In this modern era of genome sequencing and structural genomics, the fundamental question of how chromatin folds into mitotic chromosomes remains a mystery. In higher eukaryotic cells, mitotic chromosome condensation leads to an ≈10,000- to 20,000-fold compaction, as defined by the ratio of the DNA length to metaphase chromosome length (1). Compaction is achieved through at least three levels of chromatin folding (2). The first two, the coiling of DNA around the histone octamer to form nucleosomes, and the folding of regularly spaced nucleosomes into ≈30 nm “higher order” chromatin fibers, together account for only a ≈40:1 compaction ratio. This leaves the majority of mitotic chromosome compaction, ≈500-fold, the result of poorly characterized large-scale chromatin folding above the level of 30 nm chromatin fibers.
Nearly all textbooks feature the radial loop model of mitotic chromosome condensation, derived from experiments largely conducted 15–25 years ago. By a neat trick of high-salt or polyanion-driven protein extraction under conditions preserving remnants of mitotic chromosome structure, the difficult issue of higher order and large-scale chromatin folding was effectively bypassed. Striking images of extended DNA “halos” surrounding a linear protein core, roughly the shape of the native chromosome, led to a simple model in which a central protein “scaffold” constrained DNA into a set of loops, ≈50 kb in length. Yet the radial loop model has remained highly controversial. This controversy derives in large part from key assumptions that are difficult to address experimentally but implicit in the interpretation of the original experiments. Two recently published experimental approaches effectively reevaluate these questions of interpretation by testing predictions, rather than assumptions, of the radial loop model. In the first (3, 4), the in vivo dynamics of the scaffold protein topoisomerase 2 were examined by using a GFP protein. In the second, published in a recent issue of PNAS (5), micromechanical force measurements on individual, isolated chromosomes probed changes in chromosome structure produced by nuclease digestion (see Fig. 1, ref. 5). Their results prompt serious reconsideration and debate of the standard textbook model for mitotic chromosome condensation.
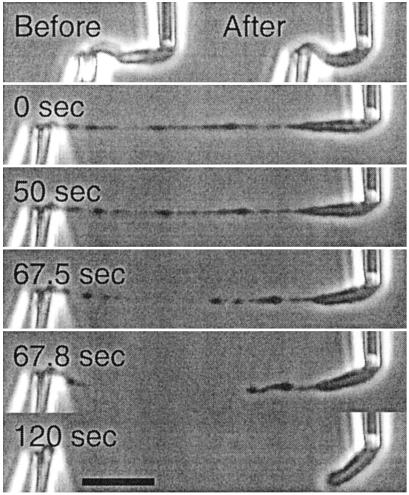
Fig 1.
Loss of chromosome integrity by nuclease treatment alone is demonstrated by a chromosome micromanipulation experiment (5). Native chromosome (Before) loses elasticity after brief nuclease digestion (After). Stretching this partially digested chromosome leads to thin fiber extensions (0 sec), which are digested and then severed by further nuclease treatment.
To appreciate the original and current appeal of the radial loop model, and the biochemical experimental approach from which it was derived, one must first understand the experimental difficulties intrinsic to a direct structural investigation of mitotic chromosome structure (6). These include the tremendous density of chromatin packing, limitations of light and electron microscopy, the sensitivity of chromatin to small changes in ionic conditions, and the lack of good DNA stains for electron microscopy. Thus the initial experiments (7, 8) showing the existence of a nonhistone protein “scaffold” through removal of dense, obscuring chromatin was very exciting.
The match between the shape and size of the extracted chromosome core and the native, unextracted chromosome led directly to the concept that that this residual structure might reflect the remnants of a nonhistone protein “scaffold.” Stability of the scaffold and DNA halo in 2 M NaCl was interpreted as consistent with the scaffold holding the DNA loops in a compact conformation. The use of nucleases to isolate the scaffold without its associated DNA halo suggested the scaffold was a structurally independent entity whose integrity was independent of DNA (8). Reversible swelling and contraction of isolated scaffolds driven by changes in ionic conditions suggested that similar conformational changes of intact chromosomes might be driven by the salt-induced changes in scaffold structure (9). In turn, this led to a speculative model in which chromosome condensation from interphase through metaphase might be driven by similar transitions in scaffold structure from a dispersed interphase network to a contracted mitotic conformation (8, 9).
In earlier work, no conclusions were made concerning the distribution of this scaffold in the native chromosome. Subsequent examination of chromosomes swollen by exposure to reduced divalent cation concentrations revealed loops of decondensed 10-nm chromatin fibers extending outwards from a dense core region of tightly packed chromatin fibers (10, 11). This finding led then to the typical textbook radial loop model of a central axial scaffold organizing radial loops of 30-nm chromatin fibers.
Since then, extensive literature has added considerable details without cleanly resolving controversy. Isolation of intact scaffolds was demonstrated to be dependent on protein cross-links stabilized by specific metal ions (12, 13), with alternative extraction conditions producing extended DNA structures without visible scaffolds (14–16) or more diffuse scaffolds without an obvious core structure (17). These results led different scientific “camps” to opposite conclusions, with one group arguing that the observed scaffolds resulted from cross-linking of chromosomal proteins during protein extraction and the other that in vivo scaffold formation depends on specific metalloprotein interactions (18).
As originally predicted, further experiments revealed retention of specific “SAR” (scaffold attachment regions) DNA in both nuclear and mitotic scaffolds (19). Essentially the same sequences, defined operationally as “MAR” (matrix attachment regions) sequences, were found to bind preferentially to isolated nuclear scaffolds, now called matrices, when added as exogenous DNA (18, 20). Again, two opposite camps arose, with one arguing that these SAR/MARs were bound in vivo to a scaffold and the other arguing that small DNA fragments containing SAR/MAR sequences produced by nuclease treatment might become bound to scaffold proteins during the experimental procedure (18). Fueling the controversy were additional experimental inconsistencies at odds with the conceptual underpinnings of the experimental design, for instance, the observation that SAR sequences were extracted by a repeated protein extraction step the same as that used for the original scaffold preparation (18).
Also, as predicted, a small number of specific proteins were demonstrated as forming the mitotic scaffold structure (12). A major advance was identification of the two major scaffold proteins, SCI and SCII, as topoisomerase 2 (21, 22) and SMC2 (23, 24), the CAP-E subunit of the condensin complex (25), respectively. A major puzzle, relative to the radial loop model, was that neither protein was an obvious candidate for a building block for a structural scaffold network. Moreover, topoisomerase 2 could be easily extracted in physiological buffers from in vitro assembled chromosomes without any obvious change in chromosome structure (26). Rather, both proteins were associated with ATP dependent activities relating to DNA topology (23, 24). Yet in isolated, intact chromosomes, both proteins showed an axial chromosome distribution (23, 27).
Previous experiments had shown a dynamic redistribution of topoisomerase 2 from chromosome to cytoplasm during mitosis in live Drosophila embyros, with peak prophase chromosomal levels dropping more than 2- and 3-fold by metaphase and telophase (28). This dynamic behavior now has been analyzed on a much finer time scale. Two different groups demonstrated a rapid recovery after photobleaching of nuclear and mitotic chromosomal bound GFP–topoisomerase 2 on a time scale of seconds. Therefore, the vast majority of toposiomerase 2 bound to mitotic chromosomes does not behave, in vivo, as an immobilized component of a static structural scaffold. Unfortunately, although using very similar experimental systems, both groups came to different conclusions concerning the in vivo chromosomal localization of topoisomerase 2, with one reporting an axial (3) and the other a uniform (4) chromosomal distribution.
The second approach monitored changes in the chromosome elasticity and deformability induced by nuclease digestion of individual chromosomes. Previously, Poirer and Marko (29–33) have described methods for mechanically isolating chromosomes from individual cells, holding them in place, and stretching the chromosomes under controlled conditions allowing force measurements. Chromosomes can be stretched reversibly to five times their normal length (30). In their present study, they now have addressed how chromosome elasticity and connectivity are altered by nuclease digestion (5).
Chromosomes were attached at one end to a large stiff pipette, and at the other end to a more flexible pipette whose deflection provides measurement of applied force. Controlled movement of the stiffer pipette allowed force/extension measurements. Chromosomes were stretched and relaxed to determine their elastic response, then held under a small, constant force while exposed to a spray of nuclease from a third pipette. When micrococcal nuclease was used, the tension generated by the elongated chromosome dropped to zero after ≈60-sec digestion, without any obvious chromosome shape changes. Additional digestion lead to thinning, severing, and then disappearance of the chromosome. Alternatively, nonstretched chromosomes were exposed to micrococcal nuclease and then stretched through several cycles of extension and relaxation. Each cycle produced a progressive drop in the required force. Further stretching, beyond the elastic regime, produced severe, nonuniform elongation with blob-like chromosome regions separated by thinner fibers that could be digested by further nuclease digestion (Fig. 1).
Similar experiments were conducted by using spraying of restriction enzymes, rather than micrococcal nuclease. Four base pair cutters produced similar results to micrococcal nuclease. In contrast, extended digestion with 5- and 6-bp cutters produced no change in chromosome elasticity. Restriction digest of chromatin typically is limited to linker DNA between nucleosomes, leading to an ≈10-fold reduction in available sites. This means that 4-bp cutters should cut chromatin at ≈1/10 (1/44) or 2.5-kb spacing. The 6-bp cutter, Cac8 I, has a degenerate recognition sequence dependent on only 4 bp, which should produce the same cutting frequency as the 4-bp cutters. Curiously, extended Cac8 I digestion led to only a partial change in chromosome elasticity converging to a force constant 40% the native value.
From these results, the authors conclude the mechanical integrity of chromosomes is largely dependent on DNA, and not on an internal protein network, behaving as a cross-linked network of 30-nm fibers. Based on a rate of force reduction with cutting 1/10 that observed for the 4-bp cutters, the authors assume this reflects a 10-fold lower number of allowed cutting sites for Cac8 I, implying a cutting frequency of 1/25 kb. Based on the 60% reduction in the force constant for stretching, they estimate ≈60% of cross-linked segments would be cut, yielding a mean distance between cross-links of 15 kb, assuming a 25-kb spacing of Cac8 I cutting. No effect on chromosome elasticity was observed even after prolonged digestion with several 5- and 6-bp cutters. Their cutting frequency should be in the 10-kb (5-bp cutters) or 40-kb (6-bp cutters) range, assuming their site accessibility is limited only by the fraction of linker DNA. Together these results suggest a distance between cross-links perhaps as low as 10–20 kb.
In the absence of direct measurements of mean DNA fragment size after cutting, these numbers must be considered soft. A more substantial issue is whether the networked behavior of the chromosome is caused by distinct, infrequently spaced protein cross-links as opposed to weaker but highly distributed chromatin fiber–fiber interactions, as previously suggested (16). Nucleosome oligomers and short chromatin fiber segments aggregate, and in some cases self-assemble into linear filaments, under physiological divalent and polycation concentrations (34, 35). Similar divalent and polycation concentrations are required for the tight packing of 30-nm fibers and compact chromosome conformation observed for native chromosomes (36). One might imagine that in a hierarchical folding model for chromosome structure, the stability of the different levels of folding will depend on large numbers of weak local interactions.
The complete digestion and disappearance of chromosomes after digestion with 4-bp cutters, with an estimated cutting frequency in chromatin of 2.5 kb, would yield chromatin fragments corresponding to ≈12 nucleosomes, just two turns of a helical nucleosome folding in a solenoid model of 30-nm chromatin fibers (37). Although this level of cutting does not support chromosome structural integrity, it is interesting that raising this to the 10- to 20-kb range may be sufficient. In the chromonema model of chromosome structure (2, 38), folding of 30-nm fibers within 100- to 130-nm diameter chromonema fibers produces tightly packed, ≈12-kb parallel stretches of 30-nm fibers (with ≈25 kb required to loop a 30-nm fiber across a chromonema fiber and back). Similarly, a revised radial loop model, which postulates that the scaffold is helically coiled in the last stage of mitotic chromosome condensation (39, 40), predicts an average loop size of ≈25 kb, assuming a ≈250 nm prophase chromatid. Both models therefore predict 30-nm chromatin fiber distributions that conceptually could be maintained in the presence of the estimated frequency of DNA cutting by either a cross-link or distributed aggregation model.
It will be important to determine the relative contribution of these two possibilities, network cross-ties versus distributed fiber-fiber interactions, to chromosome stability. One approach would be to repeat force measurements on digested chromosomes as a function of changes in ionic conditions. Buffer conditions that reduce fiber–fiber aggregation while still promoting chromatin folding into 30-nm fibers would be ideal. Characterization of the actual cutting frequency will be essential in moving from general to specific models. The current experiments exclude the possibility that the primary mechanical integrity of chromosomes is caused by a stable, structurally continuous scaffold structure running throughout the chromatid length. However, a multiprotein assembly held together by dynamic protein–protein interactions of finite lifetimes might still exist, imparting both order and stability to the chromosome. Dynamics of protein cross-linking interactions might be revealed by varying the rate of chromosome pulling to match protein on/off rates while monitoring the levels of known players in chromosome condensation (i.e., topoisomerase 2, condensins, and cohesins), and possibly manipulating their levels by buffer dilution, would be highly informative.
Where do these experiments now leave the original radial loop model? In vivo, the fluorescence recovery after photobleaching experiments now show the fraction of topoisomerase 2 bound to a static scaffold structure must be small, if not zero. Assuming a significant fraction of topoisomerase 2 isolates with the mitotic scaffold, by extension these results would imply that the originally observed stability of isolated scaffold structures was indeed the result of artifactual cross-linking. The chromosome micromanipulation experiments demonstrate that the shape, structure, and mechanical properties of chromosomes depends on the structural integrity of DNA, ruling out a primary structural role for a physically contiguous protein network embedded within the chromosome. However, these experiments do not rule out the possibility of a dynamic protein assembly that could drive chromatin folding into chromosomes of a defined shape. Nor do they yet distinguish a network model, with rare, specific protein cross-ties, from a model in which chromosomes are held together by distributed chromatin fiber–fiber interactions.
In retrospect, both biologists and physicists likely have oversimplified the process of chromosome condensation. Biologists have largely ignored the physical constraints DNA supercoiling, topology, and chromatin mechanical properties might pose for models of chromosome condensation. Physicists have tried to abstract the details of chromatin folding, concentrating on simple models without necessarily placing them in the proper biological context, which, for instance, requires models permitting rapid transitions into and out of mitosis. Moreover, both groups have focused on explaining all properties of chromosome condensation by a single structural model.
Conceptually there are several distinct aspects involved in chromosome condensation that could very well involve distinct molecular mechanisms. For instance, chromatin condensation per se may be distinct from the shaping of condensed chromatin into a linear chromosome of defined shape and size. The determinants of chromosome shape and size likewise may be different from molecular interactions determining chromosome stability and elasticity. This might explain why similar molecules, codensins, cohesins, and topoisomerase 2, are functionally implicated in chromosome condensation across species, such as budding yeast and humans, whose chromosomes vary orders of magnitude in size and levels of condensation (1, 41). The field's present challenge is to incorporate all currently known pieces of the chromosome folding problem into both a physically and biologically consistent model that can be experimentally tested and refined. The new experimental approaches described here represent a fresh start to an old problem.
See companion article on page 15393 in issue 24 of volume 99.
References
- 1.Li G., Sudlow, G. & Belmont, A. S. (1998) J. Cell Biol. 140, 975-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmont A. S. (1997) in Genome Structure and Function, ed. Nicolini, C. (Kluwer Academic, Dordrecht, The Netherlands), pp. 261–276.
- 3.Tavormina P. A., Come, M. G., Hudson, J. R., Mo, Y. Y., Beck, W. T. & Gorbsky, G. J. (2002) J. Cell Biol. 158, 23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen M. O., Larsen, M. K., Barthelmes, H. U., Hock, R., Andersen, C. L., Kjeldsen, E., Knudsen, B. R., Westergaard, O., Boege, F. & Mielke, C. (2002) J. Cell Biol. 157, 31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirier M. G. & Marko, J. F. (2002) Proc. Natl. Acad. Sci. USA 99, 15393-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belmont A. S. (1998) Methods Cell Biol. 53, 99-124. [DOI] [PubMed] [Google Scholar]
- 7.Paulson J. R. & Laemmli, U. K. (1977) Cell 12, 817-828. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli U. K., Cheng, S. M., Adolph, K. W., Paulson, J. R., Brown, J. A. & Baumbach, W. R. (1978) Cold Spring Harbor Symp. Quant. Biol. 42, 351-360. [DOI] [PubMed] [Google Scholar]
- 9.Earnshaw W. C. & Laemmli, U. K. (1983) J. Cell Biol. 96, 84-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsden M. P. F. & Laemmli, U. K. (1979) Cell 17, 849-858. [DOI] [PubMed] [Google Scholar]
- 11.Adolph K. W. (1980) Exp. Cell Res. 125, 95-103. [DOI] [PubMed] [Google Scholar]
- 12.Lewis C. D. & Laemmli, U. K. (1982) Cell 29, 171-181. [DOI] [PubMed] [Google Scholar]
- 13.Jeppesen P. & Morten, H. (1985) J. Cell Sci. 73, 245-260. [DOI] [PubMed] [Google Scholar]
- 14.Okada T. A. & Comings, D. E. (1980) Am. J. Hum. Genet. 32, 814-832. [PMC free article] [PubMed] [Google Scholar]
- 15.Goyanes V. J., Matsui, S. & Sandberg, A. A. (1980) Chromosoma 78, 123-135. [DOI] [PubMed] [Google Scholar]
- 16.Labhart P., Koller, T. & Wunderli, H. (1982) Cell 30, 115-121. [DOI] [PubMed] [Google Scholar]
- 17.Paulson J. R. (1989) Chromosoma 97, 289-295. [DOI] [PubMed] [Google Scholar]
- 18.Razin S. V. (1996) Crit. Rev. Eukaryotic Gene Expression 6, 247-269. [DOI] [PubMed] [Google Scholar]
- 19.Mirkovich J., Gasser, S. M. & Laemmli, U. K. (1988) J. Mol. Biol. 200, 101-109. [DOI] [PubMed] [Google Scholar]
- 20.Cockerill P. N. & Garrard, W. T. (1986) Cell 44, 273-282. [DOI] [PubMed] [Google Scholar]
- 21.Earnshaw W. C. & Heck, M. M. S. (1985) J. Cell Biol. 100, 1716-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasser S. M., Laroche, T., Falquet, J., Boy de la Tour, E. & Laemmli, U. K. (1986) J. Mol. Biol. 188, 613-629. [DOI] [PubMed] [Google Scholar]
- 23.Saitoh N., Goldberg, I. G., Wood, E. R. & Earnshaw, W. C. (1994) J. Cell Biol. 127, 303-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strunnikov A. V., Hogan, E. & Koshland, D. (1995) Genes Dev. 9, 587-599. [DOI] [PubMed] [Google Scholar]
- 25.Hirano T. (2002) Genes Dev. 16, 399-414. [DOI] [PubMed] [Google Scholar]
- 26.Hirano T. & Mitchison, T. J. (1993) J. Cell Biol. 120, 601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warburton P. E. & Earnshaw, W. C. (1997) BioEssays 19, 97-99. [DOI] [PubMed] [Google Scholar]
- 28.Swedlow J. R., Sedat, J. W. & Agard, D. A. (1993) Cell 73, 97-108. [DOI] [PubMed] [Google Scholar]
- 29.Houchmandzadeh B., Marko, J. F., Chatenay, D. & Libchaber, A. (1997) J. Cell Biol. 139, 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirier M., Eroglu, S., Chatenay, D. & Marko, J. F. (2000) Mol. Biol. Cell 11, 269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poirier M. G., Monhait, T. & Marko, J. F. (2002) J. Cell. Biochem. 85, 422-434. [DOI] [PubMed] [Google Scholar]
- 32.Poirier M. G., Eroglu, S. & Marko, J. F. (2002) Mol. Biol. Cell 13, 2170-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirier M. G., Nemani, A., Gupta, P., Eroglu, S. & Marko, J. F. (2001) Phys. Rev. Lett. 86, 360-363. [DOI] [PubMed] [Google Scholar]
- 34.Schwarz P. M., Felthauser, A., Fletcher, T. M. & Hansen, J. C. (1996) Biochemistry 35, 4009-4015. [DOI] [PubMed] [Google Scholar]
- 35.Widom J. & Klug, A. (1985) Cell 43, 207-213. [DOI] [PubMed] [Google Scholar]
- 36.Belmont A. S., Braunfeld, M. B., Sedat, J. W. & Agard, D. A. (1989) Chromosoma 98, 129-143. [DOI] [PubMed] [Google Scholar]
- 37.McGhee J. D., Nickol, J. M., Felsenfeld, G. & Rau, D. C. (1983) Cell 33, 831-841. [DOI] [PubMed] [Google Scholar]
- 38.Belmont A. S. & Bruce, K. (1994) J. Cell Biol. 127, 287-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rattner J. B. & Lin, C. C. (1985) Cell 42, 291-296. [DOI] [PubMed] [Google Scholar]
- 40.Boy de la Tour E. & Laemmli, U. K. (1988) Cell 55, 937-944. [DOI] [PubMed] [Google Scholar]
- 41.Guacci V., Hogan, E. & Koshland, D. (1994) J. Cell Biol. 125, 517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]