Complex traits are constructed during development by an intricate array of factors, and interactions among factors, including DNA, RNA, proteins, developmental modules, and various aspects of the biotic and abiotic environment. As a result, it is development that structures the relationship between the genotype and phenotype and thereby determines genetic architecture. The genotype–phenotype relationship plays a central role in phenotypic evolution because it determines how selection at the level of the phenotype is translated into evolutionary change at the level of the genotype (see ref. 1). Theoretical approaches to understanding phenotypic evolution have primarily focused either at the molecular genetic level, modeling evolution as changes in allele frequencies (the “population genetics” tradition), or at the gross phenotypic level, modeling evolution as changes in mean trait values by using the statistical relationship between molecular genetic variation and patterns of phenotypic variation (the “quantitative genetics” tradition). Both of these traditions have successfully advanced our understanding of phenotypic evolution, but both approaches are also inherently limited because they require an assumption of a relatively simple genotype–phenotype relationship. Thus, they are limited in their ability to incorporate the emerging data on the intricate patterns of genetic and developmental interactions that underlie the often remarkably complex genotype–phenotype relationship (2). For complex traits, this generally means that factors influencing trait expression, such as the intricate patterns of gene interactions or genotype-by-environment interactions, are either assumed absent or are included in a greatly simplified form (1).
The Rice model embraces the complexity of genetics and development, rather than avoiding complexity by invoking simplifying assumptions.
Although empirical research on the genetic architecture and developmental basis of trait expression has advanced at an extraordinary rate, formal links between the wealth of data that has been emerging and the process of phenotypic evolution have lagged behind. More recently, theoretical approaches have begun to focus at the level between the gross phenotypic and molecular genetic levels to examine the connection between patterns of development and evolution (e.g., refs. 3–7). These models have examined the impact that the developmental and epigenetic processes that underlie patterns of phenotypic variation have on a number of evolutionary processes, such as the evolution of genetic variation, canalization, and integration. Although these models have provided great insights into these processes, which have been difficult to understand using traditional population or quantitative genetic approaches, no “unifying theory” of phenotypic evolution has emerged. However, Rice (8) has produced what could prove to be the critical model that leads to this unifying theory. Rice's model is a general theory of phenotypic evolution, which provides the link between patterns of development and processes of phenotypic evolution. The model is analogous to the Fisher–Bulmer–Lande models that form the foundations of quantitative genetics (1), but it allows one to model phenotypic evolution from patterns of developmental variation instead of statistical genetic variation. The model can also be applied to discontinuous genetic data, which also makes it amenable to analyses that are usually in the realm of traditional population genetic theory. In many ways, the Rice model surpasses previous models of phenotypic evolution because it can be applied to a broader array of empirical and theoretical problems. Using an extraordinarily eloquent approach, Rice has managed to derive a model that can be applied to an arbitrarily complex pattern of development, with any number of factors (genetic, developmental, or environmental) and any form of selection acting on any number of traits. In fact, the single major simplifying assumption of the model is the form of inheritance, which assumes a linear relationship between the value of characters in parents and their offspring. However, this is unlikely to be a limiting assumption, and, regardless, will be overcome in a forthcoming extension of the theory. Perhaps what makes the Rice model particularly novel is that it embraces the complexity of genetics and development, rather than avoiding complexity by invoking simplifying assumptions.
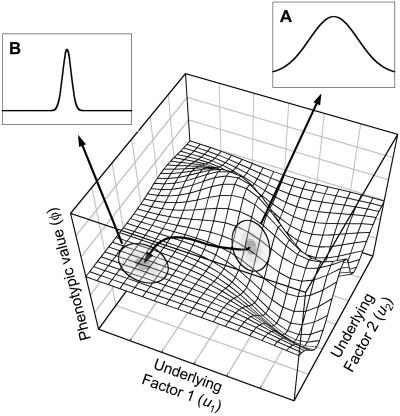
An intuitive understanding of the Rice model can best be achieved through the use of the metaphor of the phenotype landscape (see ref. 9). The surface of a phenotype landscape defines the phenotype associated with a particular combination of underlying factors (e.g., alleles present at loci, gene expression values, size of developmental modules, hormone levels, environmental variables such as temperature, etc.). When the underlying factors are genetic, the landscape represents the genotype–phenotype mapping function. The topographical features of the landscape are determined by the developmental system that governs the interactions between the underlying factors. The number of underlying factors contributing to phenotypic variation defines the number of dimensions of space in which the landscape exists. In theory there is no limit to the number of underlying factors that can influence the expression of a particular trait, and, thus, landscapes can exist in very-high-dimensional space (i.e., hyperspace). The description of the topography of a three-dimensional landscape is most intuitive, but the same descriptors can also be applied to hyperdimensional landscapes, although the intuitive interpretation of terms like “slope” or “curvature” become increasingly abstract as the dimensionality increases. However, this does not alter the usefulness of Rice's model, although it does mean that one should be cautious when interpreting the topography of hyperdimensional landscapes because descriptors like hilly or rugged, which have intuitive meanings in three-dimensional space, may be misleading when applied to higher-dimensional space (see ref. 10). Fig. 1 shows a hypothetical landscape, where the phenotype of an individual is determined by the values of two underlying factors.
Fig 1.
An example of a phenotype landscape. The expected phenotypic value of an individual (φ) is a function of the value underlying factors 1 and 2 (u1 and u2) that interact during trait development. A population is represented by the shaded ovals at two different positions on the surface. The variance in the underlying factors is the same at both locations (indicated by the size of the ovals), but the mean values of the factors are different. The population starts in a steep region of the landscape, where the underlying factors are translated into a large phenotypic variance by development (indicated by the distribution in Inset A). Canalizing selection moves the population to a much flatter region, where the underlying factors have a much smaller contribution to phenotypic variance (indicated by the distribution in Inset B). The mean phenotypic value is the same at both locations on the landscape.
The landscape provides a concise summary of the patterns of genetic effects, gene interactions, developmental interactions, environmental effects, and genotype–environment interactions that produce the relationship between variation in underlying factors and the phenotype. The location of each individual on the landscape is determined by the value of its underlying factors. In theory, the surface of the landscape can be defined for any possible combination of values of the underlying factors that define the dimensions of the landscape, even if some combinations do not actually exist. Thus, a population is expected to be limited to a local region of the landscape at any point in time. To account for this fact, the topography of the landscape is evaluated at the mean of the underlying values in the Rice model. The topography is captured in a set of derivatives that define how the phenotype changes as a function of changes in the values of the underlying factors (or combination of factors) in the local region of the landscape. This set of derivatives is represented by the tensor Dn (where n is the rank of the tensor). Although tensors may not be familiar to many people, some special types of tensors probably are. When n = 1, the tensor is rank 1, which is a vector. The vector D contains the gradient of the landscape (i.e., the steepness) associated with each of the underlying factors. The gradient vector determines the linear effects of factors on the expression of the trait. The gradient vector also determines the contribution of the underlying factors to phenotypic variation (see Fig. 1); steeper regions correspond to areas of high phenotypic variance and less steep regions correspond to lower variance (11). Genetic parameters that are used in the description of evolution in the quantitative genetics tradition can also be defined using the gradient in a local region (7). For example, the additive genetic variance corresponds directly to the gradient of the landscape when the underlying factors are genetic (7). Rice uses this relationship when describing the connection between his model and the earlier quantitative genetic models (see more below). The rank 2 tensor is a matrix of second derivatives on the surface, which can be interpreted as quadratic curvature of the surface. When a landscape is curved so that the gradient is not constant in a region, the underlying factors contribute to nonadditive components of genetic variance such as dominance and epistasis (6, 7). Rice utilizes this relationship to derive an example of the evolution of dominance.
Evolution is described by changes in the phenotype distribution associated with movement of the population on the surface of the phenotype landscape. The phenotype distribution is captured in the tensor Pk, where the rank of the tensor (k) designates which moment of the phenotype distribution that P corresponds to. When k = 1, P is a vector of trait means, and when k = 2, P is the phenotypic variance–covariance matrix. To model evolution on the landscape, a description of the relationship between phenotypes and fitness is required. The landscape metaphor can also be invoked when describing the relationship of traits and fitness (treating fitness as a trait), and in that case, the landscape is referred to as the individual selection surface (see ref. 12). The geometry of the selection surface is captured in Rice's model by partial derivatives of fitness with respect to the trait values, which includes higher-order partial derivatives that capture nonlinear relationships between traits and fitness. This model of selection jibes well with evolutionary theory because the partial derivatives of the fitness surface evaluated at the population mean are already important components of modern selection theory (13). The first-order partial derivatives are the directional selection gradients [represented by the vector β in selection theory (13)], whereas the second-order partial derivatives are the nonlinear, quadratic gradients [i.e., stabilizing, disruptive, or correlational gradients, represented by the matrix γ (13)]. However, rather than focusing on the shape of the fitness surface, Rice utilizes a tensor of selection differentials (Q), which represent the consequences of a particular fitness function in terms of how selection alters some moment of the phenotype distribution within a generation (where the rank of Q indicates which moment of the phenotype distribution it corresponds to). The within-generation changes described by Q are translated into across generational changes by using a matrix of heritabilities and genetic correlations (H), whose elements are the partial regression of the value of an underlying factor in offspring on the value of a factor in parents.
The Rice model is particularly innovative and likely to be of great utility because it allows one to examine the evolution of any moment of the phenotypic distribution, making it a far more general theory than the traditional quantitative genetic models of phenotypic evolution, which focuses almost exclusively on the evolution of the mean (the first moment). Although some quantitative genetic models have examined the evolution of phenotypic variances and covariances (i.e., second moments of the distribution; e.g., ref. 14), those models are based on very limiting assumptions. Even the previous models that have used the phenotype landscape approach (or some analog) to analyze evolution of phenotypic variance–covariance structure or other evolutionary genetic problems have invoked restrictive assumptions (e.g., ref. 7).
Fig. 1 gives an illustration of how the topography of the phenotype landscape is related to patterns of phenotypic variation. This illustration roughly corresponds to Case 2 presented in Rice's paper. A population existing at two different points in time is represented by the shaded ovals on the surface. At both locations on the landscape the variances of the underlying factors are the same, but the means are different. The population begins in a steep region of the landscape, where the phenotypic variance is relatively large (Inset A). Canalizing selection slides the population along a phenotype isocline to a flatter region of the landscape, where the phenotypic variance is much smaller (Inset B). Despite the fact that the population has the same variance in underlying factors (e.g., the same amount of molecular genetic variation) and the same mean phenotype at both locations on the landscape, the phenotypic variance changed dramatically. This example demonstrates the power of the model; a complex evolutionary problem like the evolution of canalization can be modeled using the same simple framework that can be used to predict the evolution of the mean.
Rice's model has the opportunity to do for evolutionary studies of development what Lande's multivariate quantitative genetic models (e.g., ref. 15) did for evolutionary quantitative genetics (see ref. 16). Before Lande's general multivariate model, quantitative genetic theory had primarily been limited to studies of the genetics of agricultural improvement. Lande's model brought the quantitative genetics framework to the forefront of evolutionary biology and lead to great insights into multivariate phenotypic evolution. However, the Rice model is not simply an alternative to multivariate quantitative genetic theory, it is a generalization of the theory, or, more precisely, it is a more general theory that encompasses quantitative genetics as a special case. This is clearly demonstrated in the first special case examined by Rice, where the model simplifies to the multivariate quantitative genetic model by invoking the appropriate set of assumptions. Assuming an additive (planar) phenotype landscape and a quadratic fitness function, Rice demonstrates how the model reduces to the classic “breeder's equation,” where changes in the trait means across generations are predicted from the matrix of trait heritabilities and genetic correlations (H) and the vector of selection differentials (Q).
Despite the attractiveness of Rice's model as a mathematical theory, it remains to be seen to what degree it has an impact on empirical genetic and developmental studies of evolution. Future work describing methods that can be easily adopted by empiricists will be required before researchers can embrace the model and use its framework when interpreting data. This is particularly important because the mathematics of the model are rather complex and may be unfamiliar to a large proportion of the empirically based researchers. However, this should not be seen as a shortcoming of the model. Advances in theory have generally relied on later publications to bridge the gap from formal mathematical theory to empirical applications. Bridging this gap often requires detailed methods of how statistical or mathematical computer packages can be used to estimate the relevant parameters.
Although it remains to be seen whether formal methods will emerge to combine the Rice model with empirical studies of development, it is clear that the model can interface well with the sorts of data that are derived from developmental studies, such as changes in trait values as a function of factors like morphogen concentrations (e.g., ref. 17). Developmental modules of all sorts are often useful underlying factors because their contribution to the gross phenotype is often relatively straightforward, and experimental analyses of epigenetic interactions between modules may be feasible. It is also often relatively straightforward to examine how the expression of a trait changes as a function of various environmental variables (e.g., ref. 18), allowing one to incorporate environmental factors as dimensions of the phenotype landscape. In addition, Rice discusses how the model can incorporate the sort of discontinuous data associated with allelic polymorphisms at discrete loci. Thus, the theory can also be used to model and analyze population genetic dynamics. Along these lines, Rice suggests that underlying factors such as QTL or even gene expression patterns estimated from microarrays could be used to estimate the local geometry of the landscape, providing a way to integrate theoretical studies with cutting edge genetic data. Perhaps most interesting is the fact that the model can be applied to all of these problems simultaneously, because one can combine all factors that contribute to phenotypic variation. The ability to predict evolutionary changes in all aspects of the multivariate phenotype distribution by using theory that integrates the complex developmental systems that build traits is likely to provide great insights into the evolution of complex traits, and may eventually emerge as a unifying theory of trait evolution.
See companion article on page 15518 of issue 24 of volume 99.
References
- 1.Roff D. A., (1997) Evolutionary Quantitative Genetics (Chapman & Hall, New York).
- 2.Oyama S., Griffiths, P. E. & Gray, R. D., (2001) Cycles of Contingency: Developmental Systems and Evolution (MIT Press, Cambridge, MA).
- 3.Wagner G. P., Booth, G. & Bagheri-Chaichian, H. (1997) Evolution (Lawrence, Kans.) 51, 329-347. [DOI] [PubMed] [Google Scholar]
- 4.Rice S. H. (1998) Evolution (Lawrence, Kans.) 52, 647-656. [DOI] [PubMed] [Google Scholar]
- 5.Johnson N. A. (2001) Genetica 112, 45-58. [PubMed] [Google Scholar]
- 6.Gilchrist M. A. & Nijhout, H. F. (2001) Genetics 159, 423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf J. B., Frankino, W. A., Agrawal, A. F., Brodie, E. D., III & Moore, A. J. (2001) Evolution (Lawrence, Kans.) 55, 232-245. [DOI] [PubMed] [Google Scholar]
- 8.Rice S. H. (2002) Proc. Natl. Acad. Sci. USA 99, 15518-15523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf J. B., Allen, C. E. & Frankino, W. A. (2002) in The Evolutionary Biology of Complex Phenotypes, eds. Pigliucci, M. & Preston, K. (Oxford Univ. Press, New York).
- 10.Gavrilets S. (1997) Trends Ecol. Evol. 12, 307-312. [DOI] [PubMed] [Google Scholar]
- 11.Rice S. H. (2000) in Epistasis and the Evolutionary Process, eds. Wolf, J. B., Brodie, E. D., III & Wade, M. J. (Oxford Univ. Press, Oxford).
- 12.Phillips P. C. & Arnold, S. J. (1989) Evolution (Lawrence, Kans.) 43, 1209-1222. [DOI] [PubMed] [Google Scholar]
- 13.Lande R. & Arnold, S. J. (1983) Evolution (Lawrence, Kans.) 37, 1210-1226. [DOI] [PubMed] [Google Scholar]
- 14.Tallis G. M. (1989) J. Anim. Breed. Genet. 106, 163-179. [Google Scholar]
- 15.Lande R. (1979) Evolution (Lawrence, Kans.) 33, 402-416. [DOI] [PubMed] [Google Scholar]
- 16.Arnold S. J. (1994) in Quantitative Genetic Studies of Behavioral Evolution, ed. Boake, C. R. B. (Univ. of Chicago Press, Chicago), pp. 17–48.
- 17.Strul G., Johnston, P. & Lawrence, P. A. (1992) Cell 69, 237-249. [DOI] [PubMed] [Google Scholar]
- 18.Emlen D. J. (1994) Proc. R. Soc. London 256, 131-136. [Google Scholar]