Abstract
Overexpression of the receptor tyrosine kinase (RTK) erbB2 (also designated neu or HER2) was implicated in causing a variety of human cancers, including mammary and ovarian carcinomas. Ligand-induced receptor dimerization is critical for stimulation of the intrinsic protein tyrosine kinase (PTK) of RTKs. It was therefore proposed that PTK activity is stimulated as a result of the reorientation of the cytoplasmic domains within receptor dimers, leading to transautophosphorylation and stimulation of enzymatic activity. Here, we propose a molecular mechanism for rotation-coupled activation of the erbB2 receptor. Using a computational exploration of conformation space of the transmembrane (TM) segments of an erbB2 homodimer, we found two stable conformations of the TM domain. We suggest that these conformations correspond to the active and inactive states of erbB2, and that the receptor molecules may switch from one conformation to the other without crossing exceedingly unfavorable states. This model provides an explanation for the biochemical and oncogenic properties of erbB2, such as the effects of erbB2 overexpression on kinase activity and cell transformation. Furthermore, the opposing effects of the neu* activating oncogenic point mutation and the Val-655→Ile single-nucleotide polymorphism shown to be linked to reduced risk of breast cancer are explained in terms of shifts in the equilibrium between the active and inactive states of erbB2 in vivo.
The epidermal growth factor-receptor (EGFR) family of receptor tyrosine kinases (erbB1, erbB2, erbB3 and erbB4) plays a critical role in the control of many physiological processes (reviewed in refs. 1–3). Moreover, overexpression of or dysfunction in the activity of EGFR and other members of the family has been implicated in the cause of a variety of human cancers (i.e., lung, brain, mammary, and ovarian). erbB1 and other members of the family are composed of a ligand-binding domain that is connected, via a single transmembrane (TM) helix, to a cytoplasmic domain endowed with intrinsic protein tyrosine kinase (PTK) activity flanked by regulatory sequences that are subject to autophosphorylation and phosphorylation by heterologous protein kinases. Ligand binding to the extracellular domain induces the formation of homo- and heterodimers of different members of the EGFR family, followed by stimulation of PTK activity by transautophosphorylation. In addition to their key regulatory role in the control of PTK activity, tyrosine autophosphorylation sites in RTKs serve as docking sites for recruitment and activation of cellular signaling proteins that mediate the pleiotropic responses induced by growth factor stimulation.
Despite an extensive search over more than a decade, a physiological ligand of erbB2 has not yet been identified (1, 3). It has therefore been proposed that erbB2 does not have a specific ligand, and that it functions as a preferred partner for heterodimerization with other members of the EGFR family (4–6). Indeed, strong activation of the PTK activity of erbB2 was shown to be induced by overexpression of erbB2, even without ligand stimulation (reviewed in ref. 1). Moreover, overexpression and activation of erbB2 have been detected in a large fraction of mammary and ovarian cancers. There is reliable evidence that the TM domain of erbB2 plays an active role in erbB2 dimerization and activation (7–11). A point mutation in the TM domain of the rat homologue neu (Val-664→Glu) induces PTK activation and oncogenic transformation (7, 9). The Val-664 residue is located within a consensus sequence in the TM segment's N terminus that is known to induce TM helix dimerization (11). This sequence motif is shared by the TM domains of all members of the EGFR family (Fig. 1). In addition to the N-terminal dimerization motif, erbB2 contains a second related GxxxG motif in the C terminus of its TM segment (12, 13). Each of these motifs mediate dimerization of the TM domain of erbB2 in the cell membrane (14).
Fig 1.
Multiple sequence alignment of the TM domains of the human members of the EGFR family. Highlights indicate dimerization motifs in TM domains (12, 14). Yellow corresponds to Sternberg–Gullick motifs (11) and green to motifs that are related to the GxxxG motif (12). The transforming neu* Val-664→Glu mutation in rats corresponds to a Val-659→Glu mutation in humans (shown in red). All family members except for human erbB3 contain two known dimerization motifs separated by seven positions, thus placing the two motifs on the same ridge of amino acid residues on a model α-helix (18) (Fig. 3). Position 655 in the human erbB2 (blue) exhibits a Val/Ile single-nucleotide polymorphism. The Ile variant is linked to reduced risk of contracting breast cancer (25).
In this report, we present a model for the activation of erbB2 that is based on two states of its TM domain. The conformational space of an erbB2 TM homodimer is explored by using a computational tool for predicting conformations of pairs of α-helices in TM domains of membrane proteins (15). The method is based on structural and thermodynamic considerations and consists of an exhaustive search for a structure that is likely to allow a pair of helices to pack tightly. Our computations retrieve empirical results, indicating that the TM domain of erbB2 may undergo dimerization via either one of the two dimerization motifs (14). We further show that receptor dimers are capable of switching between these two conformations. We propose that the balance between the two states may play a role in the control of the activity of erbB2 and its various mutants, both under normal conditions and in pathological states.
Methods
Calculating Scores for Helix-Pair Conformations.
A detailed explanation of the method is presented in ref. 15. The essence of the score function consists of two contributions according to the simple rule “small residues go inside:” a negative contribution from residue pairs that form contacts in the given conformation and are known to allow helix pairs to tightly pack in TM proteins; and a positive term for the burial of bulky residues in the helix pair's interface. Thus, a conformation favoring tight packing of helices is expected to have a negative score. Based on the available structural data (16), helices were assumed to be canonical. The interhelical distance was assumed to be ≈7.5 Å, corresponding to the interhelical distance in the tightly packed TM homodimer glycophorin A, which has been used as a model for the dimerization of TM domains (17).
Global Search.
A global search for an optimal conformation of the erbB2 homodimer was carried out on a five-dimensional lattice (Fig. 2 Right). To find the most optimal conformation for the helix dimer, we explored a very large part of the conformation space by modulating x between −10 and 10 Å, with a step size of 0.5 Å; z between −10 and 10 Å, with a step size of 1 Å; α and β from 0° to 360°, with a step size of 9°; and ψ between −75° and 75°, with a step size of 3.75°. We thus examined more than 50 million different conformations of the helix pair.
Fig 2.
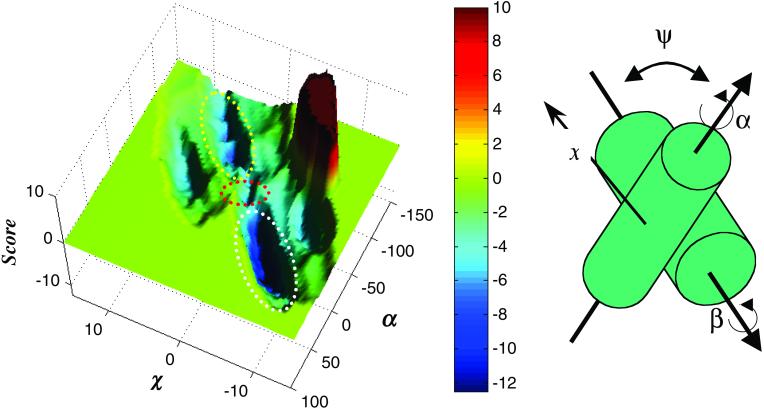
A score potential surface of a homodimer corresponding to erbB2's TM domain at a crossing angle of −35°. (Left) Each coordinate on the surface represents a unique conformation of the helix pair. Two minima are colored in deep blue, corresponding to two dimerization modes (14), in which either of the dimerization motifs mediates contact between the TM domains. The deeper minimum (white ellipse) corresponds to conformations where the C-terminal dimerization motif (Fig. 1) mediates contact, whereas the shallower minimum (yellow ellipse) corresponds to conformations where the N-terminal motif mediates dimerization (Fig. 3). The minima are not disconnected (red ellipse), and movement is likely between the two dimerization modes (Movie 1). Score is given in arbitrary units. (Right) Different conformations of the helix pair are tested by modulating α and β corresponding to rotations (°) of the monomers around their principal axes, and x to a sliding movement (Å) of one helix across the face of the other. In the global search method, the crossing angle Ψ (°) and z (Å), corresponding to movement across the face of the opposing helix along an axis perpendicular to x (not shown) are also modulated. In the restricted search method symmetry is enforced, so that α = β and z = 0. Also, Ψ is set to −35°, corresponding to a typical crossing angle for helices in the 4–4 class of helix packing (18).
Restricted Search.
We also used finer resolution to map the erbB2 homodimer's conformation space by imposing symmetry and restricting the crossing angle Ψ to −35° (Fig. 2 Right), which is typical of class 4–4 ridges-into-grooves helix packing (18). α was modulated throughout its potential range with a step size of 5°, and x was modulated between −15 and 15 Å with a step size of 0.5 Å.
Results and Discussion
We conducted a global search of the erbB2 TM homodimer's conformation space without imposing symmetry. We found a conformation, where the C-terminal GxxxG motif mediates dimerization, to have a minimal score in erbB2's TM conformation space. We therefore consider it to be optimal for tight packing of this TM helix pair.
Similar to the GxxxG motif mediating the dimerization of glycophorin A (12, 13, 19), the two dimerization motifs in erbB2 contain two critical residues that are separated by three residues in the amino acid sequence of the TM helix (Fig. 1). It is thus reasonable to assume that interactions between the motifs on two different helices are accommodated by class 4-4 ridges-into-grooves helix packing (18). We therefore conducted a restricted, although higher-resolution, search, assuming that the crossing angle (Ψ) between the two monomers is −35° (Fig. 2 Right), a value typical for this class of helix packing (18).
Our results show that the TM domains of an erbB2 homodimer are stable in either of two distinct dimerization modes. These modes correspond to two minima in the score surface shown in Fig. 2 Left. The deeper minimum (white ellipse) corresponds to dimerization of the TM domain via the C-terminal dimerization motif, and the shallower minimum (yellow ellipse) corresponds to contact formation via the N-terminal dimerization motif. Notably, the two minima are connected through a saddle-point in the score surface (red ellipse in Fig. 2 Left), indicating that the dimer is capable, in theory, of switching between the two dimerization modes without moving through excessively unfavorable conformations. The movement consists of sliding along a ridge formed by amino acid residues (18) and a large 120° rotation of each monomer with respect to the other (Figs. 2 and 3 and Movie 1, which is published as supporting information on the PNAS web site, www.pnas.org).
Fig 3.
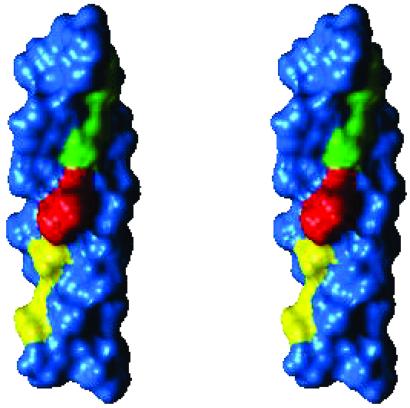
Stereoview of the ideal α-helix model of the TM domain of erbB2 used for the calculations presented in Fig. 2. Ser-656 and Gly-660 of the N-terminal dimerization motif (11) (Fig. 1) are yellow; Gly-668 and Gly-672 of the C-terminal dimerization motif (12) are green; and Val-664 is red. The monomers pack through either of the two motifs (14). The structural basis that stabilizes the two conformations is that the two motifs form relatively even surfaces on the helical face. Thus they form grooves (18) into which the other monomer may pack. Val-664 (red) is situated between the two motifs on the same ridge (18). Tight packing of this residue in the transition between the two dimerization modes (Movie 1) forms the saddle-point in Fig. 2 Left.
In light of these results, we propose a molecular-switch model for the activation of erbB2, other members of the EGFR family, and possibly other RTKs. According to the model, the structure of the TM segment in erbB2 allows the receptor dimers to exist in one of two states (Fig. 3). Our calculations show that the dimer mediated by the C-terminal dimerization motif is more stable than the dimer formed by the N-terminal motif (Fig. 2). We propose that the more stable conformation corresponds to the receptor's inactive state, which does not stimulate its PTK activity (20). At normal levels of erbB2 expression, its monomers are at equilibrium with dimers mediated by the more stable C-terminal dimerization motif, i.e., inactive dimers. Transition to the active state is caused by a conformational switch consisting of 120° rotation and movement to dimerization via the N-terminal dimerization motif (Movie 1). According to the model, contact formation via the N-terminal motif of erbB2 causes a reorientation in the cytoplasmic domains of the two juxtaposing catalytic domains of erbB2 (20, 21), resulting in transautophosphorylation and stimulation of the receptor's PTK activity.
The molecular-switch model helps explain the biochemical effects of mutations in the TM domain of erbB2 that were described in the literature (8, 14). Table 1, which is published as supporting information on the PNAS web site, summarizes the effects of various known erbB2 mutations on cell transformation and other properties of erbB2. The various mutations summarized in Table 1 modify either one or both dimerization motifs or leave them intact. Effects on focus formation and dimerization are explained in terms of the molecular-switch model.
It has been proposed that both active and inactive receptor dimers coexist on the cell surface at a normal level of receptor expression (22), and that overexpression increases the amount of active dimers resulting in enhanced PTK activity and cell transformation (1, 3). According to the model presented here, overexpression of erbB2 does not change the overall ratio of active-to-inactive receptor dimers. Rather, it suggests that the enhanced PTK activity, even in cases where an external signal is not received, is a direct consequence of the increase in the absolute number of erbB2 molecules that undergo dimerization via the active (N-terminal) dimerization motif.
The molecular-switch model for erbB2 activation may provide an explanation for seemingly contradictory properties of known erbB2 mutants or naturally occurring variants. It was proposed that the Val-664→Glu mutation in the oncogenic form of neu, known as neu*, facilitates hydrogen-bond formation between neighboring TM domains (10), resulting in enhanced neu* dimerization and autophosphorylation (23). However, it was recently demonstrated, by using an assay for receptor dimerization, that the activating mutation of neu* does not enhance receptor dimerization (14). The model presented in this report may be used to explain both phenomena. Dimerization via the C-terminal motif would result in the exposure of a polar group on the side chain of Glu-664 to the hydrophobic lipid environment, which is energetically unfavorable. Thus, the set of inactive dimeric conformations in neu (forming contact via the C-terminal dimerization motif) are, in essence, inaccessible to the mutated receptor, which in turn results in a decrease in receptor dimerization (14). However, the amount of active dimers, mediated by the N-terminal dimerization motif, will be increased because of hydrogen bond formation (23), resulting in increased autophosphorylation, PTK activation, and cell transformation.
The model may also explain why the Ile variant of the single-nucleotide polymorphism at position 655 in humans (24) (Fig. 1) exhibits a reduced risk for contracting mammary carcinomas (25). We propose that substitution of Val for a bulkier Ile residue in this position of the TM domain will destabilize the formation of active erbB2 dimers that are mediated by the N-terminal dimerization motif. Consequently, receptor activation caused by overexpression of erbB2 will be reduced even at high levels of erbB2 overexpression. In other words, the activating Val-664→Glu mutation will shift the equilibrium toward the active dimeric form, whereas the Val-655→Ile variant will destabilize the formation of the active dimeric form, resulting in reduced PTK activity, even under conditions of erbB2 overexpression.
Two evolutionary arguments were raised to explain the formation of inactive receptor dimers in the membrane (20). First, formation of inactive receptors on the cell surface would allow more rapid initiation of signal transduction as compared with activation of monomeric receptors that must undergo dimerization for activation to take place. Moreover, the higher stability of inactive compared with active dimers on the cell surface may act as a safe-lock mechanism by decreasing inadvertent dimerization and activation caused by spontaneous collisions between laterally diffusing surface receptors (20).
The mechanism proposed in this report for the activation of erbB2 may apply to other members of the EGFR family and other RTKs (26). It is noteworthy that erbB3 contains only the N-terminal dimerization motif (Fig. 1). Unlike other members of the EGFR family, erbB3 possesses an inactive PTK domain and may serve as a preferred substrate of the other members of the EGFR family (27). Therefore, it may not need the safe-lock mechanism that exists in receptors endowed with active PTK domains. A similar mechanism may also apply for the activation of insulin and insulin-like growth factor (IGF)1 receptors by insulin and IGF1, respectively. The insulin and IGF1 receptors are expressed on the cell surface as disulfide-linked inactive dimers (1, 3). Insulin binding induces a conformational change in the dimeric insulin receptor, resulting in stimulation of the intrinsic PTK activity.
It would be interesting to test and quantify the phenotypic importance of the switching mechanism we have proposed here in vivo beyond the documented mutations of Table 1. For instance, mutating the TM segment to the effect that the N-terminal motif exhibits a greater dimerization propensity than the C-terminal motif (e.g., Ser-656→Gly, Gly-668→Phe double mutant) may have a phenotypic effect similar to the Val-664→Glu substitution in neu* (Fig. 1) (7, 9). Another interesting possibility would be to leave both motifs intact and to alter the pathway between them, e.g., by deleting residues from the sequence connecting the two motifs (positions 661–667 of erbB2; Fig. 1). Our calculations show that a deletion mutation such as this would disconnect the pathway between the conformations mediated by either of the two dimerization motifs. This mutation would retain the receptor's dimerization characteristics but slow down the kinetics of the switching mechanism (data not shown).
We have used a recently developed computational method for predicting TM helix conformations (15) and combined its results with a large body of past and recent experimental information on the sequence–structure–function relationships of the erbB2 TM domains. Based on these results, we suggested a model for the activation of erbB2 receptors in molecular detail. The model clarifies previously described clinical and biochemical information on erbB2 receptors (Table 1). Finally, targeting of this mechanism by a novel class of lipid soluble inhibitors may offer new therapeutic strategies for cancers caused by overexpression of erbB2.
Supplementary Material
Acknowledgments
We acknowledge helpful discussions with Mark A. Lemmon. We thank Sharron Bransburg-Zabary for assistance in producing the accompanying animation. The reported computations were conducted on infrastructure supplied by the Bioinformatics Unit and the Computation Center at Tel Aviv University. This study was supported by a Research Career Development Award from the Israel Cancer Research Fund.
Abbreviations
EGFR, epidermal growth factor receptor
RTK, receptor tyrosine kinase
TM, transmembrane
PTK, protein tyrosine kinase
References
- 1.Prenzel N., Fischer, O. M., Streit, S., Hart, S. & Ullrich, A. (2001) Endocr. Rel. Cancer 8, 11-31. [DOI] [PubMed] [Google Scholar]
- 2.Ullrich A. & Schlessinger, J. (1990) Cell 61, 203-212. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger J. (2000) Cell 103, 211-225. [DOI] [PubMed] [Google Scholar]
- 4.Graus-Porta D., Beerli, R. R. & Hynes, N. E. (1995) Mol. Cell. Biol. 15, 1182-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graus-Porta D., Beerli, R. R., Daly, J. M. & Hynes, N. E. (1997) EMBO J. 16, 1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karunagaran D., Tzahar, E., Beerli, R. R., Chen, X., Graus-Porta, D., Ratzkin, B. J., Seger, R., Hynes, N. E. & Yarden, Y. (1996) EMBO J. 15, 254-264. [PMC free article] [PubMed] [Google Scholar]
- 7.Bargmann C. I., Hung, M. C. & Weinberg, R. A. (1986) Cell 45, 649-657. [DOI] [PubMed] [Google Scholar]
- 8.Cao H., Bangalore, L., Bormann, B. J. & Stern, D. F. (1992) EMBO J. 11, 923-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segatto O., King, C. R., Pierce, J. H., Di Fiore, P. P. & Aaronson, S. A. (1988) Mol. Cell. Biol. 8, 5570-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sternberg M. J. & Gullick, W. J. (1989) Nature 339, 587. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg M. J. & Gullick, W. J. (1990) Protein Eng. 3, 245-248. [DOI] [PubMed] [Google Scholar]
- 12.Russ W. P. & Engelman, D. M. (2000) J. Mol. Biol. 296, 911-919. [DOI] [PubMed] [Google Scholar]
- 13.Senes A., Gerstein, M. & Engelman, D. M. (2000) J. Mol. Biol. 296, 921-936. [DOI] [PubMed] [Google Scholar]
- 14.Mendrola J. M., Berger, M. B., King, M. C. & Lemmon, M. A. (2002) J. Biol. Chem. 277, 4704-4712. [DOI] [PubMed] [Google Scholar]
- 15.Fleishman S. J. & Ben-Tal, N. (2002) J. Mol. Biol. 321, 363-378. [DOI] [PubMed] [Google Scholar]
- 16.Smith S. O., Smith, C. S. & Bormann, B. J. (1996) Nat. Struct. Biol. 3, 252-258. [DOI] [PubMed] [Google Scholar]
- 17.MacKenzie K. R., Prestegard, J. H. & Engelman, D. M. (1997) Science 276, 131-133. [DOI] [PubMed] [Google Scholar]
- 18.Chothia C., Levitt, M. & Richardson, D. (1981) J. Mol. Biol. 145, 215-250. [DOI] [PubMed] [Google Scholar]
- 19.Lemmon M. A., Flanagan, J. M., Hunt, J. F., Adair, B. D., Bormann, B. J., Dempsey, C. E. & Engelman, D. M. (1992) J. Biol. Chem. 267, 7683-7689. [PubMed] [Google Scholar]
- 20.Jiang G. & Hunter, T. (1999) Curr. Biol. 9, R568-R571. [DOI] [PubMed] [Google Scholar]
- 21.Bell C. A., Tynan, J. A., Hart, K. C., Meyer, A. N., Robertson, S. C. & Donoghue, D. J. (2000) Mol. Biol. Cell 11, 3589-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burke C. L., Lemmon, M. A., Coren, B. A., Engelman, D. M. & Stern, D. F. (1997) Oncogene 14, 687-696. [DOI] [PubMed] [Google Scholar]
- 23.Weiner D. B., Liu, J., Cohen, J. A., Williams, W. V. & Greene, M. I. (1989) Nature 339, 230-231. [DOI] [PubMed] [Google Scholar]
- 24.Papewalis J., Nikitin, A. & Rajewsky, M. F. (1991) Nucleic Acids Res. 19, 5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie D., Shu, X. O., Deng, Z., Wen, W. Q., Creek, K. E., Dai, Q., Gao, Y. T., Jin, F. & Zheng, W. (2000) J. Natl. Cancer Inst. 92, 412-417. [DOI] [PubMed] [Google Scholar]
- 26.Moriki T., Maruyama, H. & Maruyama, I. N. (2001) J. Mol. Biol. 311, 1011-1026. [DOI] [PubMed] [Google Scholar]
- 27.Carraway K. L. & Cantley, L. C. (1994) Cell 78, 5-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.