Abstract
We present a combined computational and experimental method for the rapid optimization of proteins. Using β-lactamase as a test case, we redesigned the active site region using our Protein Design Automation technology as a computational screen to search the entire sequence space. By eliminating sequences incompatible with the protein fold, Protein Design Automation rapidly reduced the number of sequences to a size amenable to experimental screening, resulting in a library of ≈200,000 mutants. These were then constructed and experimentally screened to select for variants with improved resistance to the antibiotic cefotaxime. In a single round, we obtained variants exhibiting a 1,280-fold increase in resistance. To our knowledge, all of the mutations were novel, i.e., they have not been identified as beneficial by random mutagenesis or DNA shuffling or seen in any of the naturally occurring TEM β-lactamases, the most prevalent type of Gram-negative β-lactamases. This combined approach allows for the rapid improvement of any property that can be screened experimentally and provides a powerful broadly applicable tool for protein engineering.
Keywords: computational protein design, protein engineering, mutagenesis, directed evolution, β-lactamase
The increased use of enzymes and other proteins in the chemical, agricultural, and pharmaceutical industries has generated considerable interest in the design of proteins with new and improved properties. Two different but complementary technologies have been applied to this goal: (i) rational design, which relies on structural and mechanistic knowledge and human expertise; and (ii) directed evolution methods such as error-prone PCR, phage display, and DNA shuffling, which use random mutagenesis or recombination to create diversity and then experimentally screen the libraries generated for desired properties (1). Directed evolution has been successfully used on a wide range of proteins (2–7). However, this approach is limited by the number of sequences that can be screened experimentally (about 1014 for library panning and 107 for high-throughput screening). Rational design has also been applied with some success (8–10), but it was not until computational methods were developed that it could be used comprehensively.
Computational techniques use protein design algorithms to perform in silico screening of protein sequences (11–17). By taking advantage of the speed of computers, these methods allow a vast number of sequences to be screened (≈1080). The ability to search such large sequence spaces drastically increases the possibility of finding novel proteins with improved properties. Computational techniques have also been developed to enhance the efficiency of directed evolution methods (18, 19).
One computational design tool that has proven effective is Protein Design Automation (PDA) (13). PDA begins with the three-dimensional structural model of the protein to be designed and predicts the optimal sequence that will adopt this fold, allowing all or a specified set of residues to change. The fitness of sequences is scored by using physical potential functions that model the energetic interactions of protein atoms (20); stable low-energy sequences are given the best scores. By using extremely efficient search algorithms, up to 1080 sequences can be accurately screened within hours (21–23). Multiple simultaneous mutations can be made, and novel sequences that are very different from wild type can be discovered. PDA has shown tremendous success in designing proteins with improved stability and conformational specificity (13, 14, 24–28) and has even been used to engineer a catalytic site into a previously nonreactive protein (29).
In these studies, only a few optimal sequences calculated by PDA were made and tested experimentally. The utility of PDA can be extended significantly, however, if it is used to generate a library of sequences, all of which are predicted to be stable and fold into a predetermined structure. Unlike random libraries, where most of the mutations are deleterious, the mutant sequences in the PDA library are computationally screened to eliminate destabilizing mutations and sequences inconsistent with the proper fold. The selected sequences are then experimentally screened for desired properties such as improved catalytic activity, substrate specificity, or receptor binding. Therefore, PDA is a computational prescreen to decrease the sequence space many orders of magnitude, while maintaining broad diversity, to a number easily amenable to experimental screening. By coupling PDA with experimental screening, we combine the advantages of computational design with those of directed evolution: namely, access to a vast sequence space and the ability to improve any protein property that can be captured by a screen.
In this paper, we demonstrate the feasibility of this approach by using it to increase the resistance of bacteria toward the antibiotic cefotaxime by optimizing TEM-1 β-lactamase, the most prevalent plasma-encoded β-lactamase in Gram-negative bacteria.
Methods
Structure Preparation.
The crystal structure of TEM-1 β-lactamase (Protein Data Bank no. ) (30) was used as the starting point for modeling. All water molecules and the sulfate group were removed; the side chains of residues N132, N154, N170, H122, and H289 were flipped to form a better hydrogen bond network; and the disulfide bond between C77 and C123 was formed manually. The program biograf (Molecular Simulations, San Diego) was used to generate explicit hydrogens, and 50 steps of conjugate gradient minimization were performed by using the Dreiding II force field (31) without the electrostatics term. The minimization is done to make the structure compatible with our force-field parameters and results in very slight changes to the coordinates.
Construction of Mutant Library.
To facilitate introduction of the mutations into the TEM-1 gene, a pCR-Blunt (Invitrogen) vector containing the TEM-1 gene was digested with XbaI and HindIII, treated with T4 DNA polymerase, and religated. Site-directed mutagenesis was performed by using QuikChange as described by the manufacturer (Stratagene) to remove the existing XbaI and HindIII sites. New XbaI and HindIII sites were then introduced by site-directed mutagenesis at nucleotides 163 and 841, respectively, of the TEM-1 gene in the vector. A polyhistidine (6×His) sequence was then added to the 3′-end of the TEM-1 ORF to facilitate immunodetection of the proteins, thereby creating the vector pXR293. The his-tag was found to have no effect on β-lactamase activity. Escherichia coli TOP10 cells transformed with pXR293 were confirmed to grow on media containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. The β-lactamase protein expressed from this construct is termed TEM-1 in this report.
The mutated β-lactamase genes were constructed essentially as described by Prodromou and Pearl (32) and Chalmers and Curnow (33). Oligonucleotides corresponding to the gene were synthesized as 40–50 mers with ≈15-nt overlaps. At each mutational position, multiple oligonucleotides were included in the reaction, and the genes were synthesized by using recursive PCR. They were then digested with XbaI and HindIII and subcloned into pXR293. This vector was then transformed into E. coli TOP10 cells (Invitrogen) for expression.
Selection of Cefotaxime-Resistant Mutants.
E. coli cells expressing the mutant library of TEM-1 genes were grown on plates containing increasing concentrations of cefotaxime, and the minimum inhibitory concentration (MIC) for survival was determined. The cefotaxime concentrations used were: 0.01, 0.025, 0.05, 0.1, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, and 128 μg/ml. Assays were conducted at 25 and 30°C to ensure soluble expression of the designed proteins. Cells were plated at low density (<1,000 per plate) to ensure that the observed resistance was not due to confluence. Clones demonstrating the highest resistance were picked, and the β-lactamase protein was identified with immunoblot analysis (34) by using pooled 5- and 6-his polyclonal antibodies (Qiagen, Valencia, CA). The TEM-1 gene of the most resistant variants was sequenced to identify the mutations. New genes containing the mutations identified for PDA-1, -2, and -3 were constructed, and MICs were determined to confirm the initial screening results.
Results
Protein Optimization Strategy: Combining PDA with Experimental Screening.
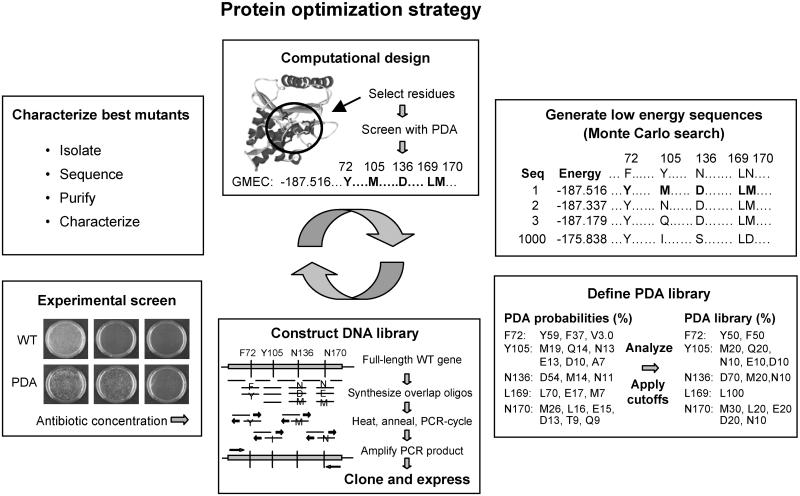
The overall strategy for protein optimization is shown in Fig. 1. PDA is used to computationally design a protein and define a library of mutant sequences at specific positions. PDA's optimization algorithms are then run to screen all possible sequences for the global optimal sequence and conformation for the target fold, the one with the lowest energy as determined by the scoring function. This conformation is termed the global minimum energy conformation (GMEC). Starting from this optimal structure, a search algorithm such as Monte Carlo (35, 36) simulated annealing is used to explore sequence space and generate a list of other near-optimal sequences. The Monte Carlo list is rank-ordered by energy score and may contain as many sequences as desired (e.g., the best 1,000). An amino acid probability table is then generated from the list by counting amino acid occurrences at each of the designed positions. Different cutoffs or weighting functions can be applied to define a library of a desired size, appropriate for experimental screening. Structure or sequence alignment information, experimental data, and diversity considerations may also be taken into account in defining the mutant library.
Fig 1.
Protein optimization strategy. A set of residues or region of the protein structure is selected to be designed. PDA is used to computationally screen the entire designed sequence space and determine the GMEC for the fold. Starting from the GMEC, the Monte Carlo search is then used to explore the sequence space and generate a list of near-optimal sequences. An amino acid probability table is obtained from the list and cutoffs are applied to define a PDA library of mutant sequences for experimental screening. The PDA library is translated into a DNA library, which is cloned and expressed. An experimental screen is used to select clones with improved properties; these are then characterized. Results can be fed into additional cycles.
Recursive PCR with overlapping oligonucleotides is then used to synthesize the genes containing all of the mutant sequences in the PDA-defined library. The genes are pooled and cloned, and the mutant proteins are expressed in an appropriate host such as E. coli. The mutant proteins are screened experimentally for desired properties, and the best mutants are isolated and characterized. These results can be used as feedback for additional rounds of computational design, library generation, and screening.
Reduction of Sequence Space.
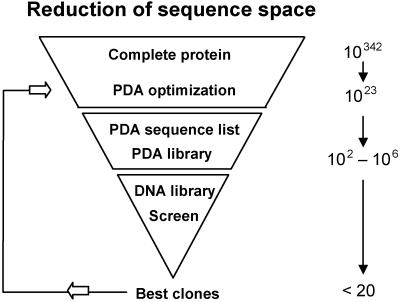
The use of PDA as a computational screen allows us to access a vast sequence space and, by eliminating sequences predicted to be destabilizing or inconsistent with the proper fold, reduce it to a size amenable to experimental screening. The reduction of sequence space obtained for TEM-1 β-lactamase, our test case, is shown in Fig. 2. If we were to consider the entire β-lactamase protein (263 residues) and allow all 20 amino acids at each position, we would need to screen 20263 or ≈1.4 × 10342 sequences. By focusing the design to a particular region (19 residues near the active site) and using a slightly restricted set of amino acids (19), we reduced this to 7 × 1023 sequences, a number that can easily be screened computationally, but not experimentally. We then chose cutoffs for the Monte Carlo list and the probability table that would define a library within the limits of experimental screening. In this case, we specified a library of ≈200,000 mutant sequences, a reduction of 18 orders of magnitude for the residues explicitly considered and an overall reduction of more than 300 orders of magnitude for the entire protein.
Fig 2.
Reduction of sequence space for PDA design of TEM-1 β-lactamase. Computational screening with PDA and judicious application of cutoffs reduced the sequence space 18 orders of magnitude for the 19 residues explicitly considered and more than 300 orders of magnitude for the entire protein. This conformational screen specified a library for experimental screening of ≈200,000 mutant sequences, enriched for structural integrity.
Computational Design of β-Lactamase.
The hydrolysis of β-lactam antibiotics, catalyzed by β-lactamase, is a common mechanism by which bacteria become resistant to antibiotics (37). The most prevalent plasmid-encoded β-lactamase in Gram-negative bacteria is the class A TEM-1 β-lactamase (38). This enzyme hydrolyzes ampicillin efficiently but is inefficient at hydrolyzing the cephalosporin cefotaxime. Our goal was to use PDA to design β-lactamase variants that confer increased resistance toward cefotaxime.
Optimizing the area around the active site is likely to have a significant effect on enzyme activity and substrate specificity (37, 39). Although more distant mutations can also be effective (40), the rationale for how to select such positions is less obvious. Designing residues around the active site also serves as a stringent test of the ability of PDA to predict nondisruptive mutations. We therefore focused our design on residues within 5 Å of the active site residues S70, K73, S130, E166, and K234. These criteria resulted in 19 positions that were allowed to change: M69, T71, F72, V74, V103, Y105, A126, I127, N132, A135, N136, L169, N170, M211, D214, K234, S235, G236, and I247. All 20 amino acids, except cysteine and proline, were considered at these positions. The catalytic residues (S70, K73, S130, and E166) were not allowed to change their amino acid identities; however, their conformations could vary. An expanded version of the backbone-dependent rotamer library of Dunbrack and Karplus (41) was used in all of the calculations, and the DEE algorithm was used to find the GMEC. The computational details, residue classification, and potential functions used are described in previous work (13, 14, 20, 42).
Definition of Mutant Library.
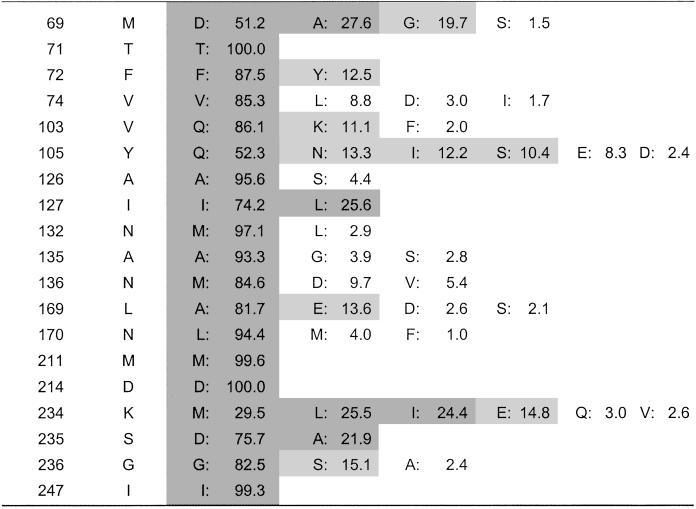
Optimization with PDA predicted an optimal sequence with nine mutations. Starting from this GMEC, we applied Monte Carlo simulated annealing to produce a rank-ordered list of the 1,000 lowest energy sequences. A probability table was generated from this list by counting the amino acid occurrences at each of the 19 designed positions (Table 1). A 10% cutoff was then applied to the probability table to define a library of mutant sequences for experimental screening; that is, for a given position, an amino acid identity was included in the library if it had a 10% or greater probability of occurrence. To ensure that the library spanned the complete sequence space from the wild-type enzyme to the most distantly related PDA mutant, we always included the wild-type identity at all designed positions, even if it did not appear in the Monte Carlo list. With a 10% cutoff, this gave us a library of 172,800 unique sequences; a 20% cutoff would have resulted in a much smaller library of 4,806.
Table 1.
PDA probability table for designed positions of TEM-1 β-lactamase
Amino acids included when applying different probability cutoffs are indicated as follows: 20% cutoff in dark gray, 10% cutoff in light gray, and 1% cutoff in white background. In defining the PDA library, the 10% cutoff was used and the wild-type amino acid identity was added if it did not appear in the Monte Carlo list. This specified a library of 172,800 unique sequences.
Construction of Genes for Mutant Library.
Recursive PCR with overlapping oligonucleotides was used to synthesize the TEM-1 β-lactamase genes containing all 172,800 mutant sequences in the PDA library. Synthetic oligonucleotides containing the designed mutations were pooled to create desired diversity at each site. Two separate reactions were performed: one that contained only a proofreading DNA polymerase (Pfu DNA polymerase), termed the nonerror prone reaction, and one that contained both Pfu DNA polymerase and TaqDNA polymerase, termed the error-prone reaction. The mutated genes were cloned and transformed into E. coli.
Validation of Mutant Library.
Sixty individual clones from the nonerror-prone library were sequenced by standard techniques. The plasmids contained intact ORFs with the desired mutations. No additional mutations were detected. With a sample size of 60, we were able to find all of the specified mutations at each designed position. It is impossible to find all combinations of the mutations within this small sample (the library contained 172,800 unique sequences), but none of the clones were identical and we were unable to detect a statistically significant bias toward any particular mutation at any position. This result indicates that we have developed an efficient method for converting a PDA-defined library into an experimental library containing all of the mutated genes required to encode the desired mutant sequences.
Experimental Screen for β-Lactamase Activity.
Experimental libraries of ≈500,000 individual E. coli colonies expressing the mutated β-lactamase genes were pooled and plated onto increasing concentrations of cefotaxime in a single round of selection, and the MIC for survival was determined. This number of colonies is about three times the size of the PDA-defined library and was used to ensure that the pooled DNA library contained at least one copy of each mutant sequence (within a 95% level of confidence) (43). Clones from the nonerror-prone library had a MIC of 64 μg/ml, which is a 640-fold increase in resistance compared with the wild-type value of 0.1 μg/ml. Clones from the error-prone library had a MIC of 128 μg/ml, a 1,280-fold increase in resistance (see PDA-1 and -2, Table 2). Because our approach allows us to assay the complete PDA library diversity in a single round, we were able to use very stringent selection conditions and directly obtain highly resistant variants. The identification of incrementally improved sequences was not necessary.
Table 2.
Antibiotic resistance of TEM-1 β-lactamase variants
| Variant
|
Mutations
|
No. of mutations
|
No. of novel mutations
|
Cefotaxime | Ampicillin | ||
|---|---|---|---|---|---|---|---|
| MIC, μg/ml | Fold increase | MIC, μg/ml | Fold decrease | ||||
| TEM-1 (WT) | — | — | — | 0.1 | — | 4,096 | — |
| PDA-1 | M69D, V103Q, Y105N | 8 | 8 | 64 | 640 | 100 | 40 |
| N132M, L169A, N170L | |||||||
| S235D, G236S | |||||||
| PDA-2 | V103Q, Y105N, I127L | 6 | 6 | 128 | 1,280 | 100 | 40 |
| L169A, S235Y, G236S | |||||||
| PDA-3 | M69D, V103Q, N132M | 6 | 6 | 64 | 640 | ND | ND |
| L169A, N170L, S235D | |||||||
| TEM-15 | E104K, G238S | 2 | — | 16 | 160 | 4,096 | 1 |
| ST-1 | E104K, G238S, M182T | 4 | — | 256 | 2,560 | ND | ND |
| A18V | |||||||
Resistance measured at 25°C unless specified otherwise. ND, MIC not determined.
Fold increase/decrease in resistance is relative to wild type (TEM-1).
Value reported by Cantu and Palzkill (44).
MIC assay done at 30°C.
Value reported by Shannon et al. (49). Boldface indicates novel mutations (not reported to significantly improve cefotaxime resistance in any TEM-β-lactamase; refs. 2, 45, and 46; Jacoby, G. and Bush, K., www.lahey.org/studies/temtable.htm).
Substrate Specificity.
We also measured resistance to ampicillin and found no growth at 100 μg/ml, significantly less than the MIC of 4,096 μg/ml reported for the wild type (43). This result suggests that our screens identified clones whose resistance to cefotaxime had dramatically improved, whereas their resistance to ampicillin was reduced at least 40-fold. The relative substrate specificity toward cefotaxime vs. ampicillin was thus enhanced 25,000- to 50,000-fold.
PDA Mutants Are Novel.
The most active mutant from each library was isolated and sequenced. PDA-1 had eight mutations (M69D, V103Q, Y105N, N132M, L169A, N170L, S235D, and G236S), all designated in the PDA library (see Table 1). PDA-2 had five PDA-designed mutations (V103Q, Y105N, I127L, L169A, and G236S) and one random mutation (S235Y). The S235Y mutation was not predicted by PDA due to steric clashes. Protein backbone motion, which is required to relieve the clash, is not considered in the computation. None of the mutations in PDA-1 or -2 have been identified by full gene random mutagenesis or DNA shuffling studies (2, 39, 45, 46) or have been observed in the 105 naturally occurring TEM β-lactamases (G. Jacoby and K. Bush, www.lahey.org/studies/temtable.htm). Orencia et al. (47) discussed the emergence of antibiotic resistances in β-lactamases and showed that there is an overlap between the mutations discovered by directed evolution and those occurring in natural evolution. PDA, however, accesses the entire designed sequence space including all possible combinations of mutations and therefore can produce multiple simultaneous mutations. PDA is therefore more likely to identify novel mutants with desired properties. The lone random mutation in PDA-2 (S235Y) was in the active site region, suggesting that the novel context of the PDA-designed mutants allowed this previously unobserved, but beneficial, mutation to emerge.
Two of the mutations in PDA-1 (V105N and G236S) were reverted to wild type to create a backcross mutant (PDA-3). This PDA-3 sequence is present in the library defined with a 20% cutoff but is absent if a 10% cutoff is used (see Table 1). PDA-3 exhibited the same cefotaxime resistance as PDA-1 (Table 2), indicating that a smaller PDA-library (4,806 vs. 172,800 sequences) can also generate mutants with significantly improved activity. Additional backcrosses were done to examine the role of the other six mutations in PDA-1. No single mutation was primarily responsible for the improved resistance, and no simple additivity was apparent, suggesting that the mutations are coupled. This conclusion is supported by extensive replacement mutagenesis studies of three-residue segments around the active site (39). They found a mutant (E168G, L169A, and N170G) that included one of our mutations (L169A), but it showed only a marginal (2-fold) improvement in cefotaxime resistance. Although they also tested most of our other mutations, no increased resistance was found for any of these. This lack of improved resistance indicates that the broader context of many simultaneous mutations provided by our approach was required to find our highly active sequences.
Comparison with Other Mutants.
To compare the activity of our PDA-designed mutants with those obtained in other studies, we introduced some previously reported mutations into our wild-type gene, including E104K/G238S (comparable to TEM-15) (2, 46) and A18V/E104K/M182T/G238S (comparable to ST-1) (2, 46). TEM-15 is a naturally occurring β-lactamase that is active against cefotaxime, and ST-1 is a highly active TEM-1 variant discovered from three rounds of DNA shuffling. We tested the ability of these mutants to confer resistance to cefotaxime. Wild type had a MIC of 0.1 μg/ml, comparable to the values reported by others; TEM-15 and ST-1 had MICs of 16 and 256 μg/ml, respectively, also in line with previously published work (Table 2) (2, 44, 46, 48–50).
Location of Mutations.
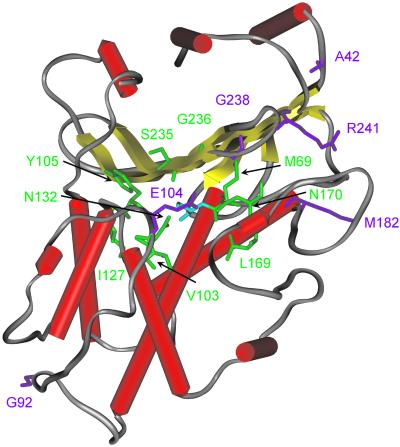
The mutations in all our variants are located in or near the active site, because our computational design restricted changes to this region. Directed evolution methods, however, tend to produce mutations spread over the entire protein structure. For example, almost all of the mutations in the best mutants obtained by DNA shuffling (2) and random hypermutagenesis (46) are located far from the active site (Fig. 3). It is possible that these techniques seldom produce mutations close to the active site, because they rely on incremental changes; a single change in the first round of screening must be beneficial to be passed to the second round. However, point mutations in the active site area are usually disruptive. Our approach, on the other hand, allows multiple simultaneous mutations in a single round, which can have compensating or even synergistic effects.
Fig 3.
Location of mutations in PDA-1 and -2 (green) vs. those obtained by DNA shuffling (2) and random hypermutagenesis (46) (magenta). The wild-type TEM-1 β-lactamase structure is illustrated, and the side chains of the mutated positions are shown. The catalytic serine (S70) is depicted in blue. The average distance between the Cα atoms and the catalytic nucleophile (OγS70) in our PDA-1 and -2 mutations was 8.0 and 8.6 Å, respectively, vs. 16.0 Å for the mutations in ST-2 and -3 (Stemmer's best mutants) (2) and 12.1 Å for 3D.5 (Zaccolo and Gherardi's best mutant) (46). This difference in distances illustrates that the mutations found by PDA are near the designed active site area, whereas those found by DNA shuffling and random hypermutagenesis are farther away.
Sequence Space Coverage.
Most of the mutations observed in our PDA variants require a minimum of two nucleotide changes, and one, M69D, can be made only by a triple nucleotide change (Table 3). Double- or triple-nucleotide changes within a single codon are very difficult to achieve by using random mutagenesis techniques such as error-prone PCR or single-gene DNA shuffling. This limitation is demonstrated by the fact that each of the mutations found in the directed evolution studies (2, 45, 46) as well as those observed in the 105 naturally occurring TEM variants (G. Jacoby and K. Bush, www.lahey.org/studies/temtable.htm) could be obtained by a single nucleotide change. If one considers all of the substitutions that are possible for each of the 20 amino acids, on average only seven can be achieved by a single nucleotide change. The sequence space coverage is further reduced by codon preferences, biases for transitions over transversions, and A ↔ T over G ↔ C mutations. These restrictions severely limit the sequence space accessible to these methods and suggest why our approach, which does not suffer from these limitations, is more likely to produce novel functional sequences.
Table 3.
Minimal number of nucleotide changes required for amino acid mutations in TEM-1 β-lactamase variants
| Variant
|
Position | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 42 | 69 | 92 | 103 | 104 | 105 | 127 | 132 | 169 | 170 | 182 | 235 | 236 | 238 | 240 | 241 | 254 | |
| TEM-1 (WT) | A | A | M | G | V | E | Y | I | N | L | N | M | S | G | G | E | R | D |
| TEM-15 | K,1 | S,1 | ||||||||||||||||
| ST-1 | V,1 | K,1 | T,1 | S,1 | ||||||||||||||
| ST-2, ST-3 | G,1 | S,1 | K,1 | T,1 | S,1 | H,1 | ||||||||||||
| ST-4 | K,1 | T,1 | S,1 | |||||||||||||||
| 3D.5 | K,1 | T,1 | S,1 | |||||||||||||||
| 3A.6 | S,1 | K,1 | H,1 | G,1 | ||||||||||||||
| PDA-1 | D,3 | Q,2 | N,1 | M,2 | A,2 | L,2 | D,2 | S,1 | ||||||||||
| PDA-2 | Q,2 | N,1 | L,1 | A,2 | Y,1 | S,1 | ||||||||||||
| PDA-3 | D,3 | Q,2 | M,2 | A,2 | L,2 | D,2 | ||||||||||||
Discussion
The purpose of this study is to show that a combined approach, using PDA as a computational screen to rationally reduce the sequence space before experimental screening, can rapidly lead to novel protein variants with improved properties. We used PDA to identify sequences compatible with the protein fold and then experimentally screened the resulting sequence library to obtain variants with novel properties. As a test case, we redesigned the active site of β-lactamase and then selected for variants with improved resistance to cefotaxime. In a single round, we obtained variants that exhibit a 1,280-fold increase in resistance and, to our knowledge, are novel.
Our approach has some key distinctions from purely experimental techniques. By using an efficient computational screen, we are able to access an extremely large region of sequence space and rapidly reduce it to a number amenable to experimental screening. Experimental libraries will always be restricted to sampling a miniscule portion of sequence space due to limitations on the sheer mass of proteins that can be physically made and tested. Computational screening, on the other hand, is scalable and can comprehensively screen enormous sequence spaces. The sequence space searched by random mutagenesis techniques is also severely limited by the fact that amino acid mutations requiring more than single nucleotide changes are extremely unlikely. Random techniques usually produce only incremental changes per round and require multiple rounds, whereas our approach creates multiple simultaneous mutations in a single round. All of these features result in the rapid discovery of novel mutants that are different from any of those observed previously.
Another difference with the PDA method is that it offers full control over the location and type of mutations, allowing incorporation of structural and experimental data and enhancing our understanding of structure–activity relationships. It results in focused designs of smaller functionally enriched mutant libraries, allowing complex and expensive screens such as mammalian cell-based assays for improved protein therapeutics, while still accessing broad sequence diversity. Our combined computational and experimental approach allows for the rapid improvement of any protein property that is amenable to experimental screening and has broad applications in the chemical, agricultural, and pharmaceutical industries.
Abbreviations
PDA, Protein Design Automation
MIC, minimum inhibitory concentration
GMEC, global minimum energy conformation
References
- 1.Arnold F. H. (2001) Nature 409, 253-257. [DOI] [PubMed] [Google Scholar]
- 2.Stemmer W. P. (1994) Nature 370, 389-391. [DOI] [PubMed] [Google Scholar]
- 3.Arnold F. H. & Moore, J. C. (1997) Adv. Biochem. Eng. Biotechnol. 58, 1-14. [DOI] [PubMed] [Google Scholar]
- 4.Crameri A., Raillard, S. A., Bermudez, E. & Stemmer, W. P. (1998) Nature 391, 288-291. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H., Giver, L., Shao, Z., Affholter, J. A. & Arnold, F. H. (1998) Nat. Biotechnol. 16, 258-261. [DOI] [PubMed] [Google Scholar]
- 6.Minshull J. & Stemmer, W. P. (1999) Curr. Opin. Chem. Biol. 3, 284-290. [DOI] [PubMed] [Google Scholar]
- 7.Pedelacq J. D., Piltch, E., Liong, E. C., Berendzen, J., Kim, C. Y., Rho, B. S., Park, M. S., Terwilliger, T. C. & Waldo, G. S. (2002) Nat. Biotechnol. 29, 927-932. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham B. C. & Wells, J. A. (1991) Proc. Natl. Acad. Sci. USA 88, 3407-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whittle E. & Shanklin, J. (2001) J. Biol. Chem. 276, 21500-21505. [DOI] [PubMed] [Google Scholar]
- 10.Bornscheuer U. T. & Pohl, M. (2001) Curr. Opin. Chem. Biol. 5, 137-143. [DOI] [PubMed] [Google Scholar]
- 11.Hellinga H. W. & Richards, F. M. (1994) Proc. Natl. Acad. Sci. USA 91, 5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desjarlais J. R. & Handel, T. M. (1995) Protein Sci. 4, 2006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahiyat B. I. & Mayo, S. L. (1996) Protein Sci. 5, 895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahiyat B. I. & Mayo, S. L. (1997) Science 278, 82-87. [DOI] [PubMed] [Google Scholar]
- 15.Street A. G. & Mayo, S. L. (1999) Struct. Folding Des. 7, R105-R109. [DOI] [PubMed] [Google Scholar]
- 16.Kraemer-Pecore C. M., Wollacott, A. M. & Desjarlais, J. R. (2001) Curr. Opin. Chem. Biol. 5, 690-695. [DOI] [PubMed] [Google Scholar]
- 17.Pokala N. & Handel, T. M. (2001) J. Struct. Biol. 134, 269-281. [DOI] [PubMed] [Google Scholar]
- 18.Voigt C. A., Mayo, S. L., Arnold, F. H. & Wang, Z. G. (2001) Proc. Natl. Acad. Sci. USA 98, 3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voigt C. A., Martinez, C., Wang, Z. G., Mayo, S. L. & Arnold, F. H. (2002) Nat. Struct. Biol. 3, 1-6. [DOI] [PubMed] [Google Scholar]
- 20.Gordon D. B., Marshall, S. A. & Mayo, S. L. (1999) Curr. Opin. Struct. Biol. 9, 509-513. [DOI] [PubMed] [Google Scholar]
- 21.Desmet J., De Maeyer, M., Hazes, B. & Lasters, I. (1992) Nature 356, 539-542. [DOI] [PubMed] [Google Scholar]
- 22.Gordon D. B. & Mayo, S. L. (1998) J. Comp. Chem. 10, 1505-1514. [Google Scholar]
- 23.Pierce N. A., Spriet, J. A., Desmet, J. & Mayo, S. L. (2000) J. Comp. Chem. 21, 999-1009. [Google Scholar]
- 24.Malakauskas S. M. & Mayo, S. L. (1998) Nat. Struct. Biol. 5, 470-475. [DOI] [PubMed] [Google Scholar]
- 25.Shimaoka M., Shifman, J. M., Jing, H., Takagi, J., Mayo, S. L. & Springer, T. A. (2000) Nat. Struct. Biol. 7, 674-678. [DOI] [PubMed] [Google Scholar]
- 26.Marshall S. A. & Mayo, S. L. (2001) J. Mol. Biol. 305, 619-631. [DOI] [PubMed] [Google Scholar]
- 27.Luo P., Hayes, R. J., Chan, C., Stark, D. M., Hwang, M. Y., Jacinto, J. M., Juvvadi, P., Chung, H. S., Kundu, A., Ary, M. L. & Dahiyat, B. I. (2002) Protein Sci. 11, 1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filikov A. V., Hayes, R. J., Luo, P., Stark, D. M., Chan, C., Kundu, A. & Dahiyat, B. I. (2002) Protein Sci. 11, 1452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolon D. N. & Mayo, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14274-14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelsch C., Mourey, L., Masson, J. M. & Samama, J. P. (1993) Proteins 16, 364-383. [DOI] [PubMed] [Google Scholar]
- 31.Mayo S. L., Olafson, B. D. & Goddard, W. A., III (1990) J. Phys. Chem. 94, 8897-8909. [Google Scholar]
- 32.Prodromou C. & Pearl, L. H. (1992) Protein Eng. 5, 827-829. [DOI] [PubMed] [Google Scholar]
- 33.Chalmers F. M. & Curnow, K. M. (2001) Biotechniques 30, 249-252. [DOI] [PubMed] [Google Scholar]
- 34.Harlow E. & Lane, D., (1988) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 471–510.
- 35.Metropolis N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H. & Teller, E. (1953) J. Chem. Phys. 21, 1087-1092. [Google Scholar]
- 36.Voigt C. A., Gordon, D. B. & Mayo, S. L. (2000) J. Mol. Biol. 299, 789-803. [DOI] [PubMed] [Google Scholar]
- 37.Matagne A., Lamotte-Brasseur, J. & Frere, J. M. (1998) Biochem. J. 330, 581-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutcliffe J. G. (1978) Proc. Natl. Acad. Sci. USA 75, 3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palzkill T. & Botstein, D. (1992) J. Bacteriol. 174, 5237-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiller B., Gershenson, A., Arnold, F. H. & Stevens, R. C. (1999) Proc. Natl. Acad. Sci. USA 96, 12305-12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunbrack R. L. & Karplus, M. (1993) J. Mol. Biol. 230, 543-574. [DOI] [PubMed] [Google Scholar]
- 42.Street A. G. & Mayo, S. L. (1998) Folding Des. 3, 253-258. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 44.Cantu C. & Palzkill, T. (1998) J. Biol. Chem. 273, 26603-26609. [DOI] [PubMed] [Google Scholar]
- 45.Vakulenko S. B., Geryk, B., Kotra, L. P., Mobashery, S. & Lerner, S. A. (1998) Antimicrob. Agents Chemother. 42, 1542-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaccolo M. & Gherardi, E. (1999) J. Mol. Biol. 285, 775-783. [DOI] [PubMed] [Google Scholar]
- 47.Orencia M. C., Yoon, J. S., Ness, J. E., Stemmer, W. P. & Stevens, R. C. (2001) Nat. Struct. Biol. 8, 238-242. [DOI] [PubMed] [Google Scholar]
- 48.Sirot D., Recule, C., Chaibi, E. B., Bret, L., Croize, J., Chanal-Claris, C., Labia, R. & Sirot, J. (1997) Antimicrob. Agents Chemother. 41, 1322-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon K., Stapleton, P., Xiang, X., Johnson, A., Beattie, H., El Bakri, F., Cookson, B. & French, G. (1998) J. Clin. Microbiol. 36, 3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stapleton P. D., Shannon, K. P. & French, G. L. (1999) Antimicrob. Agents Chemother. 43, 1881-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]