Abstract
It is well known that exposure to UV induces DNA damage, which is the first step in mutagenesis and a major cause of skin cancer. Among a variety of photoproducts, cyclobutane-type pyrimidine photodimers (CPD) are the most abundant primary lesion. Despite its biological importance, the precise relationship between the structure and properties of DNA containing CPD has remained to be elucidated. Here, we report the free (unbound) crystal structure of duplex DNA containing a CPD lesion at a resolution of 2.0 Å. Our crystal structure shows that the overall helical axis bends ≈30° toward the major groove and unwinds ≈9°, in remarkable agreement with some previous theoretical and experimental studies. There are also significant differences in local structure compared with standard B-DNA, including pinching of the minor groove at the 3′ side of the CPD lesion, a severe change of the base pair parameter in the 5′ side, and serious widening of both minor and major groves both 3′ and 5′ of the CPD. Overall, the structure of the damaged DNA differs from undamaged DNA to an extent that DNA repair proteins may recognize this conformation, and the various components of the replicational and transcriptional machinery may be interfered with due to the perturbed local and global structure.
The cis-syn pyrimidine dimer (cyclobutane-type pyrimidine photodimer, CPD) is the major photoproduct induced by UV light present in sunlight (1) and is one of the principal causes of skin cancer (2). Evidence for the formation of the thymine dimer in DNA was first obtained >40 years ago (3, 4) and a few years later for cytosine-containing CPDs (5). Because of the mutagenic and protein–DNA-disrupting properties of CPDs, many organisms have evolved enzymes to specifically recognize and repair cis-syn dimers, such as Escherichia coli photolyase (6, 7), T4 endoV (8), as well as general repair enzymes such as E. coli uvrABC (9) and human excinuclease (10). Additionally, dimers are efficiently repaired in transcription-coupled repair by virtue of their ability to block synthesis by RNA polymerases (11, 12). CPDs also block DNA replication (13) and are efficiently bypassed in a nonmutagenic manner by DNA damage bypass polymerases such as E. coli pol V (14) and the recently discovered yeast (15) and human polymerase η (16, 17). There is also evidence that the mismatch repair system can recognize mismatches opposite thymine dimers (18). Understanding the mechanism by which these proteins recognize and process CPDs will be greatly aided by a structure for CPD-containing DNA.
The efficiency of damage repair is likely to depend on the extent to which those changes alter the structure of the DNA, hence making it recognizable for the repair enzymes involved. It has been suggested that the binding affinities of the repair enzymes for the CPD-containing DNA depend on the degree of DNA unwinding or kinking caused by those lesions (19, 20). The first evidence that CPD formation causes large alterations in the structure of DNA is offered by circular dichroism studies and the sedimentation coefficient of photo-damaged sequences (21, 22). Furthermore, there are changes in the sensitivity of CPD regions to chemical modification reagents as well as nucleases both before and shortly after repair and alterations in the thermal denaturation of DNA on introduction of such lesions, indicating the distinct structural changes (23–25). CPDs were also found to unwind DNA by ≈10° (26, 27). The bending induced by CPD was first reported as 30° by a circularization assay (28) but later as 7° in a phased multimer gel electrophoretic assay (29). CPD-containing DNA duplexes have also been the subject of a number of theoretical studies (19, 30–33) that have predicted that CPDs cause little bending of DNA, to bending as high as 27° (18). Several 1H NMR studies have also been reported on duplexes containing a CPD (34–37), two of which concluded that there was little bending of the DNA (29, 34). In the most recent of these studies, it was proposed that the CPD causes narrowing of the minor groove at the CPD site and that a unique BII-type backbone linkage flanks the 3′-side of the dimer, which might be an important recognition element for the T4 endo V repair enzyme (38).
Although a lot of structural information has been obtained over the past 30 years, there has been no consensus on the effect of CPD formation on duplex DNA. Herein, we report the crystal structure of a cis-syn thymine dimer in free (unbound) duplex DNA, providing a clear picture of the effects of dimer formation on DNA structure and base pairing since it was first identified in DNA in 1960 (3, 4). Most notably, the dimer is found to bend DNA by 30°, in remarkable agreement with some of the early theoretical (19) and experimental (28) studies, and in addition has an unexpectedly wide minor groove.
Methods
Synthesis.
The oligodeoxynucleotide decamer containing a cis-syn thymine dimer was prepared on a 1-μmol scale by automated DNA synthesis on an Expedite 8909 synthesizer (Applied Biosystems) by using a cis-syn thymine dimer building block (39). A standard protocol for β-cyanoethyl phosphoramidites was used, except that the coupling of the cis-syn thymine dimer was extended to 15 min with a coupling efficiency of ≈80%. The BrdUrd-containing complementary strand was obtained from a commercial source.
Oligodeoxynucleotide Purification.
The dimer-containing decamer was purified by reversed-phase HPLC on a 250 × 4.6 mm Rainin Dynamax C18 column (5-μm particle size, 300-Å pore size) by using buffer A [0.1 M triethanolamine (TEAA), pH 7.0] and buffer B [30/70 (vol/vol) acetonitrile/0.1 M triethylammonium acetate, pH 7.0]. The dimer-containing decamer eluted at ≈27 min from a gradient of 5-min 0–20% buffer B followed by 20–50% buffer B in buffer A and was evaporated to dryness under vacuum on a Savant SpeedVac.
Preparation of the Duplex.
The dimer-containing decamer and its complementary strand were separately dissolved in D2O, evaporated to dryness, and then redissolved in D2O. The two decamers were then mixed in an approximately equal molar ratio, based on their concentrations as determined by UV absorption measurements. The two decamers were then adjusted to an equimolar ratio by monitoring the integral ratio of well-separated aromatic peaks in the 500 MHz 1H NMR spectra. Finally, the samples were evaporated to dryness and coevaporated with ddH2O three times to back exchange deuteriums for hydrogens.
Crystallization, Data Collection, and Processing.
Crystallization of native and BrdUrd DNAs were performed by using the hanging-drop vapor diffusion and microseeding method in 4°C with final 1.5 mM concentration of decamer DNAs in Tris-EDTA buffer (10 mM Tris·HCl, pH 8.0/1 mM EDTA). DNA solutions (2.5 μl) were mixed with equal volume of precipitant containing 0.2 M MgCl2, 0.1 M Hepes-Na, pH 7.5, 30% polyethylene glycol 400. Initially, clusters of crystals appeared, and these crystals were crushed and suspended in mother liquor. The slurry of mother liquor was quick-centrifuged, and 0.2 μl was inoculated to new drops with the same crystallization solution, except using half the concentrations of decamer DNAs. Single crystals appeared after 3 days of incubation and were fully grown in 10 days. Crystals were flash frozen under a nitrogen gas stream at 100 K without additional cryoprotectant. Multiple wavelength anomalous diffraction data collection was performed at Stanford Synchrotron Radiation Laboratory BL9-2 by using a BLU-ICE interface at three wavelengths, 0.92047, 0.84340, and 0.9065 Å. A total of 95 frames were collected for each wavelength. Each data set was integrated and scaled with the hkl program suite (40) (Table 1).
Table 1.
Crystal data
|
|
Native DNA
|
BrdUrd DNA | ||
|---|---|---|---|---|
| λ1 (peak) | λ2 (remote) | λ3 (edge) | ||
| Data | rigaku | ssrl | ||
| Space group | P212121 | |||
| Cell dimensions, Å | a = 21.11, b = 54.01, c = 74.19 | a = 24.50, b = 53.85, c = 75.33 | ||
| Asymmetric unit | 2 (decamer duplexes) | |||
| Resolution, Å | 20–2.3 | 50–1.9 | 50–1.9 | 50–2.1 |
| Wavelength, Å | 1.5418 | 0.9205 | 0.8434 | 0.9207 |
| Total reflections | 10,325 | 79,766 | 79,490 | 59,801 |
| Unique reflections | 4,616 | 8,062 | 7,865 | 6,101 |
| Completeness, % | 88.6 (76.4) | 96.4 (88.5) | 97.1 (97.7) | 97.6 (94.5) |
| R merge | 0.053 (0.198) | 0.051 (0.225) | 0.034 (0.228) | 0.029 (0.196) |
| Refinement | ||||
| Resolution, Å | 10–2.5 | 10–2.0 | ||
| Number of reflections | 3,220 | 6,819 | ||
| Rfree | 25.5 (30.21) | 26.4 (30.14) | ||
| Rcryst | 19.7 (25.2) | 19.8 (25.39) | ||
| rms deviation | ||||
| Bond length, Å | 0.034 | 0.023 | ||
| Bond angle, ° | 1.47 | 1.35 | ||
| Number of atoms | ||||
| DNA | 882 | 882 | ||
| Water | 56 | 62 | ||
Numbers in parentheses refer to statistics for the highest 0.1-Å resolution shell.
R merge = (ΣhΣi|〈Fh〉 − Fhi|)/ΣhFh, where 〈Fh〉 is the mean structure factor magnitude of i observations of symmetry-related reflections with Bragg index h.
Rfree is calculated with removal of 5% of the data as the test set at the beginning of refinement.
Rcryst = Σ∥Fobs| − |Fcal|/|Fobs|.
rms deviations of the bond lengths and bond angles from their respective ideal values as implemented in x-plor.
Phase Determination.
Heavy atom site location, phase calculation, and parameter refinement were performed with the automated phasing program solve (41) by using data between 50 and 3 Å, followed by solvent flattening in dm (42). There were two molecules in the asymmetric unit. The first electron density map showed clear base steps of only one DNA molecule. Performing the maximum-likelihood density modification as implemented in resolve (43) clarified the map region of the second molecule.
Model Building and Refinement.
The DNA model was built into the electron density by using the graphics program o (44). The initial model after rigid refinement had an R factor of 43%, which was improved to 26% after one round of simulated annealing by using x-plor (45). Iterative rounds of model adjustment using o and refinement using x-plor were performed to improve the model. This partially refined model from the derivative DNA crystal was transformed to native DNA by substituting two BrdUrds with two Ts followed by a subsequent round of rigid body refinement. Native data then went through a separate refinement process using x-plor. DNA helical parameters were calculated with 3DNA (46) and curves (47). The coordinates have been deposited in the Protein Data Bank.
Results
Crystal Packing and Global Structure.
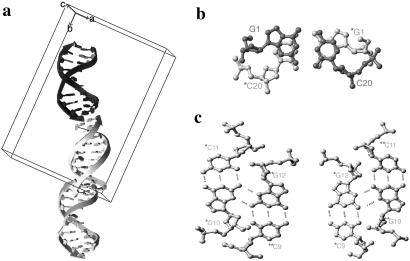
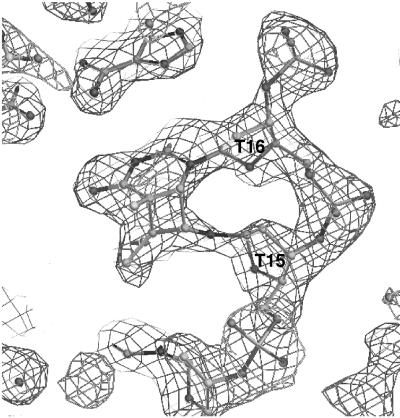
The decamer DNA containing a CPD (Fig. 1) crystallizes in the orthorhombic space group P212121, and the crystals diffract to 1.9-Å resolution. Two independent decamer duplex molecules found in the crystal lattice are stacked on top of each other through two types of contacts (Fig. 2). All atoms for both molecules have been modeled and are well defined, as indicated by the quality of the electron density maps (Fig. 3). Even though the packing environments of the two molecules are quite different, they are virtually superimposable, with a rms deviation of 0.82 Å. In one interface, two decamer molecules are stacked on each other with a base-stacking distance of 3.4 Å between the contacting terminal residues (G1-C20). One duplex molecule is rotated ≈36° relative to the other duplex in a left-handed manner, making the backbone/major-minor groove interaction discontinuous at this duplex interface. The other interface is stabilized by formation of unique intermolecular base pairs between the terminal two residues of independent duplexes, resulting in an interface where the base-paired guanines of two duplexes of the same polarity stack on top of each other. Even though two guanine bases form extra symmetric homo purine type base pairs, the local B-DNA helical structure was not perturbed by this lattice formation. We have tried to make a crystal with an undamaged DNA of the same sequence without success, thus this lattice may be possible only through a specific interaction of the damaged decamer sequence.
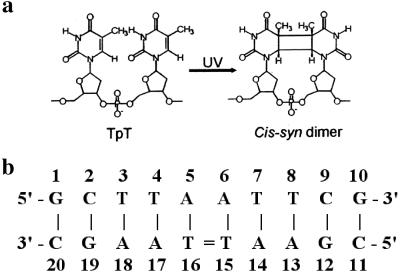
Fig 1.
Structure of the cis-syn dimer (a) and numbering of the decamer sequence (b).
Fig 2.
Diagram showing the lattice packing of molecules in the unit cell. (a) One asymmetrical unit is composed of two duplex molecules (black–light gray or light gray–dark gray). They are held together by the stacking (b) and interstrand (c) hydrogen bonds. Two guanine bases are hydrogen bonded to each other in a symmetrical way. The bases involved are labeled.
Fig 3.
Multiple-wavelength anomalous diffraction electron density around the CPD site (1-s level). Selected bases are labeled. The figure was made by the program O (44).
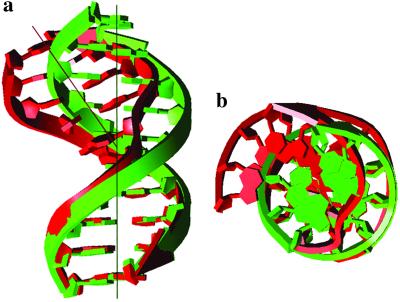
The overall structure maintains a continuous base-stacked and right-handed helical structure. It forms three distinct segments: two regular tracts of B-DNA at the top (three base pairs) and bottom (four base pairs) and the central segment containing the CPD. The overall helical axis bends ≈30° toward the major groove, caused by the presence of the CPD (Fig. 4a). This bend is greater than that of ≈10° reported for the NMR structures (34, 38) and from a gel mobility assay (34) but is consistent with that of 30° from a circularization assay (28) and close to 27° from a theoretical calculation (19). The kinked shape of the DNA is also clear from the top view of the DNA (Fig. 4b). The observed bending toward the major groove is also consistent with the periodicity and positioning of dimer formation in nucleosomes and in looped DNA (48, 49).
Fig 4.
Schematic diagram illustrating the kink of the helix. Regular B-DNA and CPD-containing DNA are depicted in green and red, respectively. (a) Side view with a helical axis. (b) Top view.
Helix Parameter and Base Pair Parameter.
Inspection of the central segment of the molecule shows considerable changes in both local base pair step parameters and helical parameters surrounding the CPD site (T15, T16) due to the formation of the covalent bonds that bring two thymine bases together. The average value of the helical twist of the decamer is 33.4° (±4.85). This average twist value corresponds to an unwinding angle of ≈9.3° (based on a helical repeat of 10.5 base pairs for B-DNA in solution), which is similar to that of −8.7° derived from experiment (26) and contrasts with the NMR solution structure (38), which is calculated to be overwound by ≈9°. However, without including the central three base pairs (A5-T16, A6-T15, and T7-A14), the value becomes 35.4°, approaching that of the ideal B-DNA, indicating that all of the unwinding induced by a CPD can be attributed to these three base pairs. The corresponding twisting angle between the central thymine base pairs of the CPD is only 26.6°, due to the fact that cyclobutane holds the two base pairs. The damaged site (T15, T16) step shows a positive roll of 22° into the major groove, which causes a localized bend at the center of the DNA.
Of particular interest is the parameter change of base pairs on the 5′ side of the CPD site. The twisting angle of the base pair (T7-A14), which is the 5′ neighbor of the CPD site, shows a reduced angle of 27°, indicating that this region of damaged DNA has a quite different conformation compared with regular B-DNA. A distinct tilt angle (−17.8°) also occurs at this base pair. In contrast, the base pair (T4-A17), which is on the 3′ side of the CPD, shows normal B-DNA-like twisting (37.1°) and tilt (−7.4°) angles.
Main Chain and χ Torsion Angles.
The glycosidic torsion angles, χ, of all of the residues except for the T15, show little deviation from that of the standard B-DNA, all falling in the anti conformation (−100 ≈ −130°). The 5′ thymine residue in the CPD site, T15, uniquely shows the syn (−52.2°) conformation, which agrees with the NMR structure (38).
The difference in the conformation of the sugar phosphate chains around the CPD site is readily observed in the backbone torsion angles. The torsion angle β [defined by P(n)-O5′(n + 1)-C5′-C4′] around the lesion adopts a lower value of 148° from its canonical value of ≈175°. The decrease in this β angle apparently facilitates the accommodation of the constrained geometry of the T15-T16 dinucleotide, creating a pinching effect in the minor groove at the CPD site as in the recent NMR structure (38). Pinching in the crystal structure is more dramatic, creating a zigzag conformation of the phosphodiester backbone (Figs. 4a and 5).
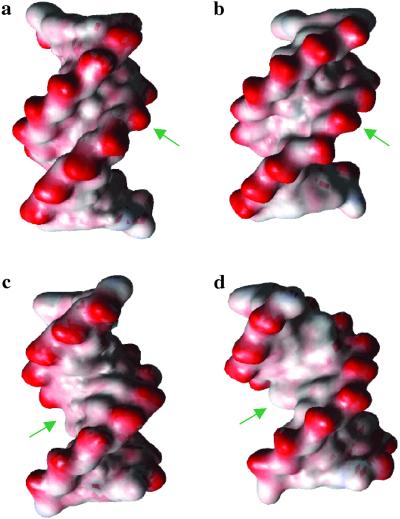
The distances between the phosphates at both minor and major grooves are 2≈3 Å wider at the damaged site than the averaged value of B-DNA (Fig. 6). Considering the report from Hizver et al. (50) that the adenine tract significantly narrows the minor groove to the value of ≈9.5 Å, the observed widening of the minor groove in our crystal structure is even more significant. Previously, the cocrystal structure of T4 endonuclease in complex with damaged DNA showed that the enzyme interacts in the minor groove of the damaged DNA (20, 51). One might expect that the wide minor groove architecture observed in our crystal could be an important enzyme recognition motif. Then enzyme widens the minor groove by an additional ≈4 Å by inserting its residues deeper into the minor groove.
Fig 6.
Electropotential surface models (a); (c) an ideal B-form decamer DNA (b); (d) crystal structure of the CPD containing decamer DNA (same sequence), with probe radii of 1.4 Å. The CPD site and corresponding site of the ideal B-form DNA are indicated by arrows. The duplex DNA molecule containing the CPD has an enlarged minor and major groove. The figure was produced by using VMD (58).
Compared with the canonical B-DNA, two backbone dihedral parameters ɛ [defined by C4′-C3′-O3′-P(n + 1)] and ζ [defined by C3′-O3′-P(n + 1)-O5′(n + 1)] between the T16-A17 residues undergo drastic transitions as shown in the NMR structure (38); i.e., the ζ value changes from ∼gauche− to a ∼trans conformation, and the ɛ value changes from the trans value to a gauche−. This BII higher-energy backbone conformation has been proposed as an energetically favorable destacking mechanism of the 3′ adenine base in duplex containing a 5′-T = TpA-3′ segment (38). But the observed increase in the rise of the neighboring base pair due to this BII conformation in the same NMR structure is not observed in this crystal structure.
Base-Pairing Interactions at the Thymine Dimer Lesion.
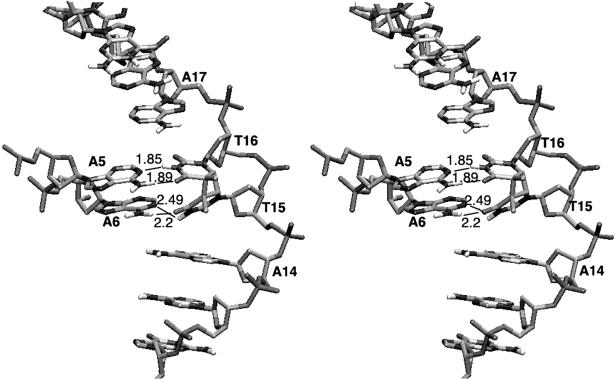
In Fig. 5, the base-pairing pattern of the CPD is shown. The central cyclobutane ring is puckered by ≈20°, and hydrogen bond patterns between the two thymines are quite different. The 3′ thymine (T16) maintains normal WC hydrogen bonds (1.85 and 1.89 Å) with the complementary base (A5). On the other hand, one hydrogen bond is lost between the 5′ thymine (T15) and the complementary adenine (A6). The hydrogen bond distance between N6H of A6 and O4 of T15 is 2.49 Å, which is longer than the normal range, and its orientation vector is not in its proper plane. The weakened H-bonding of the 5′-T is consistent with previous NMR studies of the exchangeable protons (36, 38). The crystal structure of T4 endoV complexed with thymine dimer-containing DNA has the adenine complementary to the 5′ thymine flipped out (20). Our results suggest that the weakened nature of the hydrogen bond in this base pair may facilitate the enzyme flipping out the adenine base. We could not find any significant water interactions near the CPD site.
Fig 5.
Stereo view representation of the detailed surroundings of the CPD site. Hydrogen bonds are drawn as dashed lines with distances. The view shows part of the major groove of the molecule. The hydrogen bond between N6H of A6 and O4 of T15 is significantly weakened. The phosphodeoxyribose backbone shows a sharply kinked or pinched structure. The figure was produced by using VMD (58).
Discussion
The efficiency of damage recognition and disruption of DNA–protein interactions is likely to depend on the extent to which the damage alters the structure or deformability of the DNA. Our crystal structure clearly shows a 30° bend and significant differences compared with the standard B-DNA, including the pinching effect in the 3′ side of the CPD lesion, severe change of the base pair parameter in the 5′ side, and serious widening of both minor and major groves both 3′ and 5′ of the CPD. Consequently, the CPD-containing DNA molecule presents quite different surfaces (Fig. 6) compared with B-DNA that could attract repair enzymes and interfere with normal protein–DNA interactions. Whether the crystal structure obtained represents a static structure of a CPD-containing duplex in solution or an easily accessible structure remains to be determined, but the possibility that structure is highly flexible could explain the conflicting theoretical and experimental results regarding the magnitude of the bend.
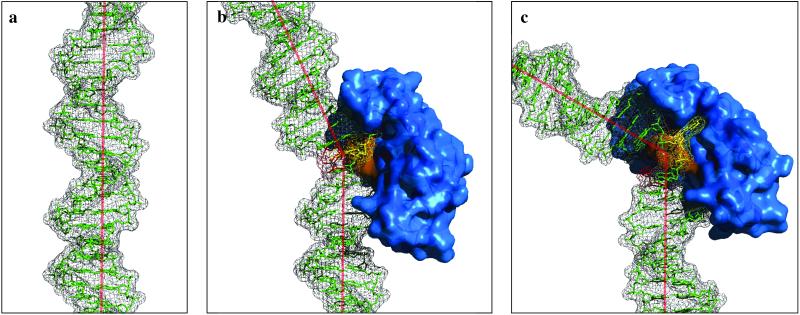
The poor H-bonding between the 5′-T of the dimer and the paired A along with the altered phosphate backbone and large bend observed in the crystal structure is consistent with a number of proposed models for dimer recognition involving base-flipping mechanisms. The best established model is for the T4 endonuclease V enzyme, which binds to a structure in which the A opposite the 5′-T is flipped out and bound by a pocket in the enzyme (51). As shown in Fig. 7, this enzyme might first recognize the dimer by detecting the bend or the bendability of the DNA and an altered electrostatic field due to the perturbed helical geometry around the dimer, and then in a concerted action flips out the base and widens the minor groove. It bends the DNA even further in the same major groove direction, probably due to binding of the enzyme to the minor groove and to the increased flexibility of the DNA due to the flipped out A.
Fig 7.
Possible recognition mechanism of CPD site. (a) B-form DNA. (b) Model of the T4 endonuclease V enzyme bound to a CPD containing DNA. (c) Crystal structure of the T4 endonuclease V enzyme complexed with a CPD containing DNA (20). The direction of kinking (or bending) in the helical axis is the same between b and c.
Another type of base-flipping mechanism has been proposed for the recognition of CPDs by E. coli photolyase (6). In this case, the crystal structure of the enzyme by itself and other experimental observations suggest that the dimer flips out of the helix into an active site pocket. This enzyme might initially recognize the 30° bend in the DNA or its bendability. In favor of such a mechanism, it has been found by atomic force microscopy that dimer-containing DNA is bent by 36° when bound by Anacystis nidulans photolyase, although the CPD-containing DNA did not appear to cause significant bending on its own (52). This enzyme may also initially recognize the altered phosphate backbone, because this and other photolyases have been found to make specific contacts with the phosphate immediately 5′ and the three phosphates immediately 3′ to the dimer as well as the phosphate across from the dimer (53). Recently, evidence that the DNA damage recognition subunit of the uvrABC excinuclease, uvrB, recognizes DNA damage by flipping of the bases opposite to the damage has been obtained, which would explain how a cis-syn dimer can be recognized (54, 55). It has also been suggested that the binding affinities of the repair enzymes for damaged DNA depend on the degree of DNA unwinding or bending caused by the lesions (51).
The local and long-range distortions of DNA conformation caused by CPDs are also expected to affect normal protein–DNA interactions important in replication and transcription (56). The altered base-pairing geometry could explain in part how CPDs arrest synthesis by DNA and RNA polymerases. Widening of the grooves coupled with bending and unwinding of the DNA could explain how CPDs in promoters interfere with transcription factor binding and consequently with the initiation of transcription. Longer-range effects that arise from kinking or displacement of the helix axis or unwinding of the DNA duplex in turn could interfere with the assembly of multicomponent transcription complexes by changing the orientation between the individual transcription factors (19, 29, 57).
Acknowledgments
Research described in this article was supported by grants to C.H.K. from the National Institutes of Health (Grant NIH-GM53651) and the Murdock Charitable Trust, and to J.-S.T. from the National Institutes of Health (Grant NIH-CA40463).
Abbreviations
CPD, cyclobutane-type pyrimidine photodimer
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes and ).
References
- 1.Cadet J. & Vigny, P., (1990) The Photochemistry of Nucleic Acids (Wiley, New York).
- 2.Cleaver J. E. & Crowley, E. (2002) Front. Biosci. 7, 1024-1043. [DOI] [PubMed] [Google Scholar]
- 3.Beukers R., Ylstra, J. & Berends, W. (1960) Rec. Trav. Chim. 79, 101-104. [Google Scholar]
- 4.Wacker A., Dellweg, H. & Weinblum, D. (1960) Naturwissenschaften 47, 477. [Google Scholar]
- 5.Setlow R. B. & Carrier, W. L. (1966) J. Mol. Biol. 17, 237-254. [DOI] [PubMed] [Google Scholar]
- 6.Sancar G. B. (2000) Mutat. Res. 451, 25-37. [DOI] [PubMed] [Google Scholar]
- 7.Park H. W., Kim, S. T., Sancar, A. & Deisenhofer, J. (1995) Science 268, 1866-1872. [DOI] [PubMed] [Google Scholar]
- 8.Lloyd R. S. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 155-175. [DOI] [PubMed] [Google Scholar]
- 9.Sancar A. (1996) Annu. Rev. Biochem. 65, 43-81. [DOI] [PubMed] [Google Scholar]
- 10.Batty D. P. & Wood, R. D. (2000) Gene 241, 193-204. [DOI] [PubMed] [Google Scholar]
- 11.Mellon I., Bohr, V. A., Smith, C. A. & Hanawalt, P. C. (1986) Proc. Natl. Acad. Sci. USA 83, 8878-8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svejstrup J. Q. (2002) Mol. Cell. Biol. 3, 21-29. [DOI] [PubMed] [Google Scholar]
- 13.Setlow R. B., Swenson, P. A. & Carrier, W. L. (1963) Science 142, 1464-1466. [DOI] [PubMed] [Google Scholar]
- 14.Tang M., Pham, P., Shen, X., Taylor, J.-S., O'Donnell, M., Woodgate, R. & Goodman, M. F. (2000) Nature 404, 1014-1018. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R. E., Prakash, S. & Prakash, L. (1999) Science 283, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 16.Masutani C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K. & Hanaoka, F. (1999) Nature 399, 700-704. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z. (2001) Mutat. Res. 486, 59-70. [DOI] [PubMed] [Google Scholar]
- 18.Liu H., Hewitt, S. R. & Hays, J. B. (2000) Genetics 154, 503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearlman D. A., Holbrook, S. R., Pirkle, D. H. & Kim, S.-H. (1985) Science 227, 1304-1308. [DOI] [PubMed] [Google Scholar]
- 20.Vassylyev D. G., Mikami, Y., Ariyoshi, M., Iwai, S., Ohtsuka, E. & Morikawa, K. (1995) Cell 83, 773-782. [DOI] [PubMed] [Google Scholar]
- 21.Shafrnovskaya N. N., Trifonov, E. N., Lazurkin, Y. S. & Frank-Kamenetskii, M. D. (1973) Nat. New Biol. 241, 58-60. [DOI] [PubMed] [Google Scholar]
- 22.Denhardt D. T. & Kato, A. C. (1973) J. Mol. Biol. 77, 479-494. [DOI] [PubMed] [Google Scholar]
- 23.Legerski R. J., Gray, H. B. & Robberson, D. L. (1977) J. Biol. Chem. 252, 8740-8746. [PubMed] [Google Scholar]
- 24.Wang T.-M. & McLaren, A. D. (1972) Biophysik 8, 237-244. [Google Scholar]
- 25.Hayes F. N., Williams, D. L., Ratliff, R. L., Varglese, A. J. & Rupert, C. S. (1971) J. Am. Chem. Soc. 93, 4940-4942. [DOI] [PubMed] [Google Scholar]
- 26.Spadari S., Sutherland, B. M., Pedrali-Noy, G., Focher, F., Chiesa, M. T. & Ciarrocchi, G. (1987) Toxicol. Pathol. 15, 82-87. [DOI] [PubMed] [Google Scholar]
- 27.Ciarrocchi G. & Pedrini, A. M. (1982) J. Mol. Biol. 155, 177-183. [DOI] [PubMed] [Google Scholar]
- 28.Husain I., Griffith, J. & Sancar, A. (1988) Proc. Natl. Acad. Sci. USA 85, 2558-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C.-I. & Taylor, J.-S. (1991) Proc. Natl. Acad. Sci. USA 88, 9072-9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooney M. G. & Miller, J. H. (1997) Nucleic Acids Res. 25, 1432-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghunathan G., Kieber-Emmons, T., Rein, R. & Alderfer, J. L. (1990) J. Biomol. Struct. Dyn. 7, 899-913. [DOI] [PubMed] [Google Scholar]
- 32.Rao S. N., Keepers, J. W. & Kollman, P. (1984) Nucleic Acids Res. 12, 4789-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miaskiewicz K., Miller, J., Cooney, M. & Osman, R. (1996) J. Am. Chem. Soc. 118, 9156-9163. [Google Scholar]
- 34.Kim J. K., Patel, D. & Choi, B. S. (1995) Photochem. Photobiol. 62, 44-50. [DOI] [PubMed] [Google Scholar]
- 35.Kemmink J., Boelens, R., Koning, T. M. G., Kaptein, R., Van der Marel, G. A. & van Boom, J. H. (1987) Eur. J. Biochem. 162, 37-43. [DOI] [PubMed] [Google Scholar]
- 36.Kemmink J., Boelens, R., Koning, R., van der Marel, G. A., van Boom, J. H. & Kaptein, R. (1987) Nucleic Acids Res. 15, 4645-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor J.-S., Garrett, D. S., Brockie, I. R., Svoboda, D. L. & Telser, J. (1990) Biochemistry 29, 8858-8866. [DOI] [PubMed] [Google Scholar]
- 38.McAteer K., Jing, Y., Kao, J., Taylor, J.-S. & Kennedy, M. A. (1998) J. Mol. Biol. 282, 1013-1032. [DOI] [PubMed] [Google Scholar]
- 39.Taylor J.-S., Brockie, I. R. & O'Day, C. L. (1987) J. Am. Chem. Soc. 109, 6735-6742. [Google Scholar]
- 40.Otwinowski Z. & Minor, W., (1997) Macromolecular Crystallography Part A (Academic, New York).
- 41.Terwilliger T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34-38. [Google Scholar]
- 43.Terwilliger T. C. (2000) Acta Crystallogr. D 56, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones T. A., Zou, J. Y., Cowan, S. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 45.Brünger A. T., (1992) X-PLOR: A System for X-Ray Crystallography and NMR (Yale Univ. Press, New Haven, CT), Ver. 3.1.
- 46.Lu X. J., El Hassan, M. A. & Hunter, C. A. (1997) J. Mol. Biol. 273, 668-680. [DOI] [PubMed] [Google Scholar]
- 47.Lavery R. & Sklenar, H. (1989) J. Biomol. Struct. Dyn. 6, 655-667. [DOI] [PubMed] [Google Scholar]
- 48.Pehrson J. R. & Cohen, L. H. (1992) Nucleic Acids Res. 20, 1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gale J. M., Nissen, K. A. & Smerdon, M. J. (1987) Proc. Natl. Acad. Sci. USA 84, 6644-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hizver J., Frolow, F., Rabinovich, D. & Shakked, Z. (2001) Proc. Natl. Acad. Sci. USA 98, 8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morikawa K. & Shirakawa, M. (2000) Mutat. Res. 460, 257-275. [DOI] [PubMed] [Google Scholar]
- 52.van Noort J., Orsini, F., Eker, A., Wyman, C., de Grooth, B. & Greve, J. (1999) Nucleic Acids Res. 27, 3875-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Husain I., Sancar, G. B., Holbrook, S. R. & Sancar, A. (1987) J. Biol. Chem. 262, 13188-13197. [PubMed] [Google Scholar]
- 54.Hsu D. S., Kim, S. T., Sun, Q. & Sancar, A. (1995) J. Biol. Chem. 270, 8319-8327. [DOI] [PubMed] [Google Scholar]
- 55.Moolenaar G. F., Hoglund, L. & Goosen, N. (2001) EMBO J. 20, 6140-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tommasi S., Swiderski, P. M., Tu, Y., Kaplan, B. E. & Pfeifer, G. P. (1996) Biochemistry 35, 15693-15703. [DOI] [PubMed] [Google Scholar]
- 57.Conconi A., Bespalov, V. A. & Smerdon, M. J. (2002) Proc. Natl. Acad. Sci. USA 99, 649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Humphrey W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14.1, 33-38. [DOI] [PubMed] [Google Scholar]