Abstract
Spleen tyrosine kinase (SYK) is a candidate tumor suppressor gene in breast. Loss of SYK expression in breast tumors as a result of DNA hypermethylation promotes tumor cell proliferation and invasion and predicts shorter survival of breast cancer patients. We previously reported that, in addition to its well-known cytoplasmic localization, the full length Syk is also present in the nucleus, and that Syk nuclear translocation is a rate-limiting step to determine Syk tumor suppressor function. Here we show that the full-length form of Syk acts as a transcription repressor in the cell nucleus. Ectopic expression of Syk down-regulates the transcription of FRA1 and cyclin D1 oncogenes. This transcription repressing activity of Syk is associated with its binding to members of the histone deacetylase family. Syk interacts with transcription factor Sp1 at the Sp1 DNA-binding site in the FRA1 promoter to repress Sp1-activated FRA1 transcription. Thus, breast tumorigenesis and progression resulting from the loss of SYK are underscored by the de-repression of Sp1-mediated oncogene transcription.
Keywords: spleen tyrosine kinase, transcription repression; Sp1; breast tumors; cyclin D1; Fra1; tumor suppressor
The de-regulation of gene transcription has a central role in the neoplastic transformation. Signaling proteins that alter gene transcription patterns are frequently targeted in tumors (1). This is exemplified by that some of the most prominent tumor suppressor genes (such as p53, SMADs, WT-1, and RB) and oncogenes (such as myc, β-catenin, STATs, AIB1, and AP1) are themselves transcription factors, co-factors, or regulators. Moreover, some non-transcription factor molecules that participate within pathways that ultimately feed to gene transcription regulation are also prone to tumorigenic alterations. For example, biallelic inactivation of APC gene in colorectal tumors leads to aberrant accumulation of β-catenin to activate downstream oncogene expression. The deregulated gene transcription resulting from these alterations is thought to contribute to tumor initiation and progression.
Spleen tyrosine kinase (SYK), a candidate tumor suppressor gene, is expressed in normal mammary epithelial cells, but is lost in >30% of breast tumors as a result of DNA hypermethylation in the SYK promoter region (2, 3). This epigenetic inactivation of SYK suggests an important role as a suppressor of breast cancer progression. Indeed, among breast cancer patients, the loss of Syk expression has been correlated with shorter survival (4, 5); overexpression of the full-length Syk [a.k.a. Syk(L)] leads to decreased proliferation, invasiveness, and mobility of breast cancer cells (6-8). These findings agree with earlier observations of a progressive decrease of SYK mRNA from normal mammary glands, to ductal carcinoma in situ, and to invasive breast carcinoma (3). Biochemically, expression of Syk(L) has been shown to lower epidermal growth factor-initiated signaling and phosphatidyliositol-3 kinase activity in mammary epithelial cells (8, 9). Whether these effects are directly related to the suppression response remains to be determined.
The role of SYK as a tumor suppressor gene is unusual in that most tyrosine kinases have instead oncogenic activity, suggesting some of the Syk biological functions is separate from its catalytic activity. In examining Syk expression in breast tumors and cell lines, we previously found a SYK variant in breast cancer cells that is created by alternative RNA splicing (6). This tumor-specific exon-skipping generates a Syk isoform, Syk(S), with an in-frame deletion of 23 residues within interdomain B (IDB) of Syk. Syk(S) differs from Syk(L) in biological activity despite their striking structural similarity. Syk(L) suppresses breast cancer invasion, while Syk(S) lacks this activity. Coincident with their differing phenotypic responses, the two Syk isoforms have different subcellular distributions. Syk(L) is present in both the cytoplasm and nucleus, while Syk(S) is localized exclusively in the cytoplasm (6, 10). Five basic amino acids within the deleted 23 residues act as an autonomous nuclear localization signal required for Syk(L) nuclear translocation. The correlation of anti-invasive action and nuclear presence of Syk(L) suggests that nuclear translocation may be a rate-limiting step in the Syk(L) biological makeup. Consistent with this notion, cells expressing nuclear translocation-deficient Syk(L) mutants do not exhibit anti-invasion phenotypes (6). These findings indicate that the nuclear activity of Syk(L) is indispensable for its tumor suppression function.
For a given protein, one potential action in the nucleus is to regulate gene transcription. In the present study, we examined the possible effect of Syk(L) on gene transcription and looked for its downstream targets.
Material and Methods
DNA Constructs
The cDNAs encoding the full-length human SYK(L) (residues 1-635), the two SH2 domains (SH2D; residues 1-259), IDB(L) (residues 259-395), and kinase domain (KD; residues 395-635) were subcloned in frame into pFA-CMV (Stratagene) to create pFA-SYK(L) (HindIII/KpnI), pFA-SH2D (BamHI/EcoRI), pFA-IDB(L) (HindIII/KpnI), and pFA-KD (BamHI/EcoRI), respectively (Fig. 1A). The cDNA encoding SYK(L) was subcloned in frame into pFlag-CMV-6a (BamHI/NotI; Sigma) to create pFlag-SYK(L). The cDNA fragments encoding SH2D, IDB(L), and KD were PCR-amplified and constructed into pFlag-CMV-6c (Sigma). To prepare the kinase-inactive Syk mutants, site-directed mutagenesis (QuickChange; Stratagene) was used to create the K402R mutation in Syk(L), using pFlag-SYK(L) and pFA-SYK(L) as templates. To prepare the GST-Syk(L) fusion protein, SYK(L) cDNA was subcloned in frame into pGEX4T2 (SmaI; GE Healthcare). The complete reading frame of all constructs generated by PCR was verified by automated sequencing.
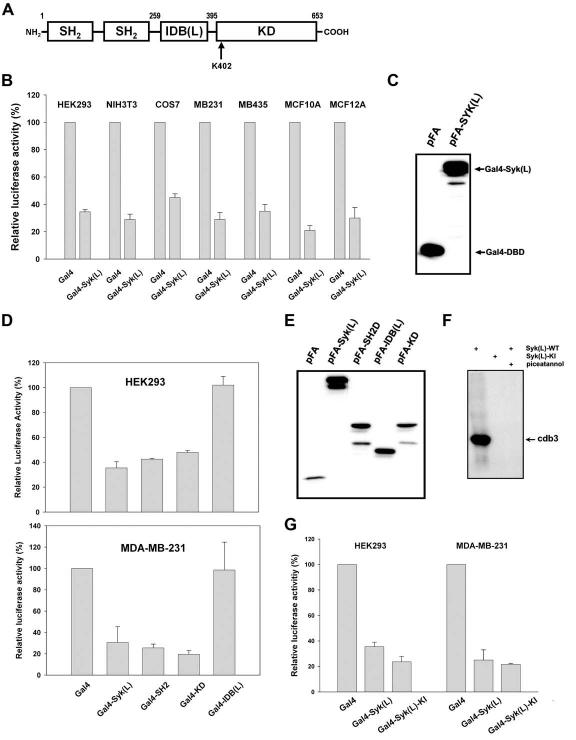
Figure 1.
Transcription repressor activity of Syk(L). A, The functional domains of the 653-amino-acid human Syk(L) protein are represented. K402 indicates the residue critical for the catalytic activity of Syk(L). B, HEK293, NIH3T3, MDA-MB-231, MDA-MB-435S, MCF10A, and MCF12A cells were transfected with pFA-SYK(L) or parental vector pFA-CMV, in combination with pRF-Luc. pCMVβ was included to normalize transfection efficiency. Gal4 luciferase activity was set at 100%. All reporter data were obtained from at least three independent experiments and are presented as means ± SE. C, Expression of Gal4-Syk(L) fusion proteins in HEK293 cells transfected with pFA or pFA-SYK(L) as detected by immunoblotting with an antibody against the Gal4 DNA-binding domain. D, Cells were transfected with pRF-Luc in combination with Gal4 constructs as described in (A). Luciferase activity of pFA was set at 100%. E, Immunodetection of Gal4 fusion proteins in HEK293 cells. F, Inactivation of kinase activity by K402R mutation of Syk. The wild-type (WT) or mutant (KI) pFlag-SYK(L) expression vectors were transfected into COS7 cells. Flag-immunoprecipitated products were incubated with cdb3 in an in vitro kinase reaction in the presence of [γ-32P]ATP. Piceatannol (100 nM) was used to control the specificity of kinase reactions. G, HEK293 and MDA-MB-231 cells were transfected with pRF-Luc and pCMVβ, together with pFA, pFA-SYK(L), or pFA-SYK(L)-KI. Reporter activity was determined as described in (A).
Cell Culture and Stable Cell Lines
All cells were purchased from American Type Culture Collection and cultured in the recommended media. To prepare Syk(L)-expressing stable cell lines, we co-transfected human MDA-MB-231 breast cancer cells with pFlag-SYK(L) and pcDNA3.1 (Invitrogen) using Fugene 6 (Roche Applied Sciences) and selected with G418 (800 μg/ml; Invitrogen). Positive Flag-Syk(L) stable clones were screened by immunoblotting using anti-Flag (M2; Sigma) and anti-Syk (4D10; Santa Cruz Biotechnology) antibodies.
Thymidine Incorporation, Chemoinvasion, and Reporter Assays
Cell proliferation rates were measured by [3H]thymidine incorporation as described previously (11). Chemoinvasion assays were performed using Boyden chambers coated with Matrigel as described previously (6).
For Gal4 reporter experiments, cells cultured in six-well dishes received 0.5 μg of pFA-CMV constructs [Syk(L), SH2D, IDB(L), or KD], 0.5 μg of RF-Luc (Stratagene), and 0.2 μg of pCMVβ. pRF-Luc that contains five copies of Gal4-response element was used as a Gal4 transactivation readout.
For the promoter response tests, Flag-Syk(L) stable and Neo control cells each received 1.5 μg of pGL3-Basic (Promega) constructs fused with FRA1 promoter regions. Forty-eight hours after transfection, cells were collected and assayed for luciferase and β-galactosidase activity.
GST Pull-Down Assays
GST-Syk(L) was purified from pGEX4T2-SYK(L) transformed E. coli using glutathione-Sepharose beads (GE Healthcare), and quantified by SDS-PAGE and Coomassie Blue staining. Radiolabeled histone deacetylases (HDACs) were synthesized by in vitro TNT reaction (Promega) from the cDNAs encoding HDACs 1-7 in the presence of [35S]methionine (Perkin Elmer). Pull-down assays were performed by incubating the TNT products with the GST beads in the presence of 20 mM Tris (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 1 mM DTT. After extensive washes, the bound proteins were eluted by boiling in 1X SDS sample buffer, separated by SDS-PAGE, and exposed for autoradiography. The HDAC cDNAs were gifts from Drs. Ronald Evans (Salk Institute, San Diego, CA), Tony Kouzarides (Wellcome Cancer Research UK Gurdon Institute, Cambridge, UK), and Jiemin Wong (Baylor College of Medicine, Houston, TX)
Adenovirus
Recombinant adenovirus expression Syk(L) or Syk(S) were prepared as described previously (11). MDA-MB-231 cells were infected for 48 h with Adeno-Syk(L), Adeno-Syk(S), or Adeno-LacZ at a multiplicity of infection of 20.
Northern Blotting
Total RNA was extracted from cells using TRIzol (MRC). Twenty micrograms of total RNA was fractionated on a 1% formaldehyde-agarose gel, transferred onto membranes, and detected by hybridization with probe that was prepared by random-primer labeling with [α-32P]dCTP (Perkin Elmer) using PCR products as templates. Gene-specific PCR products were amplified from I.M.A.G.E. clones (Invitrogen). To control for loading variability, the membrane was stripped and rehybridized with a human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe.
Microarray Analyses
DNase-digested total RNA was converted to cDNA and labeled with Cy3 or Cy5 dye. Labeled cDNAs (100 μg) were hybridized with an in-house CG11 pathway array as described previously (12). The gene list, which is available at http://www3.mdanderson.org/~genomics/, includes 1126 genes with defined tumor-related activities. All test genes on chips were spotted in duplicate. The slides were washed and scanned with a GeneTAC UC 4 laser scanner (Genomic Solutions), and the signal intensities were quantified with ArrayVision (Imaging Research) and defined as spot density minus background density divided by the SD of the background density. Hybridization procedures were carried out in the Genomics Core Facility, and the array data were analyzed by the Biostatistics and Data Management Bioinformatics Core Facility at the M. D. Anderson Cancer Center.
Immunoprecipitation and immunoblotting
For SYK(L) and HDAC1 co-transfection experiments, HEK293 cells were co-transfected with pCMX-hHDAC1-HA (from Dr. Ronald Evans) and pFlag-SYK(L) [or pFlag-SH2D, pFlag-IDB(L), and pFlag-KD]. Cells were then lysed in 1X RIPA buffer, and the soluble components were subjected to immunoprecipitation using agarose-conjugated anti-Flag antibody (M2 affinity gel; Sigma). The immunocomplex was separated by SDS-PAGE, and detected by anti-HA antibody (Y11; Santa Cruz).
To detect Syk interaction with HDAC1 at the endogenous level, cultured MDA-MB-468 cells were lysed with 1X RIPA buffer. The cleared lysates were immunoprecipitated with anti-HDAC1 antibody (Cell Signaling), using an equivalent amount of normal rabbit IgG as a control. The immunocomplex was collected by addition of protein A/G plus agarose and detected with anti-Syk antibody (4D10).
To measure binding of transfected Syk to endogenous HDACs, HEK293 cells were transfected with pFlag-SYK(L) or control pFlag-BAP (Sigma) plasmids. Cells were lysed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton-X 100, and 1X protease inhibitor cocktail (Sigma). The Flag-tagged proteins were immunoprecipitated by M2 affinity gel, and the immunocomplex was analyzed with antibodies against HDACs 1, 3, 4, 5, 6, and 7 (Cell Signaling). Similar experiments were performed to measure the binding between transfected Syk(L) and endogenous Sp1. Sp1 in the immunocomplex was analyzed by an anti-Sp1 antibody (PEP2; Santa Cruz).
Additional antibodies used were against Gal4 DNA-binding domain (RK5C1) and Syk (N19; Santa Cruz). Immunodetection was done using horseradish peroxidase-conjugated secondary antibody and Supersignal reagents (Pierce).
Measurement of HDAC
HEK293 cells transfected with pFlag-SYK(L) or pCMV-FLAG were lysed and subjected to M2-immunoprecipitation. The beads were washed extensively with the lysis buffer, once with HDAC reaction buffer, and then assayed for deacetylase activity using a fluorescence-based HDAC testing kit (BioMol Research Laboratories). The beads were incubated with 200 μM acetylased substrate in 50 μl of assay buffer, in the presence or absence of 5 μM trichostatin A (TSA). The reaction was incubated at 25°C for 90 min to allow deacetylation of the substrate. The developer for the deaceylated substrate was then added to produce a fluorophore detectable on a fluorometric reader (excitation at 360 nm and emission at 450 nm). The TSA-containing reaction was used to assess the non-HDAC background signal. The difference in signal intensity between the TSA-negative and TSA-positive reactions reflects specific HDAC activity.
In Vitro Kinase Assay
Wild-type or mutant (K402R) pFlag-SYK(L) expression vectors were transfected into COS7 cells. The M2-immunoprecipitated product was incubated for 30 min at room temperature with 2 μg of purified recombinant cdb3 (from Dr. Philip Low, Purdue University, West Lafayette, IN) in a 20-μl in vitro kinase reaction in the presence of 20 mM Tris (pH 7.5), 10 mM MnCl2, 10 mM MgCl2, 1 μM ATP, and 5 μCi [γ-32P]ATP (Perkin Elmer). The reaction supernatant was run on SDS-PAGE and subjected to autoradiography. A Syk inhibitor, piceatannol (100 nM; Sigma), was also used as a control for reaction specificity.
Chromatin Immunoprecipitation
A commercial chromatin immunoprecipitation (ChIP) assay kit (Active Motif) was used to analyze the in vivo binding of Syk(L) to the FRA1 promoter region, using MDA-MB-231/Flag-Syk(L) stable cells. Fragmented chromatin was immunoprecipitated by overnight incubation at 4°C with protein G beads and 3 μg of either anti-Syk (4D10), anti-Flag (M2), anti-TFIIB antibodies, or mouse IgG control. Recovered DNA was analyzed by semi-quantitative PCR. The primers for FRA1 were: set 1, 5′-GGCAGGAGAATCCCTTTAGC-3′ (forward) and 5″-TCTTTCCACTGGCCTTGTTT-3′ (reverse); and set 2, 5′-GGAAAGACCTCACTCCACGA-3′ (forward) and 5′-CTGACGTAGCTGCCCATACA-3′ (reverse). The control primers 5′-ATGGTTGCCACTGGGGATCT-3′ (forward) and 5′-TGCCAAAGCCTAGGGGAAGA-3′ (reverse) were used to normalize the DNA output of ChIP.
Electrophoretic Mobility Shift Assays
Approximately 4 μg of K562 nuclear extract (Active Motif) was incubated for 30 min at 4°C in binding buffer [10% glycerol, 5 mM MgCl2, 35 mM KCl, 80 mM NaCl, 1 mM DTT, 25 mM Tris–HCl (pH 7.5), 1% NP-40, and 50 μg/ml poly(dI–dC)] in a 20-μl reaction. Radiolabeled probe (20,000 cpm) was subsequently added to each reaction and incubated at room temperature for an additional 20 min. DNA-protein complexes were resolved on 4% native Tis-borate-EDTA gels that were exposed for autoradiography. For competition gel shift assays, 10- or 100-fold excess unlabeled fragment was included in reaction before the probe was added. For supershift, 1 μg of antibodies or normal IgG was incubated with nuclear extracts for 30 min at 4°C prior to probe addition. The 100-bp FRA1 probe was released from the pGL3 vector by digestion with Acc65I and HindIII and labeled by end fill-in. For experiments using the recombinant proteins, each reaction included 25 ng of recombinant Sp1 (Active Motif) or 150 ng of GST or GST-Syk(L).
Results
Transcriptional Repression Activity of Syk(L)
A Gal4-based transcription model was used to examine a potential effect of Syk(L) on gene transcription. The full-length Syk(L) was C-terminally fused in frame to the Gal4 DNA-binding domain (1-147) that is expressed under the control of a CMV promoter (pFA-CMV). This construct was transfected, together with the pRF-Luc reporter construct, into cell lines of various origins: HEK293 (human kidney cells), NIH3T3 (mouse fibroblasts), COS7 (monkey kidney cells), MDA-MB-435S, MDA-MB-231 (human breast cancer cells), MCF10A, and MCF12A (immortalized non-tumoirgenic human mammary epithelial cells). In all of the 7 cell lines tested, Gal4-Syk(L) fusion resulted in lower reporter activity than that of Gal4-DBD (Fig. 1B). The Gal4-Syk(L) fusion protein was expressed at a comparable level (Fig. 1C), indicating that the decrease in reporter activity may represent an intrinsic repressor activity of Syk(L). Notably, Syk(L) repressor activity was observed in immortalized normal mammary epithelial cells, suggesting that the deregulation of gene transcription resulting from the loss of Syk may be important in mammary tumorigenesis.
We used this reporter model to further determine which Syk domain(s) is responsible for the repression activity. Syk(L) has three major domains: two tandem SH2 domains (SH2D; residues 1-259) at the amino-terminus, the kinase domain (KD, residues 395-635) at the carboxyl-terminus, and IDB(L) (residues 259-395) in between. These domains, which cover the entire Syk sequence, were analyzed in MDA-MB-231 and HEK293 cells. Both SH2D and the KD had a repressor activity similar to that of Syk(L) (Fig. 1D), despite the comparable expression level of all Gal4 fusion proteins (Fig. 1E). In contrast, IDB(L), a domain shown to be responsible for Syk(L) nuclear localization, did not show repressor function (Fig. 1D). Thus, Syk(L) contains two distinct domains, SH2D and KD, that mediate the intrinsic repressor function of Syk(L).
Tyrosine kinase activity has been shown to be important for a variety of Syk signaling events (13), although evidence from certain experimental settings indicates that this kinase activity is not crucial for some of the downstream effects of Syk (14). To determine whether kinase activity is required for the transcription repression function of Syk(L), we tested the repressor activity of a kinase-inactive Syk mutant (K402R) in the Gal4 model. The Lys402 residue in the KD is a crucial ATP-binding site. As expected, replacing that residue with an arginine rendered the tyrosine kinase inert, as determined by in vitro kinase assay of cdb3, a classic Syk substrate (Fig. 1F). Transfection experiments in MDA-MB-231 and HEK293 cells showed that the KI mutant suppressed the Gal4 reporter at a level similar to that of the corresponding wild-type Syk (Fig. 1G), suggesting that the catalytic kinase activity is dispensable for the intrinsic transcription repression function of Syk(L).
Interaction of Syk(L) with HDACs
Many transcription repressors or co-repressors exert their functions by modulating chromatin configuration, which is largely determined by histone acetylation status (15). We tested the hypothesis that Syk(L) and HDACs are physically associated. We transfected HEK293 cells with pFlag-Syk(L) to determine whether the Flag-tagged Syk(L) forms complexes with the endogenous HDACs. Immunoprecipitation of cell lysates with anti-Flag antibody and immunoblotting of the precipitates with antibodies to HDACs 1 and 3-7 revealed an association between Syk(L) and HDACs 1, 3, 6, and 7 (Fig. 2A). HDACs 4 and 5, however, were not detectable (data not shown). We further examined whether Syk(L) bound HDACs at the endogenous expression level. A breast cancer cell line MDA-MB-468, which expresses high levels of HDAC1 and Syk, was used for this experiment. Co-immunoprecipitation experiments identified a significant interaction between the two proteins (Fig. 2B), demonstrating that Syk(L) associates with HDAC1 in vivo at the endogenous level.
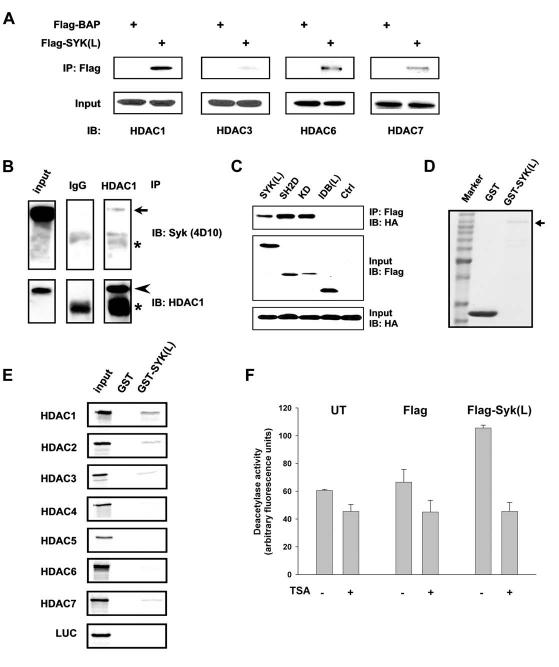
Figure 2.
In vivo and in vitro interactions of Syk(L) with HDACs. A, HEK293 cells were transfected with pFlag-SYK(L) or pFlag-BAP (bovine alkaline phosphatase; control). Two days following transfection, cells were lysed and subjected to Flag-immunoprecipitation (M2). The precipitated proteins were then probed with antibodies against HDACs 1, 3, 6, and 7. B, MDA-MB-468 cells were lysed followed by immunoprecipitation using an anti-HDAC1 antibody or rabbit IgG. The Syk protein (arrow) was detected in the precipitated materials by immunoblotting. Precipitation of HDAC1 (arrowhead) was verified by re-probing with the anti-HDAC1 antibody. Asterisks indicate the heavy chain of IgG. Input lysates were run side by side. C, pCMX-hHDAC1-HA was transfected into HEK293 cells, together with pFlag fusion proteins (pFlag-CMV was used as a control). The M2-precipitated products were measured for HA-immunoreactive substances by blotting. Input lysates were also analyzed for HA-HDAC1 and Flag-Syk expression by immunoblotting. D, Coomassie Blue staining of GST or GST-Syk(L) (arrow) fusion proteins. E, GST pull-down assays. HDACs 1-7 or an unrelated luciferase (LUC) protein were synthesized by TNT reactions in the presence of [35S]methionine. The labeled proteins were then incubated with immobilized GST or GST-Syk(L). Components bound to GST fusion were separated by SDS-PAGE and detected by autoradiography. Ten percent of the total input was run side by side to show relative binding capacity. F, HEK293 cells were transfected with pFlag-CMV (Flag), pFlag-SYK(L) [Syk(L)], or left untransfected (UT). Cell lysates were then M2-immunoprecipitated and used for HDAC activity assays. The TSA-containing reaction (5 μM) was used to determine the background fluorescence generated by non-HDAC signals. Data are means ± SE of four separate experiments.
We next investigated which Syk(L) domain is responsible for Syk(L)'s interaction with HDACs. We transfected HEK293 cells with HA-tagged HDAC1 and Flag-tagged SH2D, IDB(L), KD, or full-length Syk(L). Co-immunoprecipitation showed that, similar to the wild-type Syk(L), SH2D and KD but not IDB(L) co-immunoprecipitated with HDAC1 (Fig. 2C). These results are consistent with the intrinsic repressor activity of SH2D and KD observed in the Gal4 model (Fig. 1D), substantiating the functional relevance of Syk(L)'s binding to HDACs to transcription repression.
Transcription repressors function by forming complexes with HDAC, by either direct binding or recruitment of co-repressors (16). We used a GST pull-down assay to analyze whether Syk(L) directly interacts with HDAC. A GST-Syk(L) fusion protein attached to glutathionine beads (Fig. 2D) was incubated with radio-labeled HDACs. While HDAC5 did not appear to interact with GST-Syk(L), HDACs 1-4, 6, and 7 showed significant binding to GST-Syk(L) (Fig. 2E). The binding was specific, as the interaction did not occur with the GST control or an unrelated luciferase protein. These experiments using purified recombinant protein indicated that Syk(L) is likely to interact with HDACs directly.
The physical interaction of Syk(L) with HDACs was also reflected by Syk(L)'s association with active HDAC. HEK293 cells were transfected with pFlag-SYK(L) or the control vector, and the Flag-immunoprecipitated products were assayed for HDAC activity. Syk(L) was associated with high levels of HDAC activity (TSA-sensitive), which was marginal in the mock- or vector-transfected cells (Fig. 2F). This Syk(L)-associated HDAC activity is likely to be instrumental in the transcription repression property of Syk(L).
Downstream Genes Regulated by Syk(L)
Since nuclear translocation is crucial to the tumor suppressor function of Syk(L), we surmised that genes transcriptionally repressed by Syk(L), but not by Syk(S), are candidates responsible for Syk(L)-inducible invasion and growth suppression.
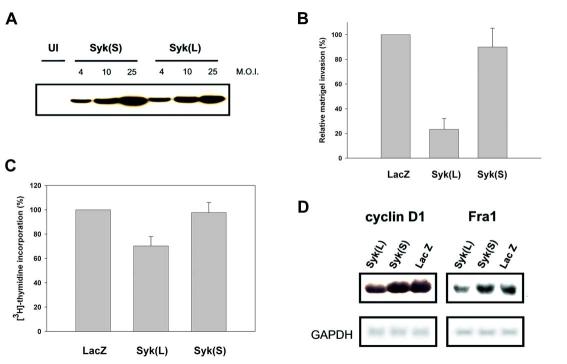
DNA microarrays were used to uncover changes in global gene expression profiles following Syk expression in the Syk-negative breast cancer cell line, MDA-MB-231. The epigenetic abrogation of Syk function in these cells (2) predicts that restoration of Syk(L) would suppress their aggressiveness. In agreement with our observations in MDA-MB-435S stable lines, we found that expression of Syk(L) brought by adenoviral delivery (Fig. 3A) led to lowered invasion (Fig. 3B). In addition, we found that Syk(L) expression led to modest but consistent inhibition of cell growth (Fig. 3C). Syk(S), however, had neither such activity, likely due to its lack of nuclear translocation property (6). To confirm that these gene expression changes were not artifacts caused by adenoviral infection other than Syk, we used Adeno-LacZ as a control to compare the relative transcript levels in Adeno-Syk- and Adeno-LacZ-infected cells. cDNA was synthesized and labeled from infected cells. Competitive hybridization was performed on an in-house CG11 oligo DNA array on which 1126 essential tumor-associated genes were spotted in duplicate, including those related to adhesion (12 genes), angiogenesis (129 genes), apoptosis (37 genes), cell cycle (250 genes), DNA repair (104 genes), immune modulation (13 genes), invasion (17 genes), metastasis (230 genes), proliferation (34 genes), tumor suppression (198 genes), and other cellular functions (102 genes), together with positive (48 spots), negative (48 spots), and blank (24 spots) controls. We reasoned that genes activated by Syk(L), but not by Syk(S), might be responsible for Syk(L)-inducible invasion suppression. We found two genes, CCND1 (a.k.a. cyclin D1) and FOSL1 (a.k.a. FRA1) that were down-regulated by the expression of Syk(L) but were not affected by Syk(S). These observations were verified by northern blotting (Fig. 3D).
Figure. 3.
Expression of Syk(L), but not Syk(S), suppresses breast cancer cell invasiveness. A, Immunoblot detection of adenovirally delivered Syk in MDA-MB-231 cells. MOI, multiplicity of infection. B, Inhibition of cell invasion by adenovirally transduced Syk(L), but not Syk(S), as measured by chemoinvasion assays in MDA-MB-231 cells. The number of cells that invaded through Matrigel was compared to that in the LacZ group (arbitrarily set as 100%). Data are means ± SE from three experiments. C, Proliferation rate of MDA-MB-231 cells, as measured by [3H]thymidine incorporation, was inhibited by Syk(L) but not by Syk(S). D, Down-regulation of cyclin D1 and FRA1 mRNA by expression of Syk(L), but not Syk(S). Total RNA was extracted from MDA-MB-231 cells infected with Adeno-SYK(L), Adeno-SYK(S), or Adeno-LacZ. Northern blotting was done with random primer-labeled gene-specific cDNA products. The membrane was stripped and re-hybridized with GAPDH probe.
Regulation of target gene promoters by Syk(L)
Syk(L) might affect its target genes by binding to their DNA regulatory sequences. We carried out a ChIP assay to determine whether Syk(L) interacts in vivo with the proximal 5′-regulatory sequences of FRA1 and CCND1, using MDA-MB-231 cells stably expressing Flag-Syk(L). The FRA1 promoter region was immunoprecipitated by either anti-Flag or anti-Syk(L) antibody, but not by the mouse IgG control (Fig. 4A), implying that Syk(L) exerts its repressor function through interaction with the FRA1 gene promoter. Binding of Syk(L) to the proximal CCND1 promoter sequence, however, was not observed (data not shown), suggesting that down-regulation of CCND1 is indirect or that Syk(L) interacts with distal regulatory sequences.
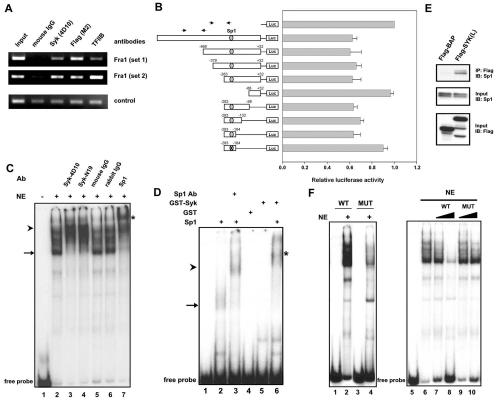
Figure. 4.
Interaction of Syk(L) with Sp1 at the Sp1-binding sequence of the FRA1 promoter. A, ChIP assays were performed using MDA-MB-231/Flag-Syk(L) stable cells. Two sets of FRA1 primers were used for PCR amplification from chromatin fragments immunoprecipitated by anti-Syk (4D10) and anti-Flag (M2) antibodies, mouse IgG (negative control), or anti-TFIIB antibody (positive control). Input represents PCR amplification of total chromatin before immunoprecipitation. A primer set of an unrelated region was used to normalize the amount of chromatin in each PCR reaction (bottom panel). B, MDA-MB-231/Flag-Syk(L) or its Neo control stable cells were transfected with the indicated reporter constructs. Bars represent means luciferase activity (± SE) relative to parental pGL3-basic (set at 100%). All luciferase measurements are from at least three independent experiments and normalized with activity in the Neo line. The relative positions of the putative Sp1 site and PCR primers (arrows) used for ChIP are shown. C, EMSA. 32P-labeled, 100-bp, Syk(L)-responsive fragment of the FRA1 promoter region was incubated with K562 cell nuclear extracts (NE) alone or in combination with anti-Syk antibodies (4D10, mouse monoclonal; or N19, rabbit polyclonal), anti-Sp1 antibody (rabbit polyclonal), or IgG controls. Ab, antibody. D, EMSA. The above-described DNA probe was incubated with recombinant Sp1 protein in combination with purified GST-Syk(L) or GST control protein in the presence or absence of Sp1. E, Lysates from HEK293 cells transfected with pFLAG-SYK(L) or pFLAG-BAP (control) were subjected to M2 immunoprecipitation and immunodetection of Sp1 (top panel). Lysate inputs were immunoblotted for Sp1 and Flag fusion proteins (center and bottom panels). F, Mutations at the putative Sp1 site disrupt DNA-protein complex formation. Left panel, EMSA was performed using K562 nuclear extracts (NE) and the 100-bp wild-type (WT) or Sp1-site-mutated (MUT) FRA1 promoter fragments as probes. Right panel, competition experiment using excess (10x and 100x) wild-type or mutant non-labeled probes.
To determine the physiological relevance of these findings, we investigated whether Syk(L) directly modulates FRA1 gene transcription by testing the response of a ∼900-bp FRA1 promoter reporter construct (nt -861 to +32) to Syk(L). This FRA1 regulatory region is critical for FRA1 induction by mitogens (17), and was shown to be highly active in MDA-MB-231 cells (∼40-fold higher activity than the promoter-less reporter) but significantly repressed in the presence of Syk(L) (Fig. 4B). Progressive 5′ deletions in this 900-bp region allowed us to narrow the Syk(L)-responsive region to a fragment containing nt -283 to -68. Testing of this fragment by nested deletion revealed a 100-bp fragment (nt -283 to -184) that responded fully to Syk(L) (Fig. 4B).
EMSA was then performed to verify Syk(L)'s physical association with the 100-bp response region, using the commercial nuclear extract from K562 cells that express high level of endogenous Syk(L) (Fig. 4C). The K562 nuclear factors resulted in multiple shifted bands (lane 2). To determine whether Syk is a component in the DNA-protein complex, we carried out an EMSA supershift experiment using two anti-Syk antibodies (4D10 and N19). Addition of either antibody led to the disappearance of a major band (arrow), accompanied by a complex supershift (arrowhead; lanes 3 and 4), whereas the respective mouse or rabbit IgG control had no such effect (lanes 5 and 6). Sequence analysis revealed a putative GC-box (5′-GGGGCGGGCC-3′; nt -238 to -229) within the 100-bp FRA1 fragment that resembled the consensus Sp1-binding sequence 5′-GGGGCGGGGC-3′ (18). We also found that the Syk-containing DNA complex was supershifted by an anti-Sp1 antibody (asterisk; lane 7), indicating that Sp1 and Syk co-exist in the same DNA complex.
To determine whether Syk(L) and Sp1 bind directly to DNA, we performed EMSA using purified recombinant proteins. The recombinant GST-Syk(L) alone did not form complexes with the 100-bp FRA1 probe (lane 5; Fig. 4D), suggesting that Syk(L) does not bind to DNA directly. This was consistent with our earlier CASTing (Cyclic Amplification and Selection of Targets) studies, in which we did not identify a consensus DNA sequence for Syk(L) (data not shown). As expected, we found that recombinant Sp1 protein created a shifted band (arrow) that was supershifted (arrowhead) by anti-Sp1 antibody (lanes 2 and 3; Fig. 4D). Interestingly, addition of GST-Syk(L), but not GST alone, led to further complex retardation (asterisk; compare lanes 2 and 6), indicative of the formation of a Syk(L)-Sp1-DNA complex. The interaction between Syk(L) and Sp1 was further verified by co-immunoprecipitation analyses (Fig. 4E). All these results collectively indicate that Syk(L) influences FRA1 gene transcription by indirect binding to DNA through the Sp1 transcription factor.
To elucidate the biological significance of the Sp1 site in Syk(L)-mediated transcription repression, we generated a version of the 100-bp fragment in which the Sp1 site was disrupted by nucleotide substitution (from 5′-GGGGCGGGCC-3′ to 5′-GGTTGGGGCC-3′). As expected, the mutation led to a markedly lower affinity for Sp1 (lanes 2 and 4) and an inability to compete with the wild-type DNA (lanes 7, 8 and lanes 9, 10), as assessed by EMSA (Fig. 4F). Consistently, the mutation rendered the 100-bp FRA1 promoter region resistant to repression by Syk(L) (bottom bar; Fig. 4B), substantiating the importance of Sp1 in mediation of Syk(L)-regulated gene transcription.
Discussions
Syk was thought to be exclusively cytoplasmic. Emerging evidence, however, suggests that the full-length form is present also in the nucleus (6, 10). We have reported that a bipartite nuclear localization signal within IDB is responsible for Syk(L) nuclear translocation, which is required for Syk(L) tumor suppression response (6). The present study provides additional evidence to confirm the expectation that Syk(L) accomplishes its tumor suppressor functions, at least in part, as a nuclear protein. We showed that Syk(L) repressor activity in the cell nucleus governs downstream gene transcription. Syk(L)-regulated gene transcription may thus be a major event underlying tumor suppression. This hypothesis is consistent with the oncogenic properties of cyclin D1 and Fra1 that are down-regulated by Syk(L) expression. Amplification of CCD1 is one of the most prominent alterations observed in breast carcinoma (19-22). Cyclin D1 is believed to stimulate proliferation by overriding the G1-to-S transition through the cyclin D-CDK4/6-Rb pathway (23, 24). Fra1, a member of the AP1 family, is primarily considered to be oncogenic (25). Over-expression of Fra1 in fibroblasts results in cell transformation, anchorage-independent growth, and tumor formation in nude mice (25-28). Over-expression of Fra1 has been consistently observed in aggressive breast cancer (29, 30) and esophageal squamous cell cancer carcinoma (31). In agreement, Fra1 has been experimentally shown to induce invasiveness and proliferation of carcinomas of the breast and other sites (32, 33).
Based on the current experimental evidence, we propose a model in which Syk counterbalances the transcription of Sp1-activated genes. By binding to Sp1, nuclear Syk(L) recruits HDACs to affect chromatin remodeling of Sp1-activated genes (Figure 5). This model resembles the pathway for Rb-mediated transcription repression of E2F-induced gene transcription (34, 35). Sp1-activated gene transcription has been to shown to be a key contributor to tumor growth, survival, invasion, and angiogenesis (36). Aberrant activation or over-expression of Sp1 is frequently observed in tumors (37, 38). We hypothesize that the loss of Syk(L) during mammary tumorigenesis promotes disease progression by de-repression of Sp1-mediated oncoprotein expression. Sp1 is a ubiquitous transcription factor that transactivates a large number of genes. It is not clear how Syk(L) affects only limited variety of transcripts. This specificity may be attributed to DNA sequences adjacent to the Sp1-binding element in a promoter context. Alternatively, other Sp1-interacting nuclear factors may also compete with Syk(L) in a gene specific fashion. These possibilities need further investigation.
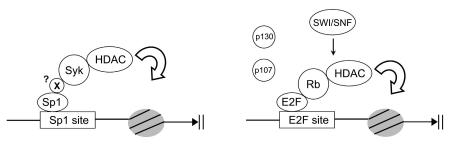
Figure 5.
Comparison of Rb- and Syk-mediated transcription repression. Rb (or its family members p107 and p130) intereacts with chromatin remodeling enzymes such as HDACs and SWI/SNF. The interaction of Rb with E2F allows the remodeling enzymes to be targeted to promoters where they affect nucleosome assembly. Similarly, the interaction of Syk(L) with HDACs is responsible for Syk(L) transcription repression activity. The binding of Syk(L) to Sp1, which is either a direct interaction or through an intermediator factor (marked as “X”), predicts Syk's ability to govern Sp1-activated gene transcription.
Syk-mediated signaling events usually involve a cascade of tyrosine phosphorylation reactions. It is thus plausible that phosphorylation of Syk(L) by upstream signals and/or Syk phosphorylation of downstream effectors influences Syk activities. This effect can be achieved by at least two means. First, Syk(L) nuclear translocation could be instigated by stimulatory signals to make Syk(L) available for nuclear activity. Second, activated Syk(L) in the nucleus may regulate gene transcription by phosphorylating its binding partners. Our results suggest that the kinase activity of Syk(L) does not influence its intrinsic repressor activity. Consistently, our in vitro kinase assays suggested that HDACs are not phosphorylated by Syk(L) (data not shown). It remains to be assessed whether DNA-binding factors such as Sp1 are phosphorylated by Syk(L) and whether this phosphorylation affects Sp1's interaction with DNA and the transcription machinery. Such regulation might also specify the downstream targets affected by nuclear Syk(L).
In summary, we have identified a distinct Syk pathway to specify gene transcription repression that is highly relevant to the suppression of breast cancer. Presumably, the shuttling of Syk(L) between the cytoplasm and the nucleus is highly regulated by upstream signals, the understanding of stimuli that trigger Syk(L) nuclear translocation is thus crucial for the development of therapeutic strategies. Moreover, it would be useful to evaluate the means to overcome the rate-limiting nature of this nuclear translocation process. The identification of chemical compounds or small peptides that would facility the Syk(L) nuclear translocation may serve as a way to rectify tumor suppression.
Acknowledgments
Grant support: NIH grant R01-CA100278.
The authors thank Drs. Ronald Evans, Tony Kouzarides, Philip Low, and Jiemin Wong for reagents; Dr. Kun-Sang Chang for critical reading of the manuscript; Drs. Wei Zhang and Keith Baggerly for help with the microarray analyses.
References
- 1.Darnell JE. Transcription factors as targets for cancer therapy. Nature Rev Cancer. 2002;2:740–9. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Yuan YF, Mendez R, Sahin A, Dai JL. Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res. 2001;61:5558–61. [PubMed] [Google Scholar]
- 3.Coopman PJ, Do MTH, Barth M, et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–7. doi: 10.1038/35021086. [DOI] [PubMed] [Google Scholar]
- 4.Toyama T, Iwase H, Yamashita H, et al. Reduced expression of the Syk gene is correlated with poor prognosis in human breast cancer. Cancer Lett. 2002;189:97–102. doi: 10.1016/s0304-3835(02)00463-9. [DOI] [PubMed] [Google Scholar]
- 5.Dejmek J, Leandersson K, Manjer J, et al. Expression and signaling activity of Wnt-5a/Discoidin Domain Receptor-1 and Syk plays distinct but decisive roles in breast cancer patient survival. Clin Cancer Res. 2005;11:520–8. [PubMed] [Google Scholar]
- 6.Wang L, Duke L, Zhang PS, et al. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res. 2003;63:4724–30. [PubMed] [Google Scholar]
- 7.Moroni M, Soldatenkov V, Zhang L, et al. Progressive loss of Syk and abnormal proliferation in breast cancer cells. Cancer Res. 2004;64:7346–54. doi: 10.1158/0008-5472.CAN-03-3520. [DOI] [PubMed] [Google Scholar]
- 8.Mahabeleshwar GH, Kundu GC. Syk, a protein-tyrosine Kinase, suppresses the cell motility and nuclear factor kappa B-mediated secretion of urokinase type plasminogen activator by inhibiting the phosphatidylinositol 3′-kinase activity in breast cancer cells. J Biol Chem. 2003;278(8):6209–21. doi: 10.1074/jbc.M208905200. [DOI] [PubMed] [Google Scholar]
- 9.Ruschel A, Ullrich A. Protein tyrosine kinase Syk modulates EGFR signalling in human mammary epithelial cells. Cell Signal. 2004;16:1249–61. doi: 10.1016/j.cellsig.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Ulanova M, Puttagunta L, Marcet-Palacios M, et al. Syk tyrosine kinase participates in {beta}1-integrin signaling and inflammatory responses in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L497–507. doi: 10.1152/ajplung.00246.2004. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Pan Y, Dai JL. Evidence of prooncogenic activity of MKK4 in breast and pancreatic tumors. Oncogene. 2004;23:5978–85. doi: 10.1038/sj.onc.1207802. [DOI] [PubMed] [Google Scholar]
- 12.Hu L, Wang J, Baggerly K, et al. Obtaining reliable information from minute amounts of RNA using cDNA microarrays. BMC Genomics. 2002;3:16. doi: 10.1186/1471-2164-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner M, Schweighoffer E, Colucci F, Di Santo JP, Tybulewicz V. Tyrosine kinase SYK: essential functions for immunoreceptor signalling. Immunol Today. 2000;21:148–54. doi: 10.1016/s0167-5699(99)01574-1. [DOI] [PubMed] [Google Scholar]
- 14.Woodside DG, Obergfell A, Leng L, et al. Activation of Syk protein tyrosine kinase through interaction with integrin beta cytoplasmic domains. Curr Biol. 2001;11:1799–804. doi: 10.1016/s0960-9822(01)00565-6. [DOI] [PubMed] [Google Scholar]
- 15.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–8. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Adiseshaiah P, Papaiahgari SR, Vuong H, Kalvakolanu DV, Reddy SP. Multiple cis-elements mediate the transcriptional activation of human fra-1 by 12-O-tetradecanoylphorbol-13-acetate in bronchial epithelial cells. J Biol Chem. 2003;278(48):47423–33. doi: 10.1074/jbc.M303505200. [DOI] [PubMed] [Google Scholar]
- 18.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 19.Buckley M, Sweeney K, Hamilton J, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–33. [PubMed] [Google Scholar]
- 20.Motokura T, Bloom T, Kim HG, et al. A novel cylin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350:512–5. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SY, Caamano J, Cooper F, Guo X, Klein-Szanto AJ. Immunohistochemistry of cyclin D1 in human breast cancer. Am J Clin Pathol. 1994;102:695–8. doi: 10.1093/ajcp/102.5.695. [DOI] [PubMed] [Google Scholar]
- 22.Weinstat-Saslow D, Merino MJ, Manrow RE, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat Med. 1995;1:1257–60. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 23.Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclin D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewen ME. The cell cycle and the retinoblastoma protein family. Cancer Metastasis Rev. 1994;13:45. doi: 10.1007/BF00690418. [DOI] [PubMed] [Google Scholar]
- 25.Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–12. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 26.Bergers G, Graninger P, Braselmann S, Wrighton C, Busslinger M. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Mol Cell Biol. 1995;15(7):3748–58. doi: 10.1128/mcb.15.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reichmann E, Schwarz H, Deiner EM, et al. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 1992;71:1103–16. doi: 10.1016/s0092-8674(05)80060-1. [DOI] [PubMed] [Google Scholar]
- 28.Saez E, Rutberg SE, Mueller E, et al. c-fos is required for malignant progression of skin tumors. Cell. 1995;82:721–32. doi: 10.1016/0092-8674(95)90469-7. [DOI] [PubMed] [Google Scholar]
- 29.Zajchowski DA, Bartholdi MF, Gong Y, et al. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 2001;61(13):5168–78. [PubMed] [Google Scholar]
- 30.Roy D, Calaf G, Hei TK. Profiling of differentially expressed genes induced by high linear energy transfer radiation in breast epithelial cells. Mol Carcinog. 2001;31:192–203. doi: 10.1002/mc.1054. [DOI] [PubMed] [Google Scholar]
- 31.Hu YC, Lam KY, Law S, Wong J, Srivastava G. Identification of differentially expressed genes in esophageal squamous cell carcinoma (ESCC) by cDNA expression array: Overexpression of Fra-1, Neogenin, Id-1, and CDC25B genes in ESCC. Clin Cancer Res. 2001;7(8):2213–21. [PubMed] [Google Scholar]
- 32.Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulate proliferation and invasiveness of breast cancer cells. Oncogene. 2005;24:1434–44. doi: 10.1038/sj.onc.1208312. [DOI] [PubMed] [Google Scholar]
- 33.Kustikova O, Kramerov D, Grigorian M, et al. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma Cells. Mol Cell Biol. 1998;18(12):7095–105. doi: 10.1128/mcb.18.12.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harbour JW, Dean DC. Chromatin remodeling and Rb activity. Curr Opin Cell Biol. 2000;12:685–9. doi: 10.1016/s0955-0674(00)00152-6. [DOI] [PubMed] [Google Scholar]
- 35.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. PNAS. 1998;95:10493–8. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–60. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 37.Yao JC, Wang L, Wei D, et al. Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin Cancer Res. 2004;10:4109–17. doi: 10.1158/1078-0432.CCR-03-0628. [DOI] [PubMed] [Google Scholar]
- 38.Kumar AP, Butler AP. Enhanced Sp1 DNA-binding activity in murine keratinocyte cell lines and epidermal tumors. Cancer Lett. 1999;137:159–65. doi: 10.1016/s0304-3835(98)00351-6. [DOI] [PubMed] [Google Scholar]