 = mouse Sept4. The regulatory sequences responsible for correlated expression are shown as squares, those responsible for anticorrelated expression are shown as diamonds. Lines delineate the relationships between the conserved regulatory sequences. Multiple control regions frequently connected the genes. Potential binding sites are: (1) LMO2COM, (2) OCT1, (3) GATA2 and GATA3, (4) MYOD, (5) LMO2COM, (6) NFAT, (7) GFI1, (8) TCF11, (9) HFH2, (10) GFI1, (11) NFAT, (12) FREAC2, (13) OCT1, (14,15) GKLF, (16) TATA, (17) SRY, (18) TH1E47, (19) SRY, (20) HFH2, (21) BARBIE, (22) HFH2, (23) HFH3, (24) HNF3B, (25) HFH2, (26,27) HFH8 and HFH3, (28) HFH-3, (29) GATA, (30) CEBPB, (31) MYOD, (32) LMO2COM, (33) E47, (34) SRY, (35) NFAT, (36) CETS1P54. (*) No known transcription factor binding site.
Gene Networks
To find control regions shared between the correlated and anti-correlated genes of the clusters shown in Figures 3D and 4A, a bioinformatics analysis was performed to look for conserved noncoding sequences (Fig. 5; Table 2). Because of the greater amounts of human genome sequence compared with the mouse, human orthologs of the relevant mouse genes were analyzed. Consequently, only known genes were investigated, and novel genes were omitted from the analysis. Gene pairs were analyzed with gene expression correlation coefficients > 0.75 or <−0.75, and the homology search was restricted to sequences 20 kb upstream, 20 kb downstream, and in all introns of the pertinent genes. BLAST was used to find homologies, but not provide reliable appraisal of their statistical significance, because the algorithm uses asymptotic statistical approximations, which are not accurate for shorter sequences (Benson et al. 2000). The resulting homology regions were additionally examined for transcription-factor-binding sites using the TRANSFAC database (Wingender et al. 2000).
Table 2.
Potential Regulatory Sequences in Coregulated Genes
| Gene paira | Homology Blocks | ||
|---|---|---|---|
| SIAT9 | t[ttgatatgt]ccacttgattgggaaaf (26/26) | ccttt[gccaatacaatgca]d gcaaatgctt (29/29) | |
| (6 exons) | (689 bp 3′ downstream exon 2) | (4358 bp 3′ downstream stop codon) | |
| HIVEP2+ | t[ttgatatat]ccaattgattaggaaac (23/26) | cctct[gccaatcacatgca]dgcaaatgctt (25/29) | |
| (6 exons) | (8331 bp 5′ upstream start codon) | (7558 bp 3′ downstream stop codon) | |
| HIVEP2 | ttttta{(t[ttcatctga)ec]fa}ga (19/19) | gataattttacttggtattt[ctttttcttga]h (31/31) | |
| (6 exons) | (16087 bp 5′ upstream start codon) | (823 bp 3′ downstream exon 3) | |
| SEPP1+ | tttttc({t[ttcatctga)ec]fa}ga (18/19) | gattatttcattttgttttt[ctttttcttga]h (26/31) | |
| (5 exons) | (16520 bp 5′ upstream start codon) | (3784 bp 3′ downstream stop codon) | |
| HIVEP2 | [agaaaatctcttccttttaa]latttct (26/26) | [atgaa]lagctctcagtatattggc (23/23) | [tttttttttttt]kgcttgtaaaa (22/22) |
| (6 exons) | (7281 bp 5′ upstream start codon) | (802 bp 5′ upstream start codon) | (17498 bp 3′ downstream stop codon) |
| NEFLmm+ | [agaaaatcttttcctattaa]iaattct (23/26) | [atgaa]jggctctcagtgtattggc (21/23) | [tttttttttttt]ktcttctaaaa (20/22) |
| (4 exons) | (19520 bp 5′ upstream start codon) | (1611 bp 3′ downstream stop codon) | (9527 bp 5′ upstream start codon) |
| HIVEP2 | aa[aaagaatgaatctgttttaa]l (22/22) | ||
| (6 exons) | (17972 bp 3′ downstream stop codon) | ||
| SCYD1+ | aa[aaaaaacgaatctgttttaa]l (20/22) | ||
| (3 exons) | (1816 bp 3′ downstream exon 1) | ||
| SEPP1 | gaatctgcaaa[gcctttcct]m (20/20) | [cttctgtttactctca]nctct (20/20) | |
| (5 exons) | (15110 bp 5′ upstream start codon) | (14403 bp 5′ upstream start codon) | |
| SCYD1+ | gaaactgtaaa[gcctttcct]m (18/20) | [cttcttgttactctca]nctct (18/20) | |
| (3 exons) | (9285 bp 3′ downstream stop codon) | (19725 bp 3′ downstream stop codon) | |
| SCYD1 | atgcata[aatatttacatata]ot (22/22) | aaattttcttctgcttagct* (20/20) | |
| (3 exons) | (5003 bp 3′ downstream 3′ exon 1) | (5235 bp 3′ downstream exon 1) | |
| NEFL+ | atgcata[tatattcacatata]ot (20/22) | aaatttttttctgcttagct* (19/20) | |
| (4 exons) | (13633 bp 3′ downstream stop codon) | (6321 bp 5′ upstream start codon) | |
| CUGBP2 | at[ggaaaatgcaaagg]pagaaaacag (25/25) | gcccccct[aggagaggcaggtg]qctgc (26/26) | |
| (13 exons) | (65490 bp 3′ downstream exon 1) | (89115 bp 3′ downstream exon 1) | |
| MBP+ | at[ggaaaaagcaaagg]gagaaatcag (23/25) | gcccacca[aggagaggcagggg]qctgc (23/26) | |
| (7 exons) | (33 bp 3′ downstream exon 1) | (17744 bp 5′ upstream) | |
| MBP | atgcaaa[tacataaaat]raa (19/19) | tcagtaacatttat[tttaacaactt]s (25/25) | |
| (7 exons) | (10021 bp 5′ upstream start codon) | (6019 bp 3′ downstream stop codon) | |
| CBLN1+ | atgtaaa[tacataaaat]raa (18/19) | tcattaaaatttat[tttaaaaactt]s (22/25) | |
| (3 exons) | (12012 bp 5′ upstream start codon) | (9085 bp 5′ upstream start codon) | |
| MBP | catgtttgcagtggagt* (17/17) | tttcttt[cttgctttctgg]t (19/19) | |
| (7 exons) | (1122 bp 5′ upstream exon 6) | (3434 bp 3′ downstream end of exon 6) | |
| NEFL− | catgtttgcagtggagt* (17/17) | tttcttt[ctagctttctgg]t (18/19) | |
| (4 exons) | (1045 bp 3′ downstream codon) | (623 bp 5′ upstream start codon) | |
| NEFL | t[gttgt{tgttgtt]ugtttt}vg (19/19) | gatac[tttcaaagcatctgg]w (20/20) | |
| (4 exons) | (17091 bp 5′ upstream start codon) | (5611 bp 5′ upstream start codon) | |
| CBLN1− | t[gttgt{tgttgtt]ugttt}vg (19/19) | gattc[tttcaaagcatctgg]w (19/20) | |
| (3 exons) | (461 bp 3′ downstream stop codon) | (18945 bp 5′ upstream start codon) | |
| NEFL | a{(a[aaaaaaaaaaaa]x)ya}zaaagcacttca (26/26) | aa[t(aaataaataaat)aa]bb,ccgtcc (19/19) | |
| (4 exons) | (9536 bp 5′ upstream start codon) | (4175 bp 3′ downstream stop codon) | |
| PNUTL2nn | a{(a[aaaaaaaaaaaa]x)ya}zagagagcttca (23/26) | aa[t(aaataaataaat)aa]bb,ccttcc (18/19) | |
| (13 exons) | (15553 bp 5′ upstream start codon) | (2140 bp 3′ downstream exon 5) | |
| NEFL | atctttgttagtt[tttttttttttt]kt (26/26) | gaggttttggtagcattct* (1919) | |
| (4 exons) | (9120 bp 5′ upstream start codon) | (412 bp 3′ downstream exon 3) | |
| USP9X− | atctttgttagga[tttttttttttt]kt (24/26) | gaggatttggtagcattct* (18/19) | |
| (44 exons) | (963 bp 3′ downstream exon 22) | (1737 bp 5′ upstream start codon) | |
| NEFL | aaacaaaa[aaaagaaaaaaat]ddtatttt (27/27) | ||
| (4 exons) | (745 bp 3′ downstream exon 1) | ||
| INA+ | aaagaaag[aaaagaaaaaaat]ddtagttt (24/27) | ||
| (3 exons) | (12338 bp 5′ upstream start codon) | ||
| CBLN1 | tagactacactccaaaattt[ggacattc]ee (28/28) | ttttggtga[aagttaagatattt]ffc (25/25) | g[gc(c{ctcatctgca}ggg)hhg]iic (17/17) |
| (3 exons) | (10346 bp 5′ upstream start codon) | (4822 bp 5′ upstream start codon) | (6168 bp 3′ downstream stop codon) |
| USP9X+ | tagactacacttcaaaagta[ggatattc]ee (24/28) | tttttggtgt[aatttaagaaattt]ffc (22/25) | g[gc(c{ctcatctgca}ggg)hhg]iic (17/17) |
| (44 exons) | (3010 bp 3′ downstream exon 34) | (4799 bp 3′ downstream stop codon) | (18415 bp 3′ downstream stop codon) |
| [ctttgttttt]jjgttttttttgg (22/22) | |||
| (8167 bp 3′ downstream stop codon) | |||
| [ctttgttttt]jjgctttttattgg (20/22) | |||
| (13160 bp 3′ downstream stop codon) | |||
| USP9X | ttttttg(tttttt[t{ttttt)kk}ll (23/23) | ||
| (44 exons) | (1572 bp 3′ downstream exon 11) | ||
| PNUTL2+ | tttgttt(tttttt[t{ttttt)kcctg]kk}ll (21/23) | ||
| (13 exons) | (625 bp 5′ upstream start codon) | ||
(+) correlated pair, (−) anti-correlated pair.
Nucleotide matches shown in parentheses, mismatches in bold. Potential binding sites (core in capitals).
LMO2COM ttGATAtat.
OCT1 gccaatcacATGCa.
GATA2 & GATA3 tcaGATGaaa (anti-sense strand).
MYOD ttCATCgac.
LMO2COMtgtCAGAtgaaa (anti-sense strand).
NFAT ntcaaGAAAaag (anti-sense strand).
GFI1 nnnnagaaAATCttttcctattaa.
TCF11 TTCAtnnnnnnnn (anti-sense strand).
HFH2 tttTTTTttttt.
GFI1 aaagaatgAATCtgttttaannnn.
NFAT nnnagGAAAggc (anti-sense strand).
FREAC2 tgagagTAAAcagaag (anti-sense strand).
ICT1 tatattcacATATa.
GKLF ggaaaaagcaAAGG.
GKLF aggagaggcaGGGG.
TATA taCATAAAAt.
SRY tttaACAActtn.
TH1E47 cttgctttCTGGnnnn.uSRY aacaACAAcaac anti-sense strand).
HFH2 tgtTGTTgtttt.
BARBIE tttcAAAGcatctgg.
HFH2 tttTTTTttttt (anti-sense strand).
HFH3 tttTTTTtttttt (anti-sense strand).
HNF3B tttttTTTTtttttt (anti-sense strand).
aaHFH2 attTATTtattt (anti-sense strand).
bb,ccHFH8 & HFH3 attTATTtattta (anti-sense strand).
ddHFH-3 attTTTTtctttt (anti-sense strand).
eeGATA gGATATtcnnn.
ffCEBPB aatttaaGAAAttt.
ggMYOD ctCATCtgca.
hhLMO2COM ctgCAGAtgagg (anti-sense strand).
iiE47 cctGCAGatgagggc (anti-sense strand).
jjSRY aaaaACAAagnn (anti-sense strand).
kkNFAT nncagGAAAaaa (anti-sense strand).
llCETS1P54 ncAGGAaaaa (anti-sense strand).
mmhuman NEFL = mouse Nfl.
nnhuman PNUTL2 = mouse Sept4.
No known transcription factor binding site.
The analysis unveiled an intricate arrangement of potential control elements shared between genes, which may play a role in their expression pattern relationships. In all cases, except for three distinct pairs (marked by asterisks in Fig. 5 and Table 2), the potential control sequences harbored known transcription-factor-binding sites. We anticipate that the exceptions may in the future be identified as binding sites for yet uncharacterized transcription factors. Nevertheless, the significance of the potential control regions must still be confirmed experimentally. One striking example that emerged from the analysis is the Nfl gene (human homolog NEFL), which had 14 putative control regions, representing binding sites for no fewer than 19 transcription factors. Interestingly, promoter sequences in the 1.7-kb region upstream of the Nfl start codon have been analyzed using reporter constructs in transgenic mice (Yaworsky et al. 1997). These studies found that the 1.7-kb region contained sequences that allowed initiation of neuronal and myogenic Nfl gene expression in embryos, but not maintenance after birth. Because the analysis reported here examines expression in adult tissues, the uncovered putative control regions may be involved in the maintenance of Nfl expression in the adult brain.
Expression Differences between Normal and MA Brains
In addition to analyses of spatial gene expression in the normal and MA brains, significant (p < 10−9) global expression differences when averaged across the 40 voxels of each brain were sought for known genes. Scatterplots were constructed that compared the expression levels for these genes between the left and right halves of the normal brain and the left and right halves of the PD brain (Fig. 6A). This analysis showed excellent replicability between the left and right halves for both the normal and diseased brains. Scatterplots were also constructed that compared the left half of the normal brain with the left half of the PD brain, and the right half of the normal brain with the right half of the PD brain (Fig. 6A). These scatterplots revealed that there was about a fourfold difference in expression between the genes repressed or induced in the MA brain. Figure 6B shows the expression level differences for the significantly differentially expressed genes. The results for the left and right halves of the brain are again shown separately and show excellent replicability. There was an approximately equal number of genes induced in the MA brain (16/36) compared with those repressed (20/36), and this did not represent a significant deviation from random (χ2 = 0.22, df = 1, p = 0.64). The differential expression of a number of genes was confirmed using real-time QRT-PCR analysis of the same RNA samples used for the microarray studies (Fig. 6C).
Figure 6.
Differentially expressed genes. (A) Scatterplots show mean expression levels across the 40 voxels in the normal and MA brains on a logarithmic scale (log2) for genes where p < 10−9. The comparisons within brains (left and right normal; left and right MA) provide another assessment of replicability (for normal brain, r = 0.90, F[1,34] = 143.98, p < 0.0001; for MA brain, r = 0.98, F[1,34] = 817.76, p < 0.0001). The comparison between brains (left normal and left MA, right normal and right MA) shows the differences in expression between genes repressed and induced in the MA brain. Red line: twofold above equivalent expression level; green line: equivalent expression level; blue line: twofold below equivalent expression level. (B) Mean expression levels across the 40 voxels of the normal and MA brains on a logarithmic scale (log2 ±SEM). (Red) Normal; (blue) MA. The genes are ranked from most (gene 1) to least significant (gene 36; p < 10−9). To allow replicability to be assessed, the data for the left and right halves of each brain were separated out as the left and right data points, respectively, for each gene. No significant differences were found comparing left normal with right normal (two-tailed t test on log2-transformed data, t = 0.012, df = 70, p = 0.99) and left MA with right MA (two-tailed t test on log2-transformed data, t = −0.33, df = 70, p = 0.75). (C) Confirmation of gene expression differences for selected genes by real-time QRT-PCR analysis of voxel RNA. Expression levels are shown relative to controls and normalized using 18S RNA. The bars indicate mean ± SEM. (***) p < 0.001, (**) p < 0.01, two-tailed t-test. (D) Gene categories in the entire 9000-gene data set (9K) and in those genes significantly different (diff) between the control and MA brains (p < 0.001) when averaged across all 40 voxels. Genes involved in cell/cell interactions (cytoskeleton, extracellular matrix, and cell adhesion) were significantly overrepresented in the regulated subset (p = 0.02), but apoptosis-related genes were not (p = 0.39). The bars show mean ± SD, (*) p < 0.05, as judged using Monte Carlo statistics. The control category represents 15 randomly chosen genes to show the validity of the Monte Carlo analysis. As expected, this category showed no significant difference in frequency between the 9K and regulated data sets (p = 0.48).
Several intriguing genes were revealed by the analysis of expression differences between the normal and MA brains (Table 3), involved in such diverse areas as transcription (e.g., Hdac5, Mxi1, Sp1), cell morphology (e.g., Cdc42, Pkcq), extracellular matrix (e.g., Eln, Lamc2), and signal transduction (e.g., Grb2, Ppp2ca). Interestingly, there appeared to be a relatively large number of genes involved in modulating cell morphology (five genes), suggesting substantial regulation of neuronal outgrowth in response to the loss of dopaminergic neurons in the PD model. Also, many genes were found to play a role in apoptosis (five genes, including Stk2, Ppp2ca, Parg, Siah1a, and qk). This category is not highly ranked in Table 3, as many of the apoptosis-related genes have more than one function and were only assigned to one category in the table.
Table 3.
Differentially Expressed Genes
| Gene # | Unigene symbol | Accession no. | Gene name (and function) |
|---|---|---|---|
| Miscellaneous (11 genes) | |||
| 7 | Abca2 | AA276158 | ATP-binding cassette, sub-family A (ABC1), member 2 (member of ABC transporter family) |
| 33 | A1p2a2 | AA222567 | ATPase, Ca2+ transporting, cardiac muscle, slow twitch 2 (endo/sacrooplasmic reticulum calcium pump) |
| 9 | C1r | AA261393 | Complement component 1, r subcomponent (activation of classical complement cascade) |
| 3 | Lrpap1 | AA253890 | Low-density lipoprotein receptor-related protein associated protein 1 (chaperone for LDL receptor family members) |
| 31 | Papss2 | AA244536 | 5′-Phosphoadenosine 5′-phosphosulfate synthase 2 (sulfation of toxins) |
| 35 | Pglyrp | AA238752 | Peplidoglycan recognition protein (innate immunity; recognizes bacterial wall peptidoglycan) |
| 11 | Psme1 | AA239485 | Protease (prosome, macropsin) 28 subunit, α (activator subunit of 205 proteasome) |
| 1 | Pura | AI894064 | Purine-rich element-binding protein A (single-stranded-DNA, and RNA-binding protein) |
| 23 | Rdh5 | AA275664 | Retinol dehydrogenase type 5 |
| 20 | Rps5 | AA240279 | Ribosomal protein S5 (translation) |
| 5 | X66 | AA249976 | Xeroderma pigmentosum, complementation group C (DNA excision repair) |
| Transcription (6 genes) | |||
| 22 | Hdac5 | AA017742 | Histone deacetylase 5 (regulation of transcription) |
| 25 | Klf1 | W97446 | Erythroid Kruppel-like factor 1 (transcriptional activator of the adult β-globin promoter) |
| 28 | Mata1l1 | AA461637 | Metastasis associated 1-like 1 (modulates histone deacetylase complex NuRD) |
| 34 | Mxl1 | AA472395 | Ma-interacting protein 1 (antagonist of c-Myc transcription factor) |
| 8 | Nr2c2 | AA501045 | Nuclear receptor subfamily 2, group H, member 2 (steroid receptor superfamily member) |
| 28 | Sp1 | AA212645 | trans-Acting transcription factor 1 (zinc-finger transcription factor) |
| Cell morphology (5 genes) | |||
| 13 | Cdc42 | AA266975 | Cell division cycle 42 homolog (ρ GTPase, cell morphology) |
| 15 | Crkas | AA240272 | v-crk associated tyrosine kinase substrate (focal adhesion docking protein p130(Cas), cell morphology, modulation cytoskeleton) |
| 18 | Mlapt/Mapl | AA028410 | Microtubule-associated protein τ (modulation cytoskeleton) |
| 27 | Pkcq | W98195 | Protein kinase C, theta (modulation of cytoskeleton, neurite outgrowth) |
| 30 | Stk2 | AA268478 | Serine/threonine kinase 2 (apoptosis, cytoskeletal remodeling) |
| Extracellular matrix/cell adhesion (3 genes) | |||
| 2 | Ecm1 | AA237378 | Extracellular matrix protein 1 (secretory glycoprotein) |
| 14 | Eln | AA239171 | Elastin (extracellular matrix component) |
| 36 | Lamc2 | W49392 | Laminin, γ2 (extracellular matrix glycoprotein; major constituent basement membranse) |
| Signal transduction (3 genes) | |||
| 16 | Grb2 | AA183927 | Growth factor receptor bound protein 2 (adaptor protein involved in signal transduction) |
| 32 | Ppp2ca | AA245165 | Protein phosphatase 2a, catalytic subunit, α isoform (signal transduction, apoptosis) |
| 29 | S100a6 | AA267952 | Calcium-binding protein A6, or calcyclin (calcium signal transduction) |
| Apoptosis (2 genes) | |||
| 19 | Parg | AA260570 | Poly(ADP-ribose) glycohydrolase (catabolism of poly(ADP-ribose), apoptosis) |
| 6 | Siah1a | AA267965 | Seven in absentia homolog 1A (apoptosis, cell cycle arrest ubiquitin-proteasome pathway) |
| Intermediary metabolism (2 genes) | |||
| 4 | Mor1 | AA266087 | Mitochondrial malate dehydrogenase (oxidative phosphorylation) |
| 17 | Mu1 | AA250181 | Methylmalonyl-coenzyme A mutase (isomerization succinyl CoA and methylmalonyl CoA) |
| RNA binding (2 genes) | |||
| 12 | Fxr2h | AA119248 | Fragile X mental retardation gene, autosomal homolog 2 (RNA binding protein) |
| 21 | qk | AA220551 | Quaking (RNA binding protein required for myelin formation, apoptosis) |
| Vesicular transport (2 genes) | |||
| 10 | Ap1b1 | AA221073 | Adaptor protein complex AP-1, β1 subunit (clathrin-coated vesicle formation) |
| 24 | Arf2 | AA266938 | ADP-ribosylation factor 2 (GTP-binding protein, formation of coated transport vesicles) |
Gene number is the rank of the gene in order of most (gene 1) to least (gene 36) (p < 10−6) significantly differentially expressed between the normal and MA brains; see Fig. 8B. For table, genes are ordered alphabetically within categories.
To more rigorously investigate the classes of genes regulated as a result of the MA treatment, the frequency of genes in the categories described above (cell morphology, extracellular matrix, cell adhesion, and apoptosis) were compared between the entire 9000-gene data set and those genes judged significantly differentially expressed between the control and MA brains (p < 0.001) when averaged across all 40 voxels (Fig. 6D). Using Monte Carlo simulations, genes involved in cell/cell interactions (cell morphology, extracellular matrix, and cell adhesion) were found to be significantly overrepresented in the regulated subset compared with the entire 9000-gene subset. Perhaps surprisingly, apoptosis-related genes were found at an equal frequency in both sets of genes, possibly reflecting the uncertain role of apoptosis in PD (Kösel et al. 1997; Banati et al. 1998; Wüllner et al. 1999). Overall, these data emphasize the importance of cell/cell interactions in response to the neuronal loss of PD.
Other gene categories were also found to be significantly regulated (p < 10–9) in the PD brains compared with controls (Fig. 6B; Table 3). Sulfation is a major pathway in the biotransformation of many drugs (Xu et al. 2000), and this may explain the induction of the Papss2 (3′-phosphoadenosine 5′-phosphosulfate synthase 2) gene in the MA brain. Interestingly, the Pon2 gene (paraoxonase2, W98586) was highly significantly induced in the MA brain (p < 10−8; data not shown), and this gene family is also involved in detoxification, as well as being linked with PD (Akhmedova et al. 2001).
The Mtapt/Mapt gene (microtubule-associated protein tau) is repressed in the MA brain. This observation may be related to the fact that tau is mutated in a genetic disorder that features parkinsonism (frontotemporal dementia with parkinsonism on chromosome 17 or FTDP-17), as well as being the constituent of neurofibrillary tangles, the defining cytological lesion for many neurodegenerative movement disorders (Spillantini et al. 2000). Perhaps related to the decreased tau transcript levels, the Ppp2ca gene (protein phosphatase 2a, catalytic subunit, α isoform) is also repressed in the MA brain. Protein phosphatase 2A (Pp2a) binds tightly to tau, and all tau mutations resulting in FTDP-17 decrease the affinity of this association (Goedert et al. 2000).
The qk (quaking) gene (Cox et al. 1999) is repressed in the MA brain and encodes a potential RNA binding protein. Interestingly, mutations in qk result in altered brain dopaminergic signaling (Nikulina et al. 1995), the neurotransmitter system most profoundly affected in PD. Another gene with a connection to dopaminergic signaling is the Sp1 transcription factor, which is repressed in the MA brain and regulates transcription of dopamine receptor genes (Yajima et al. 1998; Hwang et al. 2001).
Singular Value Decomposition Reveals Global Shifts of Gene Expression between Normal and MA Brains
Singular value decomposition (SVD) is a powerful method for parsimonious explanations of complex data sets (Hendler and Shrager 1994; Frackowiak et al. 1997; Alter et al. 2000). This statistical method reduces dimensionality while keeping the maximum possible fraction of the variance from the original data. For example, when used in biomedical imaging, SVD analysis commonly explains data sets in terms of known functional and anatomical boundaries (e.g., cortical vs. subcortical). In the area of gene expression patterns, it might be expected that SVD would show which orthogonal sets of genes (vectors) account for the major variations between the voxels. Essentially, the gene vectors would represent votes for the molecular attributes of the various brain regions in which they are manifest. SVD does not rely on preconceived ideas or hypotheses, and is entirely data-driven.
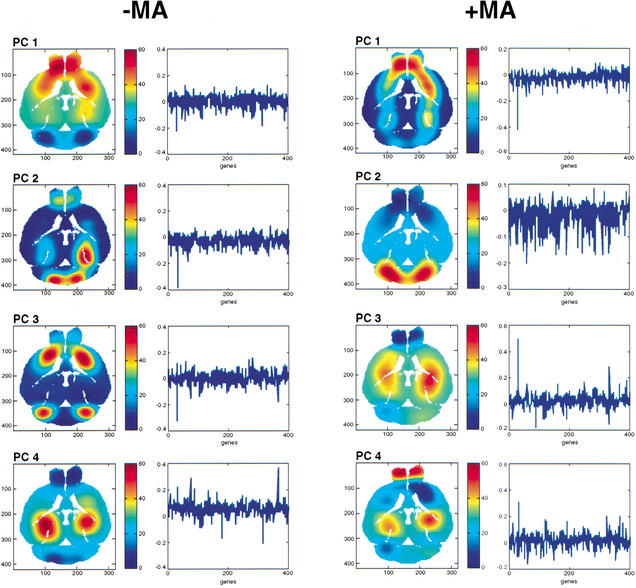
To ascertain whether SVD would illuminate the large amounts of data from the voxelation studies of the normal and MA brains, we performed an analysis on the anterior/posterior differentially expressed subset of genes. The results are shown in Figure 7, where the first four principal components (PCs), or gene vectors, resulting from the SVD are shown as pseudocolor images. These vector images, rather than representing the expression pattern of any one gene, instead represent multiple (hundreds of) genes, which may be particularly important in regional specification of the brain. The striking bilateral symmetry of these images provides vivid documentation of the excellent reproducibility of the voxelation procedure from an overall survey viewpoint.
Figure 7.
SVD delineates anatomical regions of the brain. The SVD was performed using the anterior/posterior differentially expressed subset of genes. The spatial patterns resulting from the first, second, third, and fourth PCs are shown. Alongside are the first 400 members of the corresponding gene vectors. The ordinate represents the contribution by the relevant gene to the variation of the vector spatial pattern, and the abscissa represents the genes in decreasing order of differential expression. The pseudocolor scales give the level of expression of the relevant gene vector in the spatial patterns.
The first PC is conserved in the normal and MA brains, and is most strongly expressed in the olfactory lobes and striatum. Similarly, the second PC is conserved between the two brains, and is most strongly expressed in the cerebellum. The third PC, however, is not conserved between the two brains. In the normal brain, this PC is most strongly expressed in the striatum and cerebellum, suggesting a hitherto unsuspected genetic connection between these otherwise functionally and anatomically distinct regions of the brain. In contrast, the third PC of the MA brain shows a dramatic shift away from the striatum and cerebellum toward the hippocampus. This striking alteration in the expression of the corresponding gene vector may be related to the fact that the striatum is the region of the brain most prominently affected by PD. The fourth PC is again conserved between the normal and MA brains, and is strongly expressed in the hippocampus. Together, these findings suggest that the known physiological changes in the striatum of PD brains is accompanied by prominent changes in the corresponding genetic networks responsible for maintenance of this structure.
The reliability of the SVD was assessed by repeating the analysis using a different subset of the data, consisting of those genes with a correlation coefficient between the left and right halves of the brain >0.7, and whose variance of expression through the entire brain was greater than the median of the variance for all genes. The resulting images were essentially identical to those shown in Figure 7, confirming the robustness of the SVD analysis. The bilateral symmetry of the PCs, their robustness, and their restriction to relevant anatomical regions is remarkable, especially considering the twofold uncertainty in the microarray data and the relatively crude spatial maps (40 voxels). With more comprehensive gene surveys and increased resolution, voxelation may ultimately reveal the molecular genetic relationships between various regions of the brain, as well as identifying which areas are most affected in disease. The assignation of gene expression changes to defined neuroanatomical loci may be particularly valuable in identifying responsible brain regions for neuropsychiatric disorders such as schizophrenia (Mirnics et al. 2000), where the location of such regions is presently uncertain.
DISCUSSION
The investigations reported here show that by combining high-throughput gene expression methodologies with spatial information, voxelation can provide valuable insights not easily obtainable from studies of tissue culture systems. In one example, voxelation helped define networks of anatomically coregulated genes and their relevant control regions. The overwhelming majority of the putative control regions contained potential transcription-factor-binding sites, lending credibility to the analysis. However, transcriptional control regions have considerable degeneracy, and study of genomic sequence from additional organisms may further bolster the significance of the uncovered sequences. The voxelation analysis also allowed identification of genes globally differentially expressed between normal and PD brains, whereas SVD gave insights into the principal neuroanatomical changes of PD at the molecular genetic rather than the traditional histological level.
Significant bilateral symmetry was found in the data using a number of different analytical approaches, suggesting good reproducibility of the voxelation technology. However, the symmetry may also partly reflect the relatively crude spatial maps used for the voxelation, and asymmetry might become apparent at higher resolution in certain situations, for example, as a result of sexual dimorphism (Dluzen and Kreutzberg 1996; Tabibnia et al. 1999). Several studies have used microarrays and targeted dissections to investigate gene expression in the mouse brain (Lockhart and Barlow 2001). Interestingly, in these investigations the cerebellum appeared to be the most distinct brain region in terms of gene expression (Sandberg et al. 2000), reminiscent of the finding in the present study of a posteriorly expressed cluster of coregulated genes.
It should be emphasized that some of the analytical approaches used in this study do not necessarily depend on precisely matched samples. For example, conserved networks of coregulated genes maintained across voxels and between specimens can be identified using human samples (Brown et al. 2002a), where there is an inevitable lack of exact controls. However, analysis of human brains has the decided advantage of disease validity, which may be particularly useful for entities like schizophrenia or bipolar disorder, where the etiology of the disease is obscure and the relevance of the available mouse models is in doubt. In contrast, voxelation of the mouse has the advantages of carefully controlled experiments and matched samples. In the long run, perhaps the most robust understanding will be provided by the judicious combined use of human specimens and model organisms.
Information recovery from voxelation is in principle limited by voxel inhomogeneity and the performance of the analytic tools used to investigate the voxels. Voxel inhomogeneity is a consequence of noninfinitesimal voxel size and hence finite spatial resolution, resulting in decreased signal-to-noise ratios for gene expression. However, this is a ubiquitous phenomenon in brain-imaging technologies (CT, PET, fMRI), but does not prevent these methods from providing useful information. Similarly, useful insights were obtained from the present study, despite the relatively crude spatial maps. The signal-to-noise ratio obtained from finite voxels might be improved by preselecting cells using a lineage-specific marker, such as green fluorescent protein (Peterson 2002). Analytic performance of the tools used to analyze the voxels is limited by sensitivity (qualitative performance: the ability to detect presence or absence of expression), accuracy (quantitative performance: the ability to reliably detect differences in gene expression), and throughput. Microarrays have moderate sensitivity and can only reliably discriminate between twofold differences in gene expression, but have excellent throughput.
Despite information losses in voxelation caused by voxel heterogeneity and analytic limitations, the present study shows that these deficits are overshadowed by the high throughput of the method, which allows for much greater net information recovery than is practicable with classical approaches such as in situ hybridization. This makes the use of voxelation feasible for large-scale study of gene expression in multiple disorders and models, a daunting prospect for in situ hybridization. A further advantage of voxelation is its modality independence, which will allow its use for 3D mapping of the proteome and perhaps even electrophysiology, in addition to investigation of the transcriptome. Nevertheless, an important future goal for voxelation will be to expand the amount of information it provides, both by improvements in resolution and analytic performance.
Although microarrays are a relatively cheap tool on a per gene basis, voxelation will become increasingly expensive as greater numbers of voxels are analyzed in the search for improved resolution in a wide variety of experimental circumstances. It will therefore be important to find ways to drive down costs. Using presently available technology, microarrays are sufficiently sensitive to allow construction of a 325,000-voxel human brain map (Brown et al. 2002a; Peterson 2002), but for such maps to be feasible, the cost per voxel would have to drop by ∼25-fold (Peterson 2002). For the mouse brain, microarray technology is sensitive enough to allow construction of ∼120 voxel maps, and higher resolution will require improvements in sensitivity. Real-time PCR is sufficiently sensitive to permit construction of 6000-voxel maps of the mouse brain, while still affording opportunities for scalability and automation.
All the goals of improved spatial resolution, analytic performance, and cost will provide substantial challenges. Ultimately, however, high-resolution study of human neuropsychiatric disorders, mouse models of these disorders, as well as the ability to voxelate whole model organisms, will give better comprehension of the logic of the genome, how this logic goes awry in disease, and important starting points for novel therapies.
METHODS
MA Model of PD
Adult C57BL/6J male mice (10–24 wk, 25–31 g) received four i.p. injections of MA hydrochloride (10 mg/kg per injection using 1.5 mg/mL solution) at 2-h intervals (Sonsalla et al. 1996; Melega et al. 1997). The mice were analyzed 7 d after MA treatment.
Monoamines and Metabolites in Mouse Striatum
Brain samples were weighed wet, sonicated with 250 μL of 0.1 M perchloric acid and centrifuged at 14,000 rpm for 15 min (4°C). The supernatant was filtered through a 0.2-μm PTFE filter and an aliquot diluted with water (1:2) for HPLC analysis. The solid pellet was suspended in 1.0 mL of 0.2 M NaOH for protein assay. For HPLC we used an ESA HPLC Model 580 solvent delivery module (dual-piston pump), and an ESA Coulochem II electrochemical detector with an analytical cell operating at +350 mV and 500 nA. The mobile phase consisted of acetonitrile:sodium phosphate monobasic buffer (75 mM sodium phosphate, 1.8 mM 1-octanesulfonic acid sodium salt [OSA], 12 μM ethylenediaminetetraacetic acid, disodium salt dihydrate [EDTA]) at 9.5:90.5 v/v, pH 3.1 (aqueous phase). The guard column was Adsorbosphere HS, C18, 7.5 × 4.6 mm, 5 μm, and the analytical column was Adsorbosphere HS, C18, 100 × 4.6 mm, 3 μm. Both columns were from Alltech Associates, Inc. The flow rate was 0.8 mL/min, and the injector loop volume was 20 μL.
Voxelation Procedure
Mouse brains were voxelated by first cutting along a transverse plane that included the interaural line and the anterior olfactory lobe, thus producing dorsal and ventral halves. The two halves were placed in a commercially available cutting device (ASI Instruments, RBM-2000C) that incorporates a mold to steady the brain. Machined into the mold are 14 slots separated by 1 mm for precise cutting. In the following description, slot 1 of the mold is most anterior, and slot 14 is most posterior. The brain was placed into the mold such that slice 4 of the mold (corresponding to slice C, Fig. 2) was aligned with the anterior part of the Circle of Willis. Slice 8 of the mold (corresponding to slice G, Fig. 2) was aligned with the point where the anteriormost part of the brain stem/pons could be observed emerging from the ventral surface of the brain. The first and last four slots of the mold were omitted from the voxelation scheme, so that the brain was cut into 10 approximately equal coronal sections of 1 mm thickness, while keeping the dorsal/ventral halves in register. When the blade of the device was removed after cutting each coronal section, the sections, divided into dorsal and ventral halves, remained adherent to the blade, in register, owing to surface tension. At that point each coronal section was bisected down the midline, such that each section consisted of four quarters (superior left, superior right, inferior left, inferior right).
The numbering scheme for the voxels is shown in Figure 2. Slice A was designated as most anterior and slice J the most posterior. Superior right voxels were numbered 1, superior left voxels were 2, inferior right voxels were 3, and inferior left voxels were 4. For example, the inferior right voxel from slice C would be designated voxel C3. Registration with the Mouse Brain Library (Rosen et al. 2000; Williams 2000; http://www.nervenet.org/MBL/mbl.html) was achieved by comparison of sections. Using the inbred C57BL/6J mouse strain, the cutting was found to be essentially invariant from one animal to the next. As judged using the coordinates provided by the Mouse Brain Library, the distance between sections was (mean ± SEM) 1.04 mm ± 0.17 (bregma origin) and 1.04 mm ± 0.18 (interaural origin). To provide sufficient RNA (100 μg) for microarray analysis, equivalent voxels of multiple brains were pooled. Even for the voxels with the smallest amount of tissue, a total of 29 brains provided sufficient material. The smallest voxels were the most anterior (A1–A4), encompassing the olfactory bulbs.
Microarray Analysis
For each voxel, 100 μg of Cy3-labeled voxel RNA and 100 μg of Cy5-labeled control RNA were cohybridized to a separate 9000-gene microarray, as described (Eisen and Brown 1999). The control RNA was used to facilitate interarray comparisons, and consisted of total normal C57BL/6J mouse brain RNA. For each gene, the signal-to-noise ratio was 2.5-fold above background for both the Cy3 and Cy5 channels, and the mean (±SD) signal-to-noise ratio was 7.40 ± 3.25 for Cy3 and 11.04 ± 3.96 for Cy5. The microarray data were processed using two types of normalization procedures. First, spatial trends existing in the data from chip printing were removed by nonlinear transformation of the data sets. The second normalization procedure was designed to compensate for differences in the labeling and chemical properties of the Cy3 and Cy5 dyes, by aligning the histograms of the dye signals both within, as well as between, chips.
Correlation Matrix Clustering
For Figure 3, B and D, the genes in the control omnibus correlation matrix were ordered using a similarity metric. The first row of the matrix was chosen to show a strong contrast between the highest and lowest correlation coefficient for that row. This row was denoted as the base vector, B, with respect to which the remaining rows, R, were arranged in order of decreasing similarity, using a metric consisting of ∑i(Bi − Ri)2, where i is the elements of the rows. Once the omnibus matrix for the normal brain was created, the omnibus matrix for the MA brain was created following the same order, and the data for each brain then separated into left and right halves.
Monte Carlo simulations
The Monte Carlo simulations to assess the similarity of correlation matrices for the normal left with the normal right and the MA left with the MA right (Fig. 3B) used random permutation of the columns of the matrices. The discrepancy between randomly selected pairs of permuted matrices was quantitated using the Frobenius norm of the matrix obtained by subtracting one matrix from the other. The difference between the mean of the resulting distribution and the Frobenius norm obtained from the left and right matrices was then used to assess significance.
Genes in each of the categories shown in Figure 6D were chosen using appropriate keywords (e.g., cytoskeleton, extracellular matrix, and cell adhesion for cell/cell interactions; cell death and apoptosis for apoptosis.) To assess the significance of the gene frequencies, Monte Carlo simulations were performed using two data sets: the entire 9000-gene data set and the subset consisting of those genes differentially expressed (p < 0.001) when averaged across all 40 voxels of the control and MA brains. The simulations were performed by randomly discarding the data for 30% of the genes in each data set and recalculating the mean. Significance values were assigned for each gene category by calculating the area of overlap for the two distributions from each data set. To show the validity of the Monte Carlo simulations, a control category was analyzed, consisting of 15 genes chosen randomly that were common to both data sets. As expected, no significant difference was found between the data sets for this category (Fig. 6D).
Real-Time QRT-PCR
Real-time QRT-PCR was performed using TaqMan (Gibson et al. 1996) and One-Step RT-PCR Master Mix following the manufacturer's instructions (Applied Biosystems). Reverse transcription used 100 ng of total voxel RNA for TH, and 10 ng for Ecm1, Ap1b1, Psme1, S100a6, and Stk2. Detection used an Applied Biosystems Prism 7700 Sequence Detector (Perkin-Elmer), and the data were analyzed using SEQUENCE DETECTOR software. Normalization used 18S RNA. Genomic contamination was excluded (Brown et al. 2002b) by the use of primers that cross the intron of the housekeeping gene GdX (Filippi et al. 1990) and no reverse transcriptase controls.
Images
For the anatomical images of gene expression (Fig. 4D) and SVD (Fig. 7), a midlevel transverse section was used (5.40 mm interaural, −4.60 mm bregma, section 8 of the Mouse Brain Library), and expression levels were averaged between corresponding dorsal and ventral voxels on the same side of the brain. All the images used a data set consisting of those genes that were most strongly differentially expressed between the anterior half (20 voxels) and posterior half (20 voxels) of the brain (p < 0.05). Because the cerebral cortex is featured in approximately equal amounts in both the anterior and posterior halves, it was expected a priori that the images would principally show features in parasagittal locations rather than in the periphery, where the cortex is located. Consequently, a Bayesian approach to image creation was used, based on a kernel that gave preference to the center of each voxel, and thus used prior probabilities from the known anatomy of the voxelated brain. Furthermore, a prior assumption of nearest-neighbor continuity was used, resulting in smoothed expression patterns over the voxels.
WEB SITE REFERENCES
http://www.nervenet.org/MBL/mbl.html; Mouse Brain Library.
http://www.ncbi.nlm.nih.gov/geo; Microarray data under the series accession no. GSE30.
http://www.ncbi.nlm.nih.gov/UniGene; UniGene.
http://www.pharmacology.ucla.edu/smithlab/genome_multiplex; All study results and supplementary material.
Acknowledgments
We thank Harvey Herschman for comments on the manuscript. This work was supported by grants from the Dana Foundation, Merck Genome Research Institute, W.M. Keck Foundation, National Foundation for Functional Brain Imaging, NIH (DA015802-01), NSF, Staglin Music Festival, NARSAD Young Investigator Award, and the UCLA School of Medicine.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL DSmith@mednet.ucla.edu; FAX (310) 825-6267.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.229002. Article published online before print in May 2002.
REFERENCES
- Akhmedova SN, Yakimovsky AK, Schwartz EI. Paraoxonase 1 Met–Leu 54 polymorphism is associated with Parkinson's disease. J Neurol Sci. 2001;184:179–182. doi: 10.1016/s0022-510x(01)00439-7. [DOI] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banati RB, Daniel SE, Blunt SB. Glial pathology but absence of apoptotic nigral neurons in long-standing Parkinson's disease. Mov Disord. 1998;13:221–227. doi: 10.1002/mds.870130205. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Brown VM, Ossadtchi A, Khan AH, Cherry SR, Leahy RM, Smith DJ. High-throughput imaging of brain gene expression. Genome Res. 2002a;12:244–254. doi: 10.1101/gr.204102. [DOI] [PubMed] [Google Scholar]
- Brown VM, Ossadtchi A, Khan AH, Gambhir SS, Cherry SR, Leahy RM, Smith DJ. Gene expression tomography. Physiol Genomics. 2002b;8:159–167. doi: 10.1152/physiolgenomics.00090.2001. [DOI] [PubMed] [Google Scholar]
- Chan SO, Chiu FC. The 66-kDa neurofilament protein (NF-66): Sequence analysis and evolution. Neurochem Res. 1996;21:449–455. doi: 10.1007/BF02527709. [DOI] [PubMed] [Google Scholar]
- Cox RD, Hugill A, Shedlovsky A, Noveroske JK, Best S, Justice MJ, Lehrach H, Dove WF. Contrasting effects of ENU induced embryonic lethal mutations of the quaking gene. Genomics. 1999;57:333–341. doi: 10.1006/geno.1999.5804. [DOI] [PubMed] [Google Scholar]
- Dluzen DE, Kreutzberg JD. Norepinephrine is lateralized within the olfactory bulbs of male mice. J Neurochem. 1996;66:1222–1226. doi: 10.1046/j.1471-4159.1996.66031222.x. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Filippi M, Tribioli C, Toniolo D. Linkage and sequence conservation of the X-linked genes DXS253E (P3) and DXS254E (GdX) in mouse and man. Genomics. 1990;7:453–457. doi: 10.1016/0888-7543(90)90184-v. [DOI] [PubMed] [Google Scholar]
- Frackowiak RSJ, Friston KJ, Frith CD, Dolan RJ, Mazziotta JC. Human brain function. London UK: Academic Press Ltd.; 1997. [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Gambhir SS, Barrio JR, Herschman HR, Phelps ME. Assays for noninvasive imaging of reporter gene expression. Nucl Med Biol. 1999;26:481–490. doi: 10.1016/s0969-8051(99)00021-9. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Goedert M, Satumtira S, Jakes R, Smith MJ, Kamibayashi C, White CL, III, Sontag E. Reduced binding of protein phosphatase 2A to tau protein with frontotemporal dementia and parkinsonism linked to chromosome 17 mutations. J Neurochem. 2000;75:2155–2162. doi: 10.1046/j.1471-4159.2000.0752155.x. [DOI] [PubMed] [Google Scholar]
- Hendler RW, Shrager RI. Deconvolutions based on singular value decomposition and the pseudoinverse: A guide for beginners. J Biochem Biophys Methods. 1994;28:1–33. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Hwang CK, D'Souza UM, Eisch AJ, Yajima S, Lammers CH, Yang Y, Lee SH, Kim YM, Nestler EJ, Mouradian MM. Dopamine receptor regulating factor, DRRF: A zinc finger transcription factor. Proc Natl Acad Sci. 2001;98:7558–7563. doi: 10.1073/pnas.121635798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: Mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Kavety B, Morgan JI. Characterization of transcript processing of the gene encoding precerebellin-1. Brain Res Mol Brain Res. 1998;63:98–104. doi: 10.1016/s0169-328x(98)00264-2. [DOI] [PubMed] [Google Scholar]
- Kösel S, Egensperger R, von Eitzen U, Mehraein P, Graeber MB. On the question of apoptosis in the parkinsonian substantia nigra. Acta Neuropathol. 1997;93:105–108. doi: 10.1007/s004010050590. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Barlow C. Expressing what's on your mind: DNA arrays and the brain. Nat Rev Neurosci. 2001;2:63–68. doi: 10.1038/35049070. [DOI] [PubMed] [Google Scholar]
- Louie AY, Hüber MM, Ahrens ET, Rothbächer U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- Melega WP, Raleigh MJ, Stout DB, Lacan G, Huang SC, Phelps ME. Recovery of striatal dopamine function after acute amphetamine- and methamphetamine-induced neurotoxicity in the vervet monkey. Brain Res. 1997;766:113–120. doi: 10.1016/s0006-8993(97)00548-9. [DOI] [PubMed] [Google Scholar]
- Min N, Joh TH, Kim KS, Peng C, Son JH. 5′ upstream DNA sequence of the rat tyrosine hydroxylase gene directs high-level and tissue-specific expression to catecholaminergic neurons in the central nervous system of transgenic mice. Brain Res Mol Brain Res. 1994;27:281–289. doi: 10.1016/0169-328x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Myers MW, Lazzarini RA, Lee VM, Schlaepfer WW, Nelson DL. The human mid-size neurofilament subunit: A repeated protein sequence and the relationship of its gene to the intermediate filament gene family. EMBO J. 1987;6:1617–1626. doi: 10.1002/j.1460-2075.1987.tb02409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulina EM, Skrinskaya JA, Avgustinovich DF, Popova NK. Dopaminergic brain system in the quaking mutant mouse. Pharmacol Biochem Behav. 1995;50:333–337. doi: 10.1016/0091-3057(94)00267-m. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Cardno AG, O'Donovan MC. Psychiatric genetics: Back to the future. Mol Psych. 2000;5:22–31. doi: 10.1038/sj.mp.4000702. [DOI] [PubMed] [Google Scholar]
- Peterson AS. Pixelating the brain. Genome Res. 2002;12:217–218. doi: 10.1101/gr.227802. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Williams AG, Capra JA, Connolly MT, Cruz B, Lu L, Airey DC, Kulkarni K, Williams RW. The Mouse Brain Library@www.mbl.org. Intl Mouse Genome Conf. 2000;14:166. [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, Mayford M, Lockhart DJ, Barlow C. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonsalla PK, Jochnowitz ND, Zeevalk GD, Oostveen JA, Hall ED. Treatment of mice with methamphetamine produces cell loss in the substantia nigra. Brain Res. 1996;738:172–175. doi: 10.1016/0006-8993(96)00995-x. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Van Swieten JC, Goedert M. Tau gene mutations in frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) Neurogenetics. 2000;2:193–205. doi: 10.1007/pl00022972. [DOI] [PubMed] [Google Scholar]
- Tabibnia G, Cooke BM, Breedlove SM. Sex difference and laterality in the volume of mouse dentate gyrus granule cell layer. Brain Res. 1999;827:41–45. doi: 10.1016/s0006-8993(99)01262-7. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, Goedert M, Iwatsubo T, Lee VM. Fatal attractions: Abnormal protein aggregation and neuron death in Parkinson's disease and Lewy body dementia. Cell Death Differ. 1998;5:832–837. doi: 10.1038/sj.cdd.4400432. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Williams RW. Mapping genes that modulate mouse brain development: A quantitative genetic approach. In: Goffinet AF, Rakic P, editors. Mouse brain development. New York: Springer; 2000. pp. 21–49. [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Prüss M, Reuter I, Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüllner U, Kornhuber J, Weller M, Schulz JB, Löschmann PA, Riederer P, Klockgether T. Cell death and apoptosis regulating proteins in Parkinson&rsqup;s disease—A cautionary note. Acta Neuropathol. 1999;97:408–412. doi: 10.1007/s004010051005. [DOI] [PubMed] [Google Scholar]
- Xu ZH, Otterness DM, Freimuth RR, Carlini EJ, Wood TC, Mitchell S, Moon E, Kim UJ, Xu JP, Siciliano MJ, et al. Human 3′-phosphoadenosine 5′-phosphosulfate synthetase 1 (PAPSS1) and PAPSS2: Gene cloning, characterization and chromosomal localization. Biochem Biophys Res Commun. 2000;268:437–444. doi: 10.1006/bbrc.2000.2123. [DOI] [PubMed] [Google Scholar]
- Yajima S, Lee SH, Minowa T, Mouradian MM. Sp family transcription factors regulate expression of rat D2 dopamine receptor gene. DNA Cell Biol. 1998;17:471–479. doi: 10.1089/dna.1998.17.471. [DOI] [PubMed] [Google Scholar]
- Yaworsky PJ, Gardner DP, Kappen C. Transgenic analyses reveal developmentally regulated neuron- and muscle-specific elements in the murine neurofilament light chain gene promoter. J Biol Chem. 1997;272:25112–25120. doi: 10.1074/jbc.272.40.25112. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Baird GS, Tsien RY. Recent advances in technology for measuring and manipulating cell signals. Curr Opin Neurobiol. 2000;10:416–421. doi: 10.1016/s0959-4388(00)00101-x. [DOI] [PubMed] [Google Scholar]