Abstract
Trehalose is a nonreducing disaccharide of glucose that functions as a compatible solute in the stabilization of biological structures under abiotic stress in bacteria, fungi, and invertebrates. With the notable exception of the desiccation-tolerant “resurrection plants,” trehalose is not thought to accumulate to detectable levels in most plants. We report here the regulated overexpression of Escherichia coli trehalose biosynthetic genes (otsA and otsB) as a fusion gene for manipulating abiotic stress tolerance in rice. The fusion gene has the advantages of necessitating only a single transformation event and a higher net catalytic efficiency for trehalose formation. The expression of the transgene was under the control of either tissue-specific or stress-dependent promoters. Compared with nontransgenic rice, several independent transgenic lines exhibited sustained plant growth, less photo-oxidative damage, and more favorable mineral balance under salt, drought, and low-temperature stress conditions. Depending on growth conditions, the transgenic rice plants accumulate trehalose at levels 3–10 times that of the nontransgenic controls. The observation that peak trehalose levels remain well below 1 mg/g fresh weight indicates that the primary effect of trehalose is not as a compatible solute. Rather, increased trehalose accumulation correlates with higher soluble carbohydrate levels and an elevated capacity for photosynthesis under both stress and nonstress conditions, consistent with a suggested role in modulating sugar sensing and carbohydrate metabolism. These findings demonstrate the feasibility of engineering rice for increased tolerance of abiotic stress and enhanced productivity through tissue-specific or stress-dependent overproduction of trehalose.
The explosive increase in world population, along with the continuing deterioration of arable land, scarcity of fresh water, and increasing environmental stress pose serious threats to global agricultural production and food security. Despite focused efforts to improve major crops for resistance to abiotic stresses (1) such as drought, excessive salinity, and low temperature by traditional breeding, success has been limited. This lack of desirable progress is attributable to the fact that tolerance to abiotic stress is a complex trait that is influenced by coordinated and differential expression of a network of genes. Fortunately, it is now possible to use transgenic approaches to improve abiotic stress tolerance in agriculturally important crops with far fewer target traits than had been anticipated (2).
Abiotic stresses can directly or indirectly affect the physiological status of an organism by altering its metabolism, growth, and development. A common response of organisms to drought, salinity, and low-temperature stresses is the accumulation of sugars and other compatible solutes (3). These compounds serve as osmoprotectants and, in some cases, stabilize biomolecules under stress conditions (3, 4). One such compound is trehalose, a nonreducing disaccharide of glucose, which plays an important physiological role as an abiotic stress protectant in a large number of organisms, including bacteria, yeast, and invertebrates (5). Trehalose has been shown to stabilize dehydrated enzymes, proteins, and lipid membranes efficiently, as well as protect biological structures from damage during desiccation. In the plant kingdom, most species do not seem to accumulate detectable amounts of trehalose, with the notable exception of the highly desiccation-tolerant “resurrection plants” (6). The recent discovery of homologous genes for trehalose biosynthesis in Selaginella lepidophylla, Arabidopsis thaliana, and several crop plants suggests that the ability to synthesize trehalose may be widely distributed in the plant kingdom (7). A putative plant gene for trehalose-6-phosphate synthase (TPS) can complement a Δtps1 mutant yeast strain, suggesting that the plant and yeast gene products are functionally similar (8).
In bacteria and yeast, trehalose is synthesized in a two-step process: trehalose-6-phosphate is first formed from UDP-glucose and glucose-6-phosphate in a reaction catalyzed by TPS. Trehalose-6-phosphate is then converted to trehalose by trehalose-6-phosphate phosphatase (TPP) (7). Metabolic engineering for enhanced accumulation of trehalose in plants has been the recent focus of attention in some model dicot plants (9–12). However, in these previous studies, constitutive overexpression of TPS and/or TPP genes from yeast or Escherichia coli in tobacco or potato plants resulted in undesirable pleiotropic effects, including stunted growth and altered metabolism under normal growth conditions (10–12).
Considering the importance of rice as a major crop, developing new cultivars with enhanced abiotic stress tolerance would undoubtedly have an enormous impact on global food production. We decided to improve abiotic stress tolerance by transforming rice with a trehalose-6-phosphate synthase/phosphatase (TPSP) fusion gene (13) that includes the coding regions of the E. coli otsA and otsB genes (encoding TPS and TPP, respectively). This approach has the dual advantages of necessitating only a single transformation event and producing a higher net catalytic efficiency for trehalose formation (13). Because indica rice varieties represent 80% of rice grown worldwide, we chose to transform the economically valuable indica rice, Pusa Basmati-1 (PB-1), even though transformation and regeneration are more difficult than in japonica rice varieties.
Here, we show that engineering trehalose overproduction in rice can be achieved by stress-inducible or tissue-specific expression of bifunctional TPSP fusion enzyme without any detrimental effect on plant growth or grain yield. During abiotic stress, transgenic plants accumulated increased amounts of trehalose and showed high levels of tolerance to salt, drought, and low-temperature stresses, as compared with the nontransformed plant. These results demonstrate the potential use of our transgenic approach in developing new rice cultivars with increased abiotic stress tolerance and enhanced rice productivity.
Materials and Methods
Plasmid Constructs.
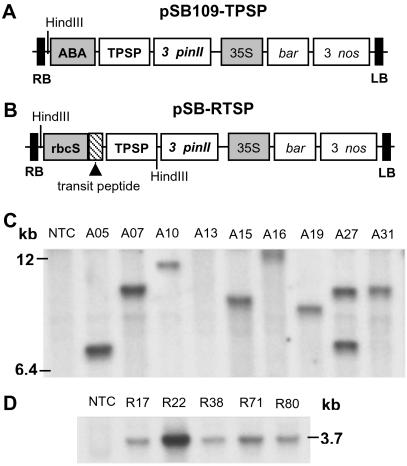
Two binary plasmids, pSB109-TPSP and pSB-RTSP, each containing a TPSP fusion gene (13), were constructed in the pSB11 vector (14) by using standard cloning and plasmid manipulation procedures. The components of the plasmid within the T-DNA region and the selected restriction enzyme sites are shown in Fig. 1 A and B. The expression cassette in pSB109-TPSP consists of an abscisic acid (ABA)-inducible promoter (15) that contains four tandem copies of ABA-inducible element ABRC1 (0.18 kb) coupled with a minimal rice actin 1 promoter (0.18 kb) and an HVA22 intron (0.24 kb). It is linked to the TPSP coding region (2.2 kb), which was constructed by fusing the otsA and otsB genes from E. coli after the stop codon of the otsA gene had been removed by PCR (13) and then ligated to the potato protease inhibitor II gene (pinII) 3′ noncoding sequence (1.0 kb). The selection cassette includes the cauliflower mosaic virus 35S promoter (0.74 kb), phosphinothricin acetyltransferase gene (bar, 0.59 kb), and the nopaline synthase gene 3′ noncoding sequence (Nos 3′, 0.28 kb). In pSB-RTSP, a 1.3-kb fragment of the rice rbcS promoter (16) with a chloroplast-targeting transit peptide (0.16 kb) is linked to the TPSP coding region; the remaining components are similar to those in pSB109-TPSP. During the cloning and ligation of an ≈3.7-kb DNA fragment containing the rbcS promoter/transit peptide and TPSP fusion gene into the plasmid pSB-RTSP, three additional restriction sites (SacI, SalI, and HindIII) were added between TPSP and 3′ pin II. Both the plasmids (pSB109-TPSP and pSB-RTSP) were separately transferred to Agrobacterium tumefaciens strain LBA4404 harboring the pSB1 vector (14) through triparental mating using the helper plasmid pRK2013. For cocultivation, the bacteria were grown from a single colony in liquid AB medium containing 50 mg/liter spectinomycin at 30°C for 3 days and were suspended at a density of 3 × 109 cells per ml in AAM medium (17) for rice transformation.
Fig 1.
Schematic representation of the expression vectors and DNA-blot hybridization analysis. Two binary plasmids, each containing the trehalose biosynthetic fusion gene (TPSP) that includes the coding regions of the E. coli otsA and otsB genes (encoding TPS and TPP, respectively), were constructed and transformed into indica rice, as described in Materials and Methods. (A) pSB109-TPSP plasmid. (B) pSB-RTSP plasmid. Shaded boxes represent promoter elements (ABA, ABA-inducible; rbcS, rice rbcS; 35S, cauliflower mosaic virus 35S); RB and LB represent T-DNA border on the right and left sides, respectively. Shown is DNA-blot hybridization analysis from nontransformed control (NTC) plant, and representative transgenic plants of nine A-lines (C) and five R-lines (D) that were transformed with the plasmid pSB109-TPSP and pSB-RTSP, respectively. The rice genomic DNA was digested with HindIII (a unique site in the plasmid pSB109-TPSP, whereas two sites are present in the plasmid pSB-RTSP) and DNA blot hybridization analysis was performed with the 2.2-kb TPSP fusion gene as the probe. Molecular sizes (kb) are indicated.
Generating Transgenic Rice Plants.
Mature seeds of indica rice variety PB-1 were dehusked and sterilized in 70% (vol/vol) ethanol for 2–3 min and then transferred into 50% (vol/vol) Clorox solution for 40 min with gentle shaking. The seeds were rinsed several times with sterile water. The sterilized PB-1 seeds were then plated for callus induction on Murashige and Skoog (MS) medium (Sigma) supplemented with 3.0 mg/liter 2,4-dichlorophenoxyacetic acid (2,4-D)/0.2 mg/liter 6-benzylaminopurine (BAP)/300 mg/liter casein hydrolysate (CH)/30 g/liter maltose/3.0 g/liter phytagel, pH 5.8 (MSCl) and grown for 21 days at 25°C in the dark. Three weeks after callus induction from the scutellar region of the rice embryo, 150 embryogenic calli were immersed in A. tumefaciens suspension for 10 min. Infected calli were cocultivated in MSCl medium supplemented with 10 g/liter glucose/100 μM acetosyringone, pH 5.2 (MSCC). After 3 days of cocultivation, calli were washed with sterile water containing 250 mg/liter cefotaxime and blotted on filter paper. The calli were immediately plated on a selection medium, MSCl medium, supplemented with 6 mg/liter bialaphos (a gift from H. Anzai, Meiji Seika Kaisha, Japan) and 250 mg/liter cefotaxime, pH 5.8 (MSS), and incubated at 25°C in the dark for 2–3 weeks. The microcalli that had proliferated after the initial selection were further subcultured for two selection cycles on fresh MSS medium every 2 weeks. The actively dividing bialaphos-resistant calli were plated on MS plant regeneration medium containing 2.5 mg/liter BAP/1.0 mg/liter kinetin/0.5 mg/liter naphthaleneacetic acid (NAA)/300 mg/liter CH/30 g/liter maltose/4 mg/liter bialaphos/250 mg/liter cefotaxime/2.0 g/liter phytagel, pH 5.8 (MSPR) and grown at 25°C for a 10-h light/14-h dark photoperiod for 3–4 weeks. The regenerated plantlets were acclimatized hydroponically in Yoshida nutrient solution (18) for 10 days. Later on, putative primary transformants (T0 generation) were transferred to pots and tested for Basta-herbicide resistance (19); the transgenic plants were grown to maturity in a greenhouse for further analysis.
DNA-Blot Hybridization Analysis.
Leaves from nontransformed control (NTC) plant, and representative (T0) transformants of nine A-lines (ABA-inducible promoter) and five R-lines (rbcS promoter) that were transformed with the plasmid pSB109-TPSP and pSB-RTSP, respectively, were ground in liquid nitrogen by using a motor and pestle. Rice genomic DNA was isolated by the guanidine-detergent lysis method by using DNAzolES (Molecular Research Center, Cincinnati) following the manufacturer's instructions. Five micrograms of the genomic DNA was digested overnight with HindIII restriction enzyme, fractionated through 0.8% agarose gel, alkali-transferred onto Hybond N+ nylon membrane (Amersham Pharmacia), and hybridized with an α-32P-labeled 2.2-kb TPSP fusion gene (13) as the probe. DNA probe preparation, hybridization, and washing of the membrane were performed as described (19). The α-32P-labeled membrane was exposed onto autoradiogram.
Detecting Trehalose and Soluble Carbohydrates.
Soluble carbohydrates were extracted as described (10). Extracts from 0.5 g of homogenized fresh leaf tissue were centrifuged (10 min at 3,220 × g); supernatants were passed through ion-exchange columns consisting of 1 ml of Amberlite IR-68 (acetate form) layered on 1 ml of Dowex 50W (hydrogen form) to remove charged compounds. After lyophilization, samples were dissolved in HPLC-grade water and subjected to high-performance anion exchange chromatography with pulsed amperometric detection by using a Dionex DX-500 series chromatograph equipped with a Carbopac PA-1 analytical column and a Carbopac PA-1 guard column (Dionex). Carbohydrates were eluted at a flow rate of 1.0 ml per min at 1,400 psi with 100 mM NaOH for 34 min. Major soluble carbohydrates present were quantified by using authentic standard sugars (Sigma). The identity of trehalose in the plant extracts was confirmed by incubating samples with porcine-kidney-derived trehalase enzyme (Sigma).
Salt Stress Tolerance and Determination of Plant Mineral Nutrients.
Ten seedlings for each T4 generation transgenic line (R22, R38, R80, A05, A07, and A27) and NTC were grown hydroponically (with modest aeration) in Yoshida nutrient solution (18) in a growth chamber at 25 ± 3°C for a 10-h light/14-h dark photoperiod (photon flux density of 280 μmol photons per m/s) and with relative humidity of 50–60%. After 5 weeks, 50% of the seedlings were subjected to 100 mM NaCl stress (conductivity of 10–12 dS/m). Nutrient solutions were replaced every week. After 4 weeks of continuous salt stress, shoot and root samples were separately harvested for fresh and dry weight determination. For mineral nutrient analysis, 150 mg of ground dry matter was digested in concentrated HNO3 overnight at 120°C. Samples then were dissolved in HNO3:HClO4 (1:1, vol/vol) at 220°C, resuspended in 5% (vol/vol) HNO3, and analyzed for elemental composition of sodium (Na+), potassium (K+), calcium (Ca2+), and iron (Fe) by means of simultaneous inductively coupled argon-plasma emission spectrometry (ICP trace analyzer; Plant, Soil, and Nutrition Laboratory, U.S. Department of Agriculture-Agriculture Research Service, Cornell University, Ithaca, NY).
Drought and Low-Temperature Stress Tolerance.
Seedlings from six independent T4 transgenic lines and nontransformed line were grown individually in 10-cm × 10-cm pots irrigated with Yoshida nutrient solution for 5 weeks before performing the drought- or low-temperature stress experiment. Drought stress (water deficit) was conducted by first withholding irrigation for 3 days to allow the soil in the pot to dry. Then, the first drought cycle of 100 h was initiated, followed by rewatering for 2 days. The drought-stress cycle was repeated for another 100 h, and the plants were allowed to recover by watering every day for 3 weeks. Low-temperature stress was conducted on five-week-old seedlings by exposing them to 10°C for 72 h under a 10-h light/14-h dark photoperiod (photon flux density of 280 μmol photons per m per s) and a relative humidity of 50–60%; the seedlings were then allowed to recover under normal growth conditions at 25 ± 3°C.
Protein Extraction and Immunoblotting.
Proteins were extracted from 0.2 g of homogenized fresh leaf tissue in protein extraction buffer (20 mM Tris⋅HCl, pH 8.0/10 mM EDTA/30 mM NaCl/2 mM phenylmethane sulfonyl fluoride for 1 h at 4°C). The homogenate was clarified by centrifugation at 12,000 × g for 15 min at 4°C. The procedure for immunoblotting was essentially the same as described (20). The anti-TPSP protein polyclonal antibody was used at a 1:1,500 dilution for Western blot analysis, using an alkaline phosphatase color reaction for detection of the protein, as per the manufacturer's instruction (Bio-Rad).
Chlorophyll Fluorescence Parameters.
Fv/Fm and φPSII were measured by using a pulse amplitude modulated fluorometer (FMS2, Hansatech Instruments, Pentney King's Lynn, U.K.) to estimate photo-oxidative damage to the Photosystem II (PS II) reaction center and the quantum efficiency of PS II photochemistry under ambient light conditions, respectively, as described (21). Measurements were made on the youngest, fully expanded leaves. Measurements of φPSII were first determined under ambient light; the same leaves were then dark-adapted for 10 min before measurement of Fv/Fm.
Results and Discussion
Transgenic Plants with Enhanced Trehalose Levels Are Phenotypically Normal and Fertile.
Two plasmid constructs, pSB109-TPSP (Fig. 1A) and pSB-RTSP (Fig. 1B), each containing the TPSP fusion gene, were introduced into indica rice cells of PB-1 by Agrobacterium-mediated gene transfer (17). In the plasmid construct pSB109-TPSP, an ABA and stress-inducible promoter (15) drives the fusion gene for cytosolic expression. In the other plasmid, pSB-RTSP, the light-regulated promoter (16) of the Rubisco small subunit gene, rbcS, from Oryza sativa with a transit peptide drives the fusion gene for chloroplast targeting in the leaf mesophyll cells. A large number of putative transgenic PB-1 plants (T0 generation) were regenerated (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org); these plants included 28 A-lines (ABA-inducible promoter) and 76 R-lines (rbcS promoter). Integration of the TPSP transgene was confirmed by DNA-blot hybridization analysis (Fig. 1 C and D). Based on the T-DNA junction fragment analysis, ≈40% of the transgenic plants transformed with either of the plasmids harbor a single copy, and 35–45% of plants harbor two or three copies of the transgene.
Most of the 90 independent primary transformants (T0) that contained a low copy number of the transgene showed a normal phenotype and were completely fertile. In contrast to previous reports that used constitutive promoters driving individual TPS and/or TPP genes, the use of stress-inducible or tissue-specific promoters in this work appears to minimize the negative effects of the transgene on plant growth. The T0 plants were self-pollinated to obtain segregating T1 progeny for genetic and HPLC analysis. Forty-five transgenic lines showed a segregation pattern of 3:1 for the basta-herbicide resistance marker gene. HPLC analysis of leaf extracts showed that transgenic lines had a trehalose content that was between three times and eight times that of the nontransgenic plants (17 ± 5 μg of trehalose per g of fresh weight). The identity of trehalose in the plant tissue extracts was confirmed by incubating samples in porcine trehalase followed by chromatographic analysis of the monosaccharide products (Fig. 6, which is published as supporting information on the PNAS web site). Physiological experiments were conducted for abiotic stress tolerance on homozygous plants through the T4 generation, because gene silencing has been reported to occur in the T3 generation, even though T2 and T1 generation plants were not silenced (22). The results from many independent transgenic lines were consistent for salt- and drought-stress tolerance in each generation, except in few transgenic lines which had multiple copies of the transgene (data not shown).
Transgenic Plants Are Salt Tolerant and Maintain Balanced Mineral Nutrition.
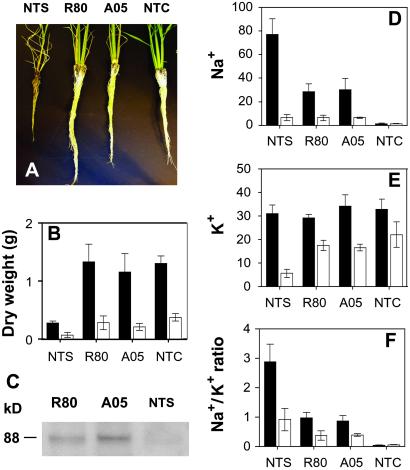
The T4 transgenic plants with either one or two copies of the transgene showed markedly enhanced salt tolerance during and subsequent to 4 weeks of 100 mM NaCl treatment under hydroponic growth conditions. Six independent transgenic plant lines (three A-lines and three R-lines) were analyzed in detail. For clarity of presentation, results from two representative transgenic lines (R80 and A05) are shown (Fig. 2); results for the other four lines were very similar to the two lines presented. After prolonged exposure to salt stress, almost all of the transgenic plants survived and displayed vigorous root and shoot growth. In contrast, all of the nontransformed stressed (NTS) plants were either dead or nearly dead because of severe salt damage to the leaves and concomitant loss of chlorophyll. Transgenic plants developed longer and thicker roots than NTS plants after salt stress (Fig. 2A). Salt stress severely inhibited the growth of shoot and roots of NTS plants, as indicated by their lower dry weights compared with NTC plants. Shoot and root dry weights of both the transgenic lines (Fig. 2B) approached those of NTC plants, and after removal of salt stress, the transgenic plants were able to grow, flower, and set normal viable seeds. To determine whether the TPSP gene product was present in the salt-stressed plants, total protein was isolated from the leaf samples for Western blot analysis. Immunoblot analysis using polyclonal antibodies raised against the fusion protein showed the presence of a protein with the expected apparent molecular mass of 88 kDa only in the transgenic plants (Fig. 2C).
Fig 2.
Salt tolerance of rice plants and changes in mineral nutrition caused by salt stress. (A) Plant roots after 4 weeks of continuous 100 mM NaCl stress; the plants were not stressed in NTC. (B) Dry weight of shoots (black bars) and roots (white bars) of plants grown under salt stress (NTS, R80, and A05) or no stress (NTC) conditions. (C) Western blots of leaf extracts (20 μg of proteins) immediately after salt stress of plants. (D–F) Plant mineral nutrient content in shoots (black bars) and roots (white bars) under salt stress (NTS, R80, and A05) or no stress (NTC) conditions. (D) Na+. (E) K+. (F) Na+/K+ ratio. The ionic concentration is presented as mg/g dry weight. Values are the means ± SD (n = 5).
To assess how trehalose accumulation in transgenic rice affected plant mineral nutrition during salt stress, shoot and root mineral content for the six independent transgenic lines and two nontransgenic lines were determined by using inductively coupled plasma emission spectrometry (Table 2, which is published as supporting information on the PNAS web site). After continuous salt stress (100 mM NaCl) for 4 weeks, NTS plants showed a very large increase in Na+ content in both shoots and roots compared with NTC, whereas the increase in the shoots of all of the transgenic plants was much smaller (Fig. 2D). The Na+ content of transgenic plant shoots was only 30–35% of the NTS plants after salt stress. The observed differences in shoot Na+ content between transgenic and NTS plants could be caused in part by a growth dilution because of the much faster growth rate of the transgenic plants under salt stress. Alternatively, trehalose might have played a direct or indirect role in maintaining ion selectivity and, thus, facilitating cellular Na+ exclusion. This possibility is consistent with the report that in salt-stressed rice seedlings, the accumulation of Na+ in leaf tissues was not prevented by exogenous proline. In contrast, treatment with exogenous trehalose significantly reduced the salt-induced accumulation of Na+ in the leaves (23).
Transgenic lines R80 and A05 maintained shoot to root K+ homeostasis both under nonstress and salt-stress conditions (Table 2). After salt stress, the levels of shoot and root K+ content in transgenic plants was similar to the nonstressed controls, while a fourfold decrease in root K+ in the NTS plants was seen (Fig. 2E). Thus, the transgenic plants were able to maintain a higher level of selectivity for K+ over Na+ uptake in the roots and Na+ exclusion from the shoots compared with the NTS plants. The maintenance of the Na+/K+ ratio in both shoot and roots of transgenic plants (Fig. 2F) correlated with nearly normal plant growth and may be the basis for minimizing Na+ toxicity under salt stress. It is generally accepted that the maintenance of Na+/K+ homeostasis is an important aspect of salt tolerance (24, 25).
Several other changes in plant mineral status that may have played indirect roles in stress tolerance were seen in the transgenic lines compared with the NTCs. It was found that salt stress led to a significant increase in root and shoot Ca2+ content in the NTS lines, whereas in the transgenic lines, this Na-mediated increase in Ca2+ content was only observed in the shoots and not the roots (Table 2). This rise in Ca2+ may be caused by alterations in the ion selectivity of the transporters at high concentrations of Na+ (25). We also found significantly higher levels of shoot Fe ion content in the transgenic lines compared with the NTCs (Table 2). It has been well documented that Fe, Cu, and Zn ions are essential for the function of critical antioxidant enzymes such as the superoxide dismutases that play a role in scavenging reactive oxygen species during a number of abiotic stresses (25, 26). In general, the relationship between salt stress and plant mineral content is complex, and the links between elevated trehalose content and improved mineral status during salt stress are not known.
Transgenic Plants Are Drought Tolerant.
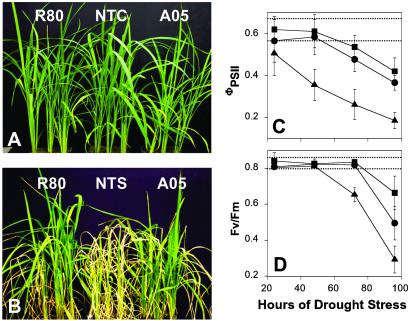
To study drought tolerance, 5-week-old nontransformed and transgenic seedlings grown in soil were subjected to two cycles of 100 h of drought stress. After the drought treatments, all 15 plants of each line showed wilting and drought-induced rolling of the young leaves. Nontransgenic plants exhibited rolling of leaves within 48 h of the stress as compared with considerably fewer visual symptoms in transgenic plants during the same time period. After two cycles of 100 h of drought stress and subsequent watering for 3 weeks, the growth of both the transgenic lines, R80 and A05 (Fig. 3B), were almost identical to nonstressed control plant (Fig. 3A). In contrast, the growth of the drought-stressed NTS was severely inhibited (Fig. 3B).
Fig 3.
Appearance of plants and chlorophyll fluorescence parameters during drought stress. Five-week-old nontransformed and T4 generation transgenic (R80 and A05) seedlings grown in soil were subjected to two cycles of 100 h of drought stress followed by watering for 3 weeks. (A) Plants grown under well watered conditions (NTC, nontransgenic plants). (B) Plants of the same age after two cycles of drought-stress treatment (NTS, nontransgenic plants after drought stress). (C and D) Chlorophyll fluorescence measurements on young, fully expanded leaves during the first cycle of 100 h of continuous drought stress. (C) ΦPSII, a measure of the efficiency of PS II photochemistry under ambient growth conditions. (D) Decreases in Fv/Fm are a measure of photooxidative damage to PS II. ▴, nontransformed plants; ▪, R80; •, A05. Dotted lines represent the range of values for nonstressed control plants of all lines. Data represent means ± SD (n = 5) from independent plants.
Transgenic Plants Produced Increased Amounts of Trehalose and Other Soluble Carbohydrates.
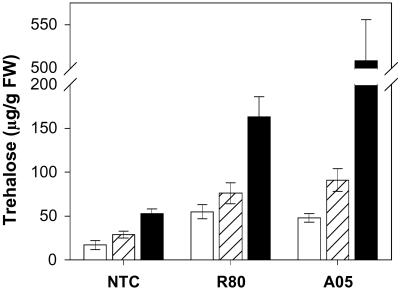
To evaluate our hypothesis that trehalose accumulation in plants might act as a positive regulator of stress tolerance, we measured the levels of trehalose and other soluble carbohydrates (Table 3, which is published as supporting information on the PNAS web site). A low but significant amount of trehalose was detected in the shoots (17 μg/g fresh weight) of NTC plants; these levels increased significantly under salt or drought stresses. The transgenic plants grown under control conditions exhibited trehalose levels comparable with the NTS plants (Fig. 4). After salt stress, the transgenic lines (R80 and A05) showed 2.5–3 times higher shoot trehalose levels compared with NTS plants, whereas after drought stress, trehalose levels in the transgenic lines increased 3- to 9-fold (Fig. 4). Despite the similarities in tolerance levels exhibited by transgenic plants engineered to increase trehalose synthesis in either the cytosol or chloroplast, R-lines showed considerable protection at much lower trehalose concentrations during drought stress (Table 3). In general, there was no obvious relationship between trehalose accumulation and stress tolerance among the transgenic lines evaluated (data not shown). On the other hand, the difference in trehalose levels between the transgenic and nontransgenic lines clearly correlates with increased tolerance to abiotic stress.
Fig 4.
Trehalose content in shoots of transgenic (R80 and A05) and nontransgenic plants with or without stress. Trehalose accumulation under nonstressed (white bars), salt-stressed (100 mM NaCl for 4 weeks, hatched bars), or drought-stressed (100 h, black bars) conditions.
Transgenic Plants Show Improved Photosystem II Function.
During many different abiotic stresses, a reduction in photosynthesis and the subsequent production of reactive oxygen species are thought to be a major contributor to decreased plant performance and photooxidative damage. The effects of increased trehalose accumulation on photosynthesis during drought stress were assessed by determination of the quantum yield of PS II photochemistry (ΦPSII) by using in vivo chlorophyll fluorescence techniques (21). ΦPSII is a measure of the photosynthetic performance of the plant under ambient light conditions. After the first cycle of 100 h of drought stress, the quantum yield of PS II photochemistry in NTS plants decreased by ≈68%, whereas the activity of the two best-performing transgenic lines (R80 and A05) only decreased by 29–37% compared with the nonstressed controls (Fig. 3C). Similarly, drought-induced decreases in the fluorescence parameter Fv/Fm, which is a measure of accumulated photo-oxidative damage to PS II, were considerably smaller in the transgenic lines than in the NTS plants (Fig. 3D). In other independent experiments, similar results were obtained for both low-temperature stress (Fig. 7, which is published as supporting information on the PNAS web site) and salt stress (data not shown), indicating the common role that maintenance of photosynthetic capacity plays in tolerance to these stresses.
Transgenic Plants Have Increased Photosynthetic Capacity Under Nonstress Conditions.
Improved photosynthesis under abiotic stress conditions is known to limit photo-oxidative damage and permit continued growth (27) and is clearly suggested by the data in Fig. 3. Under the same conditions, transgenic plants exhibited soluble carbohydrate levels that were ≈20% higher than those of corresponding NTC plants, including subtle changes in levels of glucose, fructose, and sucrose (Table 3). Both of these results are consistent with the suggestion that trehalose may be involved in sugar sensing and modulating carbon metabolism (7, 28). The ability of trehalose to modulate photosynthetic capacity has been demonstrated recently (29) in transgenic tobacco plants expressing E. coli trehalose biosynthetic genes. Plants with enhanced TPS expression exhibited a higher photosynthesis per unit of leaf area than nontransgenic controls, whereas those over-expressing TPP showed diminished rates of photosynthesis. These data lead them to conclude that it is trehalose-6-P and not trehalose that is modulating photosynthetic capacity (29).
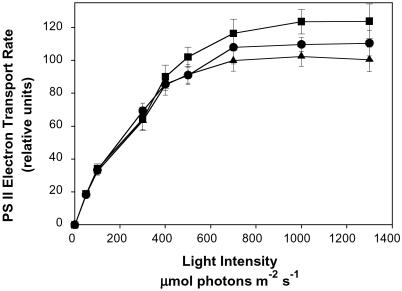
Fig. 5 shows the light intensity dependence of PS II electron transport rates, as determined by φPSII measurements (21) for nontransgenic rice and transgenic lines R80 and A05 measured under control (nonstress) conditions. Although the differences in photosynthesis are small at limiting light intensities, at light saturation, the rates of photosynthesis in the transgenic plants are 5–15% higher than in the NTCs. At light saturation, photosynthetic rate is limited by the capacity of the dark reactions, in particular the Calvin cycle and triose phosphate utilization in the cytoplasm (27). Together with the observed higher levels of soluble carbohydrate under both stress and nonstress conditions (Table 3), the elevated levels of light-saturated photosynthesis in the transgenic plants supports the suggestion that in plants, trehalose acts as a regulator of sugar sensing and, thus, the expression of genes associated with carbon metabolism (29). The presence of a higher capacity for photosynthesis before stress provides a larger sink for the products of photosynthesis during stress, thus limiting the extent of excess-light-induced photooxidative damage and accounting, in part, for the more vigorous growth of the transgenic lines during stress. Interestingly, the higher efficiency of trehalose synthesis by the TPSP fusion gene product (13) would suggest that trehalose, rather than trehalose-6-P is leading the enhanced capacity for photosynthesis.
Fig 5.
Photosystem II electron transport rate in nontransformed and two independent, fifth generation transgenic plants grown under control conditions. The electron transport rate under increasing irradiance was calculated from chlorophyll fluorescence measurements on the youngest fully expanded leaf of NTC (▴), R80 (▪), and A05 (•) at 360 ppm of CO2, 25°C, and 50% relative humidity after 10 weeks of growth. Values are the means ± SD (n = 9). Data are normalized to the average light-saturated rate of the nontransgenic control plants.
Conclusions
We have demonstrated that regulated overexpression of trehalose biosynthetic genes in rice has considerable potential for improving abiotic stress tolerance and, at the same time, augmenting productivity under both stress and nonstress conditions. This work showed successful conferment of tolerance to multiple abiotic stresses by means of overexpression of trehalose biosynthesis without the negative pleiotropic effects seen in previous studies. The modest increase in trehalose levels in transgenic lines, using either the tissue-specific or stress-dependent promoters, resulted in a higher capacity for photosynthesis and a concomitant decrease in the extent of photo-oxidative damage during stress. In addition, trehalose must be interacting with other physiological processes to account for changes in ion uptake and partitioning during salt stress. Because other cereal crops, like rice, are also sensitive to abiotic stresses, it is likely that overexpression of trehalose biosynthetic genes in maize and wheat may also confer high levels of abiotic stress tolerance.
Supplementary Material
Acknowledgments
We thank A. Jagendorf, M. Hanson, W. B. Miller, and T. L. Setter for critical review of the manuscript. We also thank J. Lee, H. Manslank, and A. S. Stolfi for technical assistance. This research was supported in part by Rockefeller Foundation Grant RF 98001-606 (to R.J.W.), by a postdoctoral fellowship (to A.K.G.) from the Rockefeller Foundation, and by grants from the Ministry of Science and Technology of Korea through the Crop Functional Genomics Center (to J.-K.K. and Y.D.C.).
Abbreviations
TPS, trehalose-6-phosphate synthase
TPP, trehalose-6-phosphate phosphatase
TPSP, TPS/phosphatase
PB-1, Pusa Basmati-1
ABA, abscisic acid
NTC, nontransgenic control
NTS, nontransgenic stressed
PS II, Photosystem II
References
- 1.Boyer J. S. (1982) Science 218, 443-448. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H. X., Hodson, J. N., Williams, J. P. & Blumwald, E. (2001) Proc. Natl. Acad. Sci. USA 98, 12832-12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hare P. D., Cress, W. A. & van Staden, J. (1998) Plant Cell Environ. 21, 535-553. [Google Scholar]
- 4.Yancey P. H., Clark, M. E., Hand, S. C., Bowlus, R. D. & Somero, G. N. (1982) Science 217, 1214-1222. [DOI] [PubMed] [Google Scholar]
- 5.Crowe J. H., Hoekstra, F. A. & Crowe, L. M. (1992) Annu. Rev. Physiol. 54, 579-599. [DOI] [PubMed] [Google Scholar]
- 6.Wingler A. (2002) Phytochemistry 60, 437-440. [DOI] [PubMed] [Google Scholar]
- 7.Goddijn O. J. & van Dun, K. (1999) Trends Plant Sci. 4, 315-319. [DOI] [PubMed] [Google Scholar]
- 8.Zentella R., Gallardo, J. O. M., Van Dijck, P., Mallol, J. F., Bonini, B., Van Vaeck, C., Gaxiola, R., Covarrubias, A. A., Sotelo, J. N., Thevelein, J. M. & Iturriaga, G. (1999) Plant Physiol. 119, 1473-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmstrom K. O., Mantyla, E., Welin, B., Mandal, A. & Palva, E. T. (1996) Nature 379, 683-684. [Google Scholar]
- 10.Goddijn O. J., Verwoerd, T. C., Voogd, E., Krutwagen, R. W., de Graaf, P. T., van Dunn, K., Poels, J., Ponstein, A. S., Damm, B. & Pen, J. (1997) Plant Physiol. 113, 181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero C., Belles, J. M., Vaya, J. L., Serrano, R. & Culianez-Macia, F. A. (1997) Planta 201, 293-297. [DOI] [PubMed] [Google Scholar]
- 12.Pilon-Smits E. A. H., Terry, N., Sears, T., Kim, H., Zayed, A., Hwang, S., van Dun, K., Voogd, E., Verwoerd, T. C., Krutwagen, R. W. & Goddijn, O. J. M. (1998) J. Plant Physiol. 152, 525-532. [Google Scholar]
- 13.Seo H. S., Koo, Y. J., Lim, J. Y., Song, J. T., Kim, C. H., Kim, J. K., Lee, J. S. & Choi, Y. D. (2000) Appl. Environ. Microbiol. 66, 2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komari T., Hiei, Y., Saito, Y., Murai, N. & Kumashiro, T. (1996) Plant J. 10, 165-174. [DOI] [PubMed] [Google Scholar]
- 15.Su J., Shen, Q., David Ho, T. H. & Wu, R. (1998) Plant Physiol. 117, 913- 922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyozuka J., McElroy, D., Hayakawa, T., Xie, Y., Wu, R. & Shimamoto, K. (1993) Plant Physiol. 102, 991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiei Y., Ohta, S., Komari, T. & Kumashiro, T. (1994) Plant J. 2, 271-282. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida S., Forno, D. A., Cook, J. H. & Gomez, K. A., (1976) Laboratory Manual for Physiological Studies of Rice (International Rice Research Institute, Philippines), pp. 61–66.
- 19.Roy M. & Wu, R. (2001) Plant Sci. 160, 869-875. [DOI] [PubMed] [Google Scholar]
- 20.Xu D., Duan, X., Wang, B., Hong, B., Ho, T. & Wu, R. (1996) Plant Physiol. 110, 249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saijo Y., Hata, S., Kyozuka, J., Shimamoto, K. & Izui, K. (2000) Plant J. 23, 319-327. [DOI] [PubMed] [Google Scholar]
- 22.Iyer L. M., Kumpatla, S. P., Chandrasekharan, M. B. & Hall, T. C. (2000) Plant Mol. Biol. 43, 323-346. [DOI] [PubMed] [Google Scholar]
- 23.Garcia A. B., de Engler, J. A., Iyer, S., Gerats, T., van Montagu, M. & Caplan, A. (1997) Plant Physiol. 115, 159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rus A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B. H., Matsumoto, T. K., Koiwa, H., Zhu, J. K., Bressan, R. A. & Hasegawa, P. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epstein E. (1998) Science 280, 1906-1907. [DOI] [PubMed] [Google Scholar]
- 26.Alscher R. G., Erturk, N. & Heath, L. S. (2002) J. Exp. Bot. 53, 13311341. [PubMed] [Google Scholar]
- 27.Owens T. G. (1996) in Photosynthesis and the Environment, ed. Baker, N. R. (Kluwer, Dordrecht, The Netherlands), pp. 1–23.
- 28.Thevelein J. M. & Hohmann, S. (1995) Trends Biochem. Sci. 20, 3-10. [DOI] [PubMed] [Google Scholar]
- 29.Paul M., Pellny, T. & Goddijn, O. (2001) Trends Plant Sci. 6, 197-200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.