Abstract
The RFN element is a highly conserved domain that is found frequently in the 5′-untranslated regions of prokaryotic mRNAs that encode for flavin mononucleotide (FMN) biosynthesis and transport proteins. We report that this domain serves as the receptor for a metabolite-dependent riboswitch that directly binds FMN in the absence of proteins. Our results also indicate that in Bacillus subtilis, the riboswitch most likely controls gene expression by causing premature transcription termination of the ribDEAHT operon and precluding access to the ribosome-binding site of ypaA mRNA. Sequence and structural analyses indicate that the RFN element is a natural FMN-binding aptamer, the allosteric character of which is harnessed to control gene expression.
The expression of certain genes is controlled by mRNA elements that form receptors for target metabolites. Selective binding of a metabolite by such an mRNA “riboswitch” permits a shift in the conformation to an alternative structure that results ultimately in the modulation of protein synthesis. For example, the btuB gene of Escherichia coli carries a highly conserved sequence element termed the B12 box (1) in the 5′ UTR of the mRNA that directly binds coenzyme B12 with high selectivity (2). This binding event seems to operate via an allosteric mechanism that represses expression of a reporter gene to ≈1% of that observed in cells grown in the absence of added coenzyme B12. Similarly, many organisms carry a highly conserved “thi box” sequence (3) in mRNAs that are required for the biosynthesis of the coenzyme thiamine pyrophosphate. An expanded region of RNA that encompasses the thi box also has been shown (4) to function as a riboswitch by directly binding thiamine pyrophosphate, resulting in reduced translation of thiM and thiC mRNAs in E. coli. It is conceivable that riboswitches also could modulate transcription termination, although only proteins (5, 6) or RNAs are currently known to regulate transcription termination in trans (7).
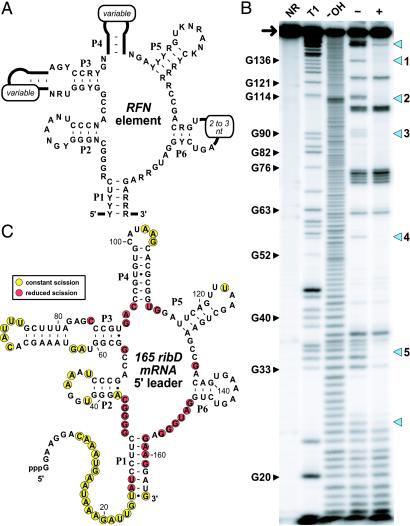
Another highly conserved but distinct RNA domain, termed the RFN element, has been identified in mRNAs of prokaryotic genes required for the biosynthesis of riboflavin and FMN (8, 9). It is known (10, 11) that FMN is required for down-regulation of the ribDEAHT operon (hereafter termed ribD) of Bacillus subtilis, which encodes several FMN biosynthetic enzymes. Furthermore, mutations within the RFN element of ribD eliminate FMN-mediated regulation (12, 13). Sequence comparisons of RFN elements from various riboflavin biosynthesis genes and ypaA (a putative riboflavin transport protein) (8, 14) have been used to generate a secondary structure model (Fig. 1A) and to establish the conserved nucleotides of this domain (8, 9). The structural model is composed of a six-stem junction wherein extensive sequence conservation exists at the bases of the stem elements and among the intervening nucleotides that form the core of the junction (Fig. 1A).
Fig 1.
FMN-induced structure modulation of an RFN element. (A) Structural model and conserved sequences of the RFN element derived from a phylogenetic analysis of prokaryote mRNA sequences. Nucleotides defined by letters are present in >90% of the RNAs examined. The letters R, Y, K, and N represent purine, pyrimidine, G or U, and any nucleotide identity, respectively. The six stem–loop structures are labeled P1–P6. The original model and the sequence data used to create this adaptation are described in ref. 9. (B) Structural probing of the 165 ribD RNA (arrow) in the presence (+) and absence (−) of 10 μM FMN. Also, 5′ 32P-labeled ribD RNA precursors were not reacted (NR) or were subjected to partial digest with RNase T1 (T1) or alkali (-OH) as indicated. Blue arrowheads indicate regions of FMN-dependent modulation of spontaneous cleavage. Regions 1 (nucleotide 136), 2 (nucleotides 113 and 114), 3 (nucleotides 89–91), 4 (nucleotide 56), and 5 (nucleotides 33 and 34) were subsequently used to establish an apparent KD value for FMN binding (Fig. 2). See Materials and Methods for experimental details. (C) Secondary-structure model of the 165 ribD RNA and the nucleotide positions of spontaneous cleavage in the presence (yellow) and absence (yellow and red) of FMN as identified from B. Cleavage occurs 3′ relative to the nucleotides highlighted in color.
It has been proposed that either an unidentified FMN-dependent protein effector (15) or perhaps FMN alone (8, 9) binds to the RFN element to repress ribD expression in B. subtilis. To explore the possibility that the RFN element serves as a component of an FMN-dependent riboswitch, we prepared RNAs corresponding to the 5′-UTR sequences of B. subtilis ribD and ypaA mRNAs by in vitro transcription. The RNAs were found to undergo FMN-dependent structural alterations in a protein-free mixture. Initial analyses suggest that FMN binding by the ribD leader causes transcription termination, whereas FMN binding by the ypaA leader results in sequestration of an adjacent ribosome-binding site. These results suggest that the RFN element serves as a component of a metabolite-dependent riboswitch that can bind FMN in the absence of proteins. In addition, FMN binding is likely to be responsible for down-regulating the expression of ribD and ypaA via distinct genetic control mechanisms.
Materials and Methods
Oligonucleotides and Chemicals.
Synthetic DNAs were purchased from The Keck Foundation Biotechnology Resource Center at Yale University, purified by denaturing PAGE, and eluted from the gel by crush-soaking in 10 mM Tris⋅HCl (pH 7.5 at 23°C)/200 mM NaCl/1 mM EDTA. The DNA was recovered from the crush-soak solution by precipitation with ethanol. FMN, FAD, and riboflavin were acquired from Sigma. The radiolabeled nucleotides [α-32P]UTP and [γ-32P]ATP were purchased from Amersham Pharmacia.
Cloning of B. subtilis ribD and B. subtilis ypaA Regulatory Regions.
The nucleotide sequence from −61 to 302 of the B. subtilis ribDEAHT operon (16, 17) was amplified by PCR from B. subtilis strain 168 (a gift from D. Söll, Yale University) as an EcoRI–BamHI fragment. The DNA was ligated into EcoRI-, BamHI-digested pGEM4Z (Promega) to generate pGEM4Z-ribD. Similarly, the sequence spanning nucleotides −94 to 347 of B. subtilis ypaA was used to generate pGEM4Z-ypaA. The plasmids were transformed into E. coli Top10 cells (Invitrogen) for all other manipulations. All sequences were verified by DNA sequencing (United States Biochemical Thermosequenase).
In Vitro Transcription.
DNA templates for preparative in vitro transcription of ribD and ypaA RNAs were produced by PCR amplification of the corresponding regions of the pGEM4Z-ribD and pGEM4Z-ypaA plasmids, respectively, with the appropriate DNA primers. The DNA primers were designed to replace the endogenous B. subtilis promoter sequence with that of a T7 promoter sequence and to introduce a CC dinucleotide into the DNA templates such that both RNAs carry a 5′-terminal GG sequence. RNAs were prepared by in vitro transcription as described (17) or through the use of the RiboMAX transcription kit (Promega) according to the manufacturer's directions. The transcription products were resolved by PAGE and examined by PhosphorImager (Molecular Dynamics). RNA products were isolated by denaturing 6% PAGE and 5′ 32P-labeled as described (18).
The addition or deletion of U residues to alter the ribD RNA at positions 261–264 was achieved by using mutant oligonucleotides during PCR amplification of the pGEM4Z-ribD template DNA. The resulting templates each carry the 20 nucleotides immediately downstream of position 264 in unaltered form, whereas the poly U-encoding region beginning at position 258 varies from three to seven nucleotides.
In-Line Probing of RNA Constructs.
The B. subtilis 165 ribD and 349 ypaA leader mRNAs were subjected to in-line probing (which reveals the structural context of nucleotides based on their relative susceptibility to in-line attack from the adjacent 2′-hydroxyl group) by using a protocol adapted from those described (2, 4, 19, 20). Specifically, ≈1 nM 5′ 32P-labeled RNA was incubated for ≈40 h at 25°C in 20 mM MgCl2/50 mM Tris⋅HCl (pH 8.3 at 25°C)/100 mM KCl in the presence or absence of added ligand (FMN, FAD, or riboflavin) at concentrations that are indicated for each experiment. All ligand stock solutions and reactions were protected from light by wrapping each tube in aluminum foil. RNA cleavage products of >200 nt in length were resolved by subjecting samples to PAGE for extended time periods (7–21 h) with frequent buffer exchanges. The apparent KD values for each ligand were established by plotting the normalized fraction of RNA cleaved for each site (relative to the maximum and minimum cleavage values measured) against the logarithm of the concentration of ligand used.
Transcription Termination Assays Using T7 RNA Polymerase.
Transcription termination assays were conducted by using RiboMAX transcription kits. Each 10-μl reaction was incubated at 37°C for 2 h and contained 1× buffer (supplied by the manufacturer), 175 μM each NTP, 5 μCi of [α-32P]UTP (1 Ci = 37 GBq), and 0.3 μl of T7 RNAP (supplied by the manufacturer). FMN, riboflavin, or FAD was added to individual transcription reactions as indicated for each experiment. Reaction products were separated by using denaturing 6% PAGE. Bands corresponding to full-length and terminated RNAs were visualized by PhosphorImager, and the yields were established by using IMAGEQUANT software. The fraction of terminated templates was adjusted for the difference in specific activity between the terminated and full-length products. Similarly, transcription termination assays with DNA templates carrying U-insertion or -deletion mutations were incubated as described above for 30 min.
Mapping the Transcription Termination Site.
Run-off and FMN-induced termination products for the 304 ribD RNA were prepared and separated as described for the transcription termination assays. The resulting RNAs were recovered from the gel by crush-soaking followed by precipitation with ethanol. Both the transcription termination product and run-off transcript were digested separately with a 10–23 deoxyribozyme (21) that was engineered to target ribD mRNA for cleavage after position A222. Approximately 40 pmol of RNA were incubated in a 20-μl mixture for 1.5 h at 37°C under conditions similar to those described (22). The 3′ fragments of the deoxyribozyme-cleaved RNAs were separated by denaturing 10% PAGE, recovered from the gel, and 5′ 32P-labeled as described above. The 3′ fragment of the run-off transcript (FL*) was subjected to partial digestion with RNase T1 or alkali as indicated. The resulting RNA fragments were compared with the 3′ fragment of the FMN-induced termination products (T*) by denaturing 10% PAGE and analyzed by PhosphorImager.
Results
The RFN Element Binds FMN in the Absence of Protein.
To determine whether the RFN element serves as part of a metabolite-specific riboswitch, we subjected a portion (nucleotides 1–163) of the ribD 5′ UTR of B. subtilis to an in vitro structure-probing assay (2, 4, 19, 20) in the absence of proteins. This “in-line” probing assay relies on the fact that spontaneous RNA cleavage by internal transesterification typically occurs faster within regions of the RNA chain that are structurally unconstrained, whereas constrained (2°- or 3°-structured) portions of the chain generally undergo spontaneous cleavage less frequently. We find that incubation of 5′ 32P-labeled 165 ribD RNA (5′-GG plus the ribD 5′-UTR fragment) for 40 h in the presence of 10 μM FMN results in a fragmentation pattern that is distinct from that observed when no effector is added to the incubation (Fig. 1B). In contrast, incubation of the 165 ribD RNA with 10 μM riboflavin results in a similar but less dramatic change in RNA fragmentation pattern (data not shown), suggesting that riboflavin is bound by the RNA less tightly.
The structure-probing data are consistent also with the secondary structure that has been proposed (8, 9) for the RFN element based on phylogenetic sequence comparisons (Fig. 1A). For example, the B. subtilis 165 ribD RNA exhibits fragmentation patterns after incubation in the presence and absence of FMN that are consistent with the formation of stems P1–P5 (Fig. 1C), because the nucleotides involved in these structural elements exhibit a low level of spontaneous cleavage. In contrast, many of the highly conserved nucleotides that form the internal bulges of the six-stem RNA junction experience a high level of spontaneous cleavage in the absence of FMN but strongly resist fragmentation when FMN is present. These results indicate that the RNA adopts a structural state wherein elements P2–P6 are preorganized and the junction nucleotides adopt a precise FMN-binding fold that is stabilized after ligand association.
The ribD RFN Element Discriminates Against Riboflavin and FAD.
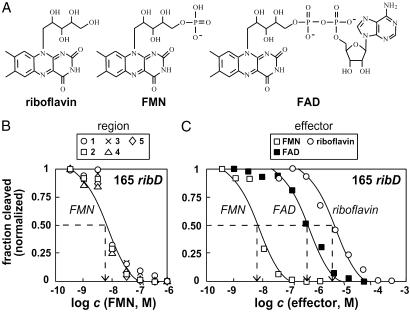
It has been shown (10) that a disruptive mutation in the riboflavin kinase (FAD synthase) gene causes derepression of riboflavin biosynthetic genes and overproduction of riboflavin. Although FMN and its biologically relevant analogs riboflavin and FAD share the riboflavin moiety (Fig. 2A), only FMN and/or FAD serve as the modulating effector(s) for genetic control of the ribD RNA. The molecular recognition characteristics of 165 ribD RNA were assessed by establishing the apparent KD value for FMN (Fig. 2B) and comparing this value to those determined for riboflavin and FAD (Fig. 2C).
Fig 2.
Affinities and specificity of the 165 ribD RNA. (A) Chemical structures of riboflavin, FMN, and FAD. (B) Determination of the apparent KD value for FMN binding to the 165 ribD RNA. The extent of spontaneous RNA cleavage (normalized relative to the highest and lowest cleavage values measured for each region) was plotted for five regions (Fig. 1B) that exhibit FMN-dependent modulation. The dashed line identifies the apparent KD value or the concentration of FMN required for conversion of half the RNAs into the ligand-bound form. RNAs were subjected to in-line probing as described for Fig. 1 by using various concentrations of ligand as indicated. (C) The normalized fraction of spontaneous cleavage at region two for FMN (from B) and for the related compounds FAD and riboflavin as indicated. Details are as described for B.
The apparent KD for FMN binding to the 165 ribD RNA was established by identifying the concentration of ligand needed to cause ≈50% reduction in the spontaneous cleavage of RNA at each of five regions along the RNA chain. Each of these sites exhibits half-maximal modulation of spontaneous cleavage when ≈5 nM FMN is added to an in-line probing assay. This result is consistent with the hypothesis that the RFN element is a tightly binding receptor for FMN and that structural modulation of the RNA is a concerted process that is induced by the presence of ligand.
Interestingly, we find that riboflavin, which differs from FMN by the absence of a single phosphate group (Fig. 2A), is bound by the 165 ribD RNA less tightly (KD ≈3 μM) by almost 3 orders of magnitude. This finding is similar to that observed for the thiamine pyrophosphate-specific riboswitch, which discriminates against thiamine and thiamine monophosphate ligands to a similar extent. Thus, the RFN element of ribD provides another example of an RNA structure that presumably creates productive binding interactions with a negatively charged phosphate moiety despite the fact that RNA itself is polyanionic. These RNAs must form a binding pocket, perhaps by exploiting divalent metal ions, to make productive binding interactions with phosphate.
Furthermore, our in-line probing experiments suggest that FAD is bound by the RNA with an apparent KD of ≈300 nM. However, we have not eliminated the possibility that our ligand sample might have a small percentage of FMN present, which is expected because of the spontaneous hydrolytic breakdown of the pyrophosphate linkage of FAD. Although our data indicate that the 5′ UTR of ribD discriminates between FMN and FAD by at least 60-fold, it is possible that the RNA might achieve a much greater level of discrimination that could be observed only if trace FMN were removed from the FAD sample.
An FMN-Dependent Riboswitch That Causes Transcription Termination.
The metabolite-binding domain of a riboswitch must bring about a structural change in the mRNA that modulates gene expression in some defined manner. In a previous study (4) it was shown that a thiamine pyrophosphate-dependent riboswitch from E. coli thiM RNA is comprised of an aptamer domain that binds the effector and an “expression platform” that is directly responsible for altering gene expression. We speculate that different riboswitches will make use of expression platforms that provide distinct mechanisms for genetic control and that these expression platforms might have a modular character that can be dissected to establish their mode of operation. Accordingly, we conducted a series of experiments to establish the mechanism of the expression platform for the FMN-dependent riboswitch of ribD.
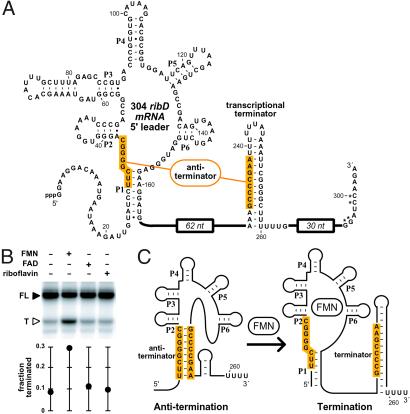
It has been proposed (9) that certain RFN elements, including the one present in the ribD RNA, might use a transcription termination mechanism for controlling gene expression. Genes from many prokaryotes carry an RFN element that is followed by complementary domains that can form transcription terminator or antiterminator stem structures. Thus, the metabolite-dependent control of transcription termination could result if the binding of FMN modulates the formation of these structural elements. In preparation for testing this hypothesis, we created a DNA template that encodes the larger 304-nt leader sequence of the ribD RNA (termed 304 ribD; Fig. 3A). This construct carries the RFN element and includes additional nucleotides that encompass the putative terminator and antiterminator elements. Also present in the construct are two domains of six U residues, the first beginning at position 239 and the second beginning at position 258. This second U-rich domain resides immediately 3′ relative to the terminator stem, and together these elements resemble bacterial intrinsic terminators (24, 25). Therefore, this second U-rich domain was identified as the most likely location for transcription termination.
Fig 3.
FMN causes transcription termination of the ribD RNA in vitro. (A) Sequence and secondary structure of the 304 ribD RNA. Shaded regions identify nucleotides that are complementary and might serve as an antiterminator structure. Nucleotides denoted with an asterisk have been altered from the wild-type sequence to generate a restriction site. The nucleotide sequence comprising the 62- and 30-nt regions (not shown) are presented in refs. 16 and 17. (B) In vitro transcription termination assays. In vitro transcriptions were conducted by using T7 RNA polymerase and a double-stranded DNA construct that serves as a template for the synthesis of 304 ribD RNA. Reactions were incubated in the absence (−) or presence (+) of 100 μM of the compounds as indicated for each lane. FL and T denote full-length and terminated RNA transcripts, respectively. The fraction of total transcripts that are termination products is plotted in the graph. See Materials and Methods for experimental details. (C) Model for the FMN-dependent riboswitch. Shaded regions identify the putative antiterminator structure that is disrupted after binding of FMN and formation of the P1 structure.
Using an in vitro transcription assay based on the action of bacteriophage T7 RNA polymerase, we transcribed the extended DNA template in the absence or presence of 100 μM FMN, FAD, or riboflavin (Fig. 3B). Although ≈10% of the transcripts terminate within this second U-rich domain in most instances, the addition of FMN selectively enhances transcription termination to ≈30%. Similar results are obtained when in vitro transcription is carried out with E. coli RNA polymerase by using a related DNA template (data not shown). Therefore, we believe that T7 RNA polymerase serves as a functional surrogate for a bacterial RNA polymerase in our transcription termination assays. Our data also indicate that binding of FMN to the nascent RNA transcript results in the formation of a structure that promotes transcription termination with unrelated RNA polymerases.
In the 165 ribD construct, the nucleotides between P1 and P2 undergo a significant reduction in spontaneous cleavage, which indicates that this region is involved in forming an ordered structure that is necessary for FMN binding. These same nucleotides are predicted also to participate in the formation of the antiterminator structure (9). Therefore, these data are consistent with the proposed riboswitch mechanism (Fig. 3C) in which the structures of the effector-bound RFN element and the antiterminator stem are mutually exclusive.
Mapping the FMN-Induced Transcription Termination Site.
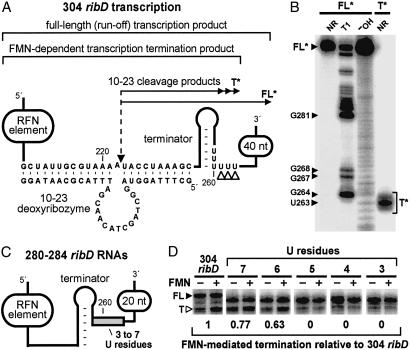
The transcription termination site for the 304 ribD RNA construct was identified by comparison of RNAs that correspond to the FMN-induced transcription termination product with the run-off transcript. Because the original transcripts from the 304 ribD template are too large to obtain single-nucleotide resolution by PAGE, we used a strategy wherein the transcription products were digested with a deoxyribozyme that catalyzes site-specific cleavage of the RNAs between nucleotides 222 and 223 (Fig. 4A). The subsequent 5′ 32P labeling of the 3′-digestion fragments of the full-length and terminated transcripts (to generate FL* and T*, respectively) permitted the high-resolution mapping of the termination site by PAGE (Fig. 4B). We find that transcription termination occurs between nucleotides U261 and U263, which are the last U residues in the U-rich domain that immediately follows the terminator stem.
Fig 4.
Mapping of the transcription termination site and the importance of the U-rich domain. (A) The 304 ribD construct (see also Fig. 3A) and processed fragments used to map the transcription termination site. The nucleotides presented depict the interaction with a 10–23 deoxyribozyme and the U-rich region where transcription termination was expected to occur. The dashed arrow identifies the location of deoxyribozyme-mediated cleavage. The portions of the construct that correspond to the full-length product, the FMN-induced transcription product and the deoxyribozyme digestion products FL* and T* that are derived from the full-length and terminated RNAs, respectively, are also identified. The open arrowheads identify the site of FMN-modulated transcription termination. (B) PAGE analysis of the 5′ 32P-labeled FL* and T* RNAs. RNAs were subjected to no additional reaction (NR) or were subjected to partial digestion with RNase T1 (T1) or alkali (-OH) as indicated for each lane. Bands corresponding to the FL* and T* RNAs, along with several T1 digestion products, are identified. The T* RNA corresponds to termination at nucleotides U261–U263, the mobility of which differs from the markers by 1 nt equivalent due to the absence of a 2′,3′-cyclic phosphate (unpublished data). (C) Schematic representation of ribD RNAs ranging from 280 to 284 nucleotides that carry three through seven U residues, respectively, in the U-rich domain (shaded box). Other than the U insertion or deletions, the RNA is identical in sequence to that of nucleotide 1–284 of the 304 ribD RNA. (D) Transcription termination assays with templates that encode the 304 ribD RNA and for 280–284 ribD variants that carry three through seven U residues, respectively, as indicated for each lane. Transcription reactions were conducted as described for Fig. 3B and incubated in the absence (−) or presence (+) of 100 μM FMN. FL and T denote full-length and terminated RNA transcripts, respectively. The numbers below each gel image reflect the relative level of FMN-induced termination compared with the nonmutated 304 ribD construct.
To explore the importance of the second U-rich domain in greater detail, we constructed a series of ribD constructs in which the number of U residues comprising this domain were varied from three to seven (Fig. 4C). In vitro transcription of the DNA templates that encode either seven or six U residues yielded near wild-type levels of FMN-induced transcription termination (Fig. 4D). In sharp contrast, constructs that carry fewer than six U residues do not exhibit FMN-dependent modulation of transcription termination. These observations indicate that transcription termination occurs at positions near the end of the second U-rich domain and that a stretch of at least six U residues is required for FMN-modulated termination.
The ypaA mRNA Riboswitch Uses a Shine–Dalgarno (SD) Sequestration Mechanism.
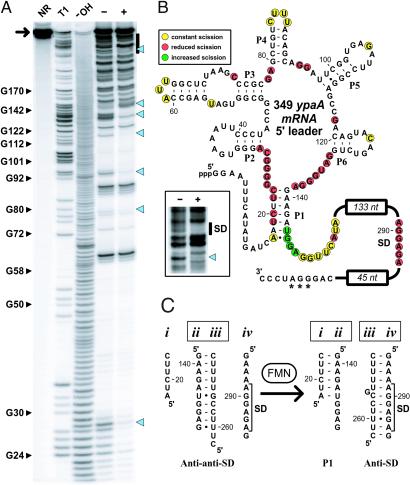
Comparative sequence analysis (9) indicates that a second type of expression platform also can be brought under the control of RFN-based riboswitches. For example, the ypaA gene, which encodes a riboflavin transport protein, carries an RFN element (Fig. 5) that is followed by putative anti-SD and anti-anti-SD elements. In-line probing of a 349-nt construct that encompasses the 5′ UTR of ypaA (Fig. 5A) demonstrates that the RNA undergoes significant structural modulation after the addition of FMN. Nearly all structure modulation within the 349 ypaA RNA (Fig. 5B) occurs at positions corresponding to those that undergo FMN-induced change in ribD RNA.
Fig 5.
FMN-induced structure modulation of the ypaA riboswitch. (A) Structural probing of the 349 ypaA RNA (arrow) in the presence (+) and absence (−) of 100 μM FMN. The bar identifies the region of cleavage products that is expanded in B Inset. Additional details are as described for Fig. 1B. (B) Secondary-structure model of the 349 ypaA RNA and the nucleotide positions of spontaneous cleavage in the presence (yellow and green) and absence (yellow and red) of FMN as identified from A. Nucleotides marked with an asterisk have been altered from the wild-type sequence to generate a restriction site. Sequences not depicted in the 133- and 45-nt domains can be obtained from the B. subtilis genome sequence (23). (Inset) An image of an extended PAGE separation of the region encompassing the SD element. (C) Proposed mechanism for the FMN-modulated formation of structures within the expression platform that control translation. RNA domains ii and iii form in the absence of FMN, thus exposing the SD element for ribosome binding. In contrast, FMN binding requires the formation of P1, which occupies RNA domain ii and permits the sequestration of the SD element by RNA domain iii.
In addition, we noticed that the 349 ypaA RNA exhibits structural modulation in the region occupied by the SD element (Fig. 5B Inset). This observation indicates that the expression platform indeed might be exploiting the FMN-dependent formation of a structure that restricts ribosome access to the SD element. A model for the mechanism of this expression platform is depicted in Fig. 5C. In the absence of FMN, the anti-anti-SD element (domain ii) is free to base-pair with the anti-SD element (domain iii), thus presenting the SD element (domain iv) in an unencumbered structural context. However, FMN-binding to the RFN element requires formation of the P1 stem between domains i and ii, which permits the anti-SD to pair with and sequester the SD element. Consistent with this model is the fact that the 5′-most nucleotides (139–143) within domain ii do not undergo significant modulation of spontaneous cleavage after FMN addition, whereas the 3′-most nucleotides (positions 144–147) exhibit an increase in spontaneous cleavage (Fig. 5B). This observation is expected if the 5′ portion of domain ii is base-paired in the absence (anti-anti-SD structure) or presence (P1 structure) of FMN, whereas the 3′ portion of domain ii is unpaired only when FMN is bound.
Discussion
RNA structural probing studies with the 5′ UTRs of ribD and ypaA RNAs confirm that the highly conserved RFN element is a natural FMN-binding aptamer. This RNA motif exhibits an exceptionally high affinity for its target ligand (apparent KD of <10 nM). As with the two natural metabolite-binding aptamers reported (2, 4), the FMN-binding domain of ribD exhibits a high level of discrimination against closely related compounds. Furthermore, both the thiamine pyrophosphate- and the FMN-dependent aptamers require the presence of phosphate groups on their respective ligands to bind with the highest affinity, which is a somewhat surprising achievement for a polyanionic receptor molecule. These aspects of molecular recognition are of particular importance in a biological setting, where the promiscuous binding of closely related biosynthetic intermediates would interfere with proper regulation of genetic expression.
The role of the natural FMN aptamer in these two instances most likely is to serve as the recognition domain for FMN-dependent riboswitches that control gene expression of the riboflavin biosynthetic operon and the riboflavin transporter in B. subtilis. Preliminary investigations into the mechanisms used by the associated expression platforms in both cases are consistent with those proposed from comparative sequence analysis data (9). Specifically, ribD RNA undergoes an increased frequency of transcription termination after the addition of FMN. This is perhaps the most efficient riboswitch mechanism for controlling the expression of large operons, because termination of transcription in the 5′ UTR prevents the synthesis of long mRNAs when their translation is unnecessary. Indeed, regulation by transcription termination may be common among many bacterial species (26). A search for sequences in the B. subtilis genome that could form terminator–antiterminator elements revealed nearly 200 candidates (27). In contrast, the riboswitch in the ypaA mRNA leader seems to control ribosome access to the SD element. Although the entire sequence of this smaller mRNA would be produced, this genetic control mechanism permits the organism to respond rapidly to declining concentrations of FMN.
Our findings provide additional evidence in support of earlier speculation (3, 8, 28–31) that mRNAs have the ability to play an active role in sensing metabolites for the purpose of genetic control. It seems likely that new riboswitches will be discovered that respond to other metabolites and exhibit more diverse mechanisms of genetic control. The riboswitches examined in this study provide examples of two mechanisms for expression platform function for the down-regulation of gene expression. However, we speculate that there will be instances where gene expression might be increased in response to metabolite binding to mRNAs. For example, certain enzymes that make use of the riboswitch effectors coenzyme B12 (2), thiamine pyrophosphate (4), and FMN might make use of expression platforms that permit gene activation. If the occurrence of these or other riboswitches extends across the phylogenetic landscape, or if they are used to control the expression of more than just biosynthetic and transport genes, then it is likely that a greater diversity of genetic control mechanisms will be discovered.
Acknowledgments
We thank members of the R.R.B. laboratory for comments on the manuscript. This work was supported by National Institutes of Health Grant GM 559343, National Science Foundation Grant EIA-0129939, and a fellowship (to R.R.B.) from The David and Lucile Packard Foundation.
Abbreviations
SD, Shine–Dalgarno
References
- 1.Lundrigan M. D., Köster, W. & Kadner, R. J. (1991) Proc. Natl. Acad. Sci. USA 88, 1479-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nahvi A., Sudarsan, N., Ebert, M. S., Zou, X., Brown, K. L. & Breaker, R. R. (2002) Chem. Biol. 9, 1043-1049. [DOI] [PubMed] [Google Scholar]
- 3.Miranda-Rios J., Navarro, M. & Soberón, M. (2001) Proc. Natl. Acad. Sci. USA 98, 9736-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler W., Nahvi, A. & Breaker, R. R. (2002) Nature 419, 952-956. [DOI] [PubMed] [Google Scholar]
- 5.Henkin T. M. (2000) Curr. Opin. Microbiol. 3, 149-153. [DOI] [PubMed] [Google Scholar]
- 6.Stülke J. (2002) Arch. Microbiol. 177, 433-440. [DOI] [PubMed] [Google Scholar]
- 7.Grundy F. J., Winkler, W. C. & Henkin, T. M. (2002) Proc. Natl. Acad. Sci. USA 99, 11121-11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelfand M. S., Mironov, A. A., Jomantas, J., Kozlov, Y. I. & Perumov, D. A. (1999) Trends Genet. 15, 439-442. [DOI] [PubMed] [Google Scholar]
- 9.Vitreschak A. G., Rodionov, D. A., Mironov, A. A. & Gelfand, M. S. (2002) Nucleic Acids Res. 30, 3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack M., van Loon, A. P. G. M. & Hohmann, H.-P. (1998) J. Bacteriol. 180, 950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.-M., Zhang, S., Saha, S., Santa Anna, S., Jiang, C. & Perkins, J. (2001) J. Bacteriol. 183, 7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gusarov I. I., Kreneva, R. A., Podcharniaev, D. A., Iomantas, I. V., Abalakina, E. G., Stoinova, N. V., Perumov, D. A. & Kozlov, I. I. (1997) Mol. Biol. 31, 446-453. [PubMed] [Google Scholar]
- 13.Kil Y. V., Mironov, V. N., Gorishin, I. Y., Kreneva, R. A. & Perumov, D. A. (1992) Mol. Gen. Genet. 233, 483-486. [DOI] [PubMed] [Google Scholar]
- 14.Kreneva R. A., Gelfand, M. S., Mironov, A. A., Yomantas, J. A., Kozlov, Y. I., Mironov, A. S. & Perumov, D. A. (2000) Russ. J. Genet. [Transl. of Genetika (Moscow)] 36, 972-974. [Google Scholar]
- 15.Perkins J. B. & Pero, J. (2002) in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, eds. Sonenshein, A. L., Hoch, J. A. & Losick, R. (Am. Soc. Microbiol., Washington, DC), pp. 271–286.
- 16.Azevedo V., Sorokin, A., Ehrlich, S. D. & Serror, P. (1993) Mol. Microbiol. 10, 397-405. [DOI] [PubMed] [Google Scholar]
- 17.Mironov V. N., Perumov, D. A., Krayev, A. S., Stepanov, A. I. & Skryabin, K. G. (1990) Mol. Biol. 24, 256-260. [PubMed] [Google Scholar]
- 18.Seetharaman S., Zivarts, M., Sudarsan, N. & Breaker, R. R. (2001) Nat. Biotechnol. 19, 336-341. [DOI] [PubMed] [Google Scholar]
- 19.Soukup G. A. & Breaker, R. R. (1999) RNA 5, 1308-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soukup G. A., DeRose, E. C., Koizumi, M. & Breaker, R. R. (2001) RNA 7, 524-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoro S. W. & Joyce, G. F. (1997) Proc. Natl. Acad. Sci. USA 94, 4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyle A. M., Chu, V. T., Jankowsky, E. & Boudvillain, M. (2000) Methods Enzymol. 317, 140-146. [DOI] [PubMed] [Google Scholar]
- 23.Kunst F., Ogasawara, N., Moszer, I, Albertini, A. M., Alloni, G., Azevedo, V., Bertero, M. G., Bessières, P., Bolotin, A., Borchert, S., et al. (1997) Nature 390, 249-256. [DOI] [PubMed] [Google Scholar]
- 24.Wilson K. S. & von Hippel, P. H. (1995) Proc. Natl. Acad. Sci. USA 92, 8793-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gusarov I. & Nudler, E. (1999) Mol. Cell 3, 495-504. [DOI] [PubMed] [Google Scholar]
- 26.Henkin T. M. & Yanofsky, C. (2002) BioEssays 24, 700-707. [DOI] [PubMed] [Google Scholar]
- 27.Merino E. & Yanofsky, C. (2002) in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, eds. Sonenshein, A. L., Hoch, J. A. & Losick, R. (Am. Soc. Microbiol., Washington, DC), pp. 323–336.
- 28.Gold L., Brown, D., He, Y., Shtatland, T., Singer, B. S. & Wu, Y. (1997) Proc. Natl. Acad. Sci. USA 94, 59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold L., Singer, B., He, Y. & Brody, E. (1997) Curr. Opin. Genet. Dev. 7, 848-851. [DOI] [PubMed] [Google Scholar]
- 30.Nou X. & Kadner, R. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stormo G. D. & Ji, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 9465-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]