Abstract
Mitochondrial biogenesis is a critical adaptation to chronic energy deprivation, yet the signaling mechanisms responsible for this response are poorly understood. To examine the role of AMP-activated protein kinase (AMPK), an evolutionarily conserved fuel sensor, in mitochondrial biogenesis we studied transgenic mice expressing a dominant-negative mutant of AMPK in muscle (DN-AMPK). Both DN-AMPK and WT mice were treated with β-guanidinopropionic acid (GPA), a creatine analog, which led to similar reductions in the intramuscular ATP/AMP ratio and phosphocreatine concentrations. In WT mice, GPA treatment resulted in activation of muscle AMPK and mitochondrial biogenesis. However, the same GPA treatment in DN-AMPK mice had no effect on AMPK activity or mitochondrial content. Furthermore, AMPK inactivation abrogated GPA-induced increases in the expression of peroxisome proliferator-activated receptor γ coactivator 1α and calcium/calmodulin-dependent protein kinase IV (both master regulators of mitochondrial biogenesis). These data demonstrate that by sensing the energy status of the muscle cell, AMPK is a critical regulator involved in initiating mitochondrial biogenesis.
Keywords: β-guanidinopropionic acid, calcium, calmodulin-dependent protein kinase IV, peroxisome proliferator-activated γ receptor coactivator-1α, AMP-activated protein kinase
Long-term exercise training increases oxidative capacity in muscle by promoting mitochondrial biogenesis (1–3). This type of training is associated with chronic metabolic stress and energy deprivation. However, the signaling pathways linking metabolic stress to mitochondrial biogenesis are poorly understood. In this regard, AMP-activated protein kinase (AMPK) is an attractive potential candidate, and its role in this process has been suggested by recent correlative studies demonstrating that AMPK activation is associated with increased mitochondrial enzyme content (4) and mitochondrial biogenesis (5) in rat skeletal muscle. AMPK is the mammalian homologue of the yeast SNF1 gene, which in turn is required for the yeast survival response to glucose starvation (6–9). AMPK is activated by a low ATP/AMP ratio, and in this manner is thought to serve as a fuel gauge for mammalian cells to protect against energy deprivation (10). Acute activation of this enzyme in muscle helps defend against energy deficiency by promoting increased glucose transport and fatty acid oxidation through increased GLUT4 translocation (11–13) and inhibition of acetyl CoA carboxylase respectively (4, 6, 14–16). Because of its localization in the nuclei of many cells, it is also thought to be involved in the regulation of gene expression (17, 18). Other factors that are thought to play a role in mitochondrial biogenesis include peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) (19, 20) and calcium/calmodulin-dependent protein kinase IV (CaMK IV) (21). Evidence for this comes from the observation of increased mitochondrial density in cultured myoblasts where PGC-1α was overexpressed by retrovirus transfection (19), and in transgenic mice studies where a constitutively active form of CaMK IV was overexpressed in skeletal muscle (21). However, the mechanisms controlling PGC-1α and CaMK IV expression in muscle are not known. To definitively examine the role of AMPK in mitochondrial biogenesis and its effect on other known signaling molecules in this process, we examined mice with a dominant-negative mutant of AMPK expressed in muscle (DN-AMPK). Here we show that AMPK is required for increased muscle mitochondrial biogenesis in response to chronic energy deprivation. Furthermore, we demonstrate that AMPK activation is required for increased expression of PGC-1α and CaMK IV, indicating that AMPK is an important proximal signaling step in regulating their expression in response to chronic energy deficiency.
Methods
Animals.
Transgenic mice expressing DN-AMPKα2 in skeletal muscle (DN-AMPK) and WT littermate mice (25–40 g) were created as described (22). Mice were housed in individual cages in a temperature-controlled environment with a 12 h light/12 h dark cycle and given standard chow and water freely. WT and transgenic mice were subsequently divided into two groups [control and β-guanidinopropionic acid (GPA) treated]. GPA was given by i.p. injections once daily for 8 weeks (0.2 ml, 0.5 M). The control group received daily injections of a comparable volume of saline for 8 weeks. Body weight and food consumption of the mice were measured every week. Overnight-fasted mice were anesthetized with pentobarbital sodium (50 mg/kg body weight), and different muscle groups were rapidly dissected and frozen in liquid nitrogen. All studies were approved by the Yale Animal Care and Use Committee.
Measurement of Nucleotides and Creatine Phosphate.
Superficial white gastrocnemius muscles were extracted by 6% perchloric acid. Approximately 4 mg of extracted sample was assayed for adenine nucleotide content by high-pressure liquid chromatography (23), and ≈8 mg of extracted sample was assayed for creatine phosphate content as described (24).
Measurement of Enzyme Activities.
To measure AMPK activity in skeletal muscles, superficial white gastrocnemius muscle was homogenized in buffer (20 mM Tris, pH 7.5/1 mM EDTA/100 mM mannitol/50 mM sodium fluoride/5 mM sodium pyrophosphate/1 mM DTT/1 mM benzamidine/0.04% trypsin inhibitor/0.03% Pefabloc/0.5% Triton X-100) and submitted to ultracentrifugation (48,000 × g for 30 min). After extraction, 2 μg of supernatant protein was assayed for AMPK activity by measuring incorporation of 32P into a synthetic peptide (SAMS) as described (25).
Western Blot Analysis.
Muscle was homogenized in homogenization buffer (50 mM Hepes/150 mM NaCl/1 mM EDTA/2 mM sodium vanadate/20 mM sodium pyrophosphate/100 mM sodium fluoride/1% Triton X-100/20 μg/ml aprotinin). Protein extracts from superficial white gastrocnemius were submitted to SDS/PAGE (4–12% gradient gel) and transferred to nitrocellulose membrane by using a semidry electroblotter (Owl Separation System, Portsmouth, NH). The membranes were immunoblotted with anti-cytochrome c monoclonal antibody (BioSource International, Camarillo, CA) and anti-CaMK II and IV monoclonal antibody (BD Biosciences), and detected using ECL chemiluminescence (Amersham Pharmacia Biotech). The blots were quantified by NIH image software. The blot was then stripped and immunoblotted for actin (Santa Cruz Biotechnology).
Preparation of cDNA Probes.
Generation of the actin probe was performed as described (5). The δ-aminolevulinate synthase (ALAS) (GenBank accession no. M63245) probe was amplified from mouse muscle total RNA and reverse transcribed using the Superscript One Step RT-PCR kit (Invitrogen) with an annealing temperature of 52°C (forward primer, 5′-AGATGATGCCAGGCTGTGAA-3′; reverse primer, 5′-GCAATGTATCCTCCAACACAG-3′). The COXII (accession no. AF3788230) probe was amplified from a rat muscle cDNA library by denaturing for 2 min at 95°C, and then amplifying 30 times (45 sec at 95°C, 1 min at 58°C, 2 min at 72°C) with a final extension of 5 min at 72°C (forward primer, 5′-GCACAATAGTGCACAAGAAGTTGAA-3′; reverse primer, 5′-ACCATTTCTAGGACAATGGGCATAA-3′). The PGC-1α probe was a kind gift from Clay Semenkovich (Washington University, St. Louis); 36B4 (acidic ribosomal phosphoprotein PO) was purchased from American Type Culture Collection.
RNA Isolation and Northern Blot.
Total RNA was isolated from the superficial white gastrocnemius muscle by TRIzol reagent (Invitrogen). Twenty micrograms of total RNA sample was loaded on a denaturing agarose gel and transferred to a supported nitrocellulose membrane (Hybond-N, Amersham Pharmacia Biotech), and then hybridized to different [32P]dCTP-labeled cDNA probes by using random priming labeling kit (Invitrogen). Northern blots were quantified using a phosphoimager and imagequant software (Molecular Dynamics). The blot was then stripped and hybridized to cDNA for actin.
DNA Isolation and Southern Blot.
Total DNA was isolated from the superficial white quadriceps muscle as described (26). Total DNA (≈25 μg) was digested by NcoI and subjected to Southern blot analysis using COX II cDNA probes (19). Southern blots were quantified using a phosphoimager and imagequant software. The blot was then stripped and hybridized to cDNA for 36B4 (acidic ribosomal phosphoprotein PO).
Transmission Electron Microscopy.
For electron microscopic examination, individual muscle samples were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at 4°C overnight, postfixed in 1% osmium tetroxide in the same buffer for 1 h at room temperature, stained in 2% uranyl acetate, dehydrated in a graded series of ethanol, and embedded in epoxy resin (Embed-812, Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections (60 nm) were stained with 2% uranyl acetate and lead citrate, and examined in a Philips 410 electron microscope. For each individual mouse and muscle, two random sections were examined and, from each section, six random pictures were taken at a magnification of 7,100 and printed at a final magnification of 18,250. The volume density of mitochondria was estimated using the point counting method (27). Counting was carried out by someone with no previous knowledge of the experiment (blind method). For each set of six pictures, average volume density was calculated and the mean of both values used to estimate the volume density for each individual mouse and muscle. These values were in turn used to calculate the average volume density for each treatment.
Statistical Analysis.
Results were presented as mean ± SEM. Differences between groups were examined for statistical significance by using Student's t test or ANOVA with Fischer's probable least-squares difference (PLSD) post hoc testing.
Results
Mouse Characteristics.
There were no differences between food intake (WT-NS, 4.7 ± 0.7 g; WT-GPA, 4.6 ± 0.48 g; DN-AMPK-NS, 4.8 ± 0.36 g; DN-AMPK-GPA, 4.7 ± 0.42 g) or body weight (WT-NS 32.2 ± 1.6 g vs. WT-GPA 31.7 ± 1.1 g; DN-AMPK-NS 32 ± 1.5 g; DN-AMPK-GPA, 31.2 ± 1.1 g) in any of the groups following 8 weeks of saline or GPA treatment (NS, normal saline). Randomly fed plasma concentrations of glucose (WT-NS, 217 ± 2 mg/dl; WT-GPA, 193 ± 4 mg/dl; DN-AMPK-NS, 210 ± 2 mg/dl; DN-AMPK-GPA, 257 ± 2 mg/dl) and insulin (WT-NS, 38 ± 6 microunits/ml; WT-GPA, 31 ± 5 microunits/ml; DN-AMPK-NS, 44 ± 12 microunits/ml; DN-AMPK-GPA, 37 ± 3 microunits/ml) were also similar in all four groups at time of sacrifice.
Effect of GPA on Muscle High-Energy Phosphate Metabolites and AMPK Activity.
GPA is a creatine analogue that competes for the transport of creatine in the skeletal muscle (28) and inhibits creatine kinase activity (29). As a result, chronic GPA treatment is a pharmacologic method to mimic endurance exercise training by chronically depleting intracellular phosphocreatine and ATP concentrations (5, 28, 29). This pharmacologic approach has a major advantage in these DN-AMPK transgenic mice, which do not tolerate exercise (22). GPA caused a 40–60% decrease in muscle creatine phosphate, ATP, and ADP content in both WT mice and DN-AMPK transgenic mice (Table 1). The ATP/AMP ratio, which is an important regulator of AMPK activity, was decreased by ≈60% in muscle of both GPA-treated WT mice and GPA-treated DN-AMPK transgenic mice (Table 1). AMPK activity was increased by 94% (P < 0.001) in WT mice injected with GPA. In contrast, no increase in AMPK activity was observed in DN-AMPK transgenic mice injected with GPA (Table 1).
Table 1.
Muscle high-energy phosphate metabolite concentrations and AMPK activity
| WT-NS | WT-GPA | DN-AMPK | DN-AMPK-GPA | |
|---|---|---|---|---|
| Creatine phosphate | 14.48 ± 1.04 | 6.42 ± 0.68 | 15.37 ± 0.92 | 5.6 ± 0.46 |
| ATP | 4.91 ± 0.77 | 2.81 ± 0.69 | 4.56 ± 0.67 | 1.97 ± 0.71 |
| ADP | 0.73 ± 0.11 | 0.45 ± 0.10 | 0.91 ± 0.11 | 0.42 ± 0.14 |
| AMP | 0.08 ± 0.01 | 0.09 ± 0.02 | 0.10 ± 0.01 | 0.10 ± 0.02 |
| ATP/AMP | 59.37 ± 3.84 | 23.74 ± 4.45 | 46.58 ± 6.67 | 19.00 ± 3.37 |
| AMPK activity | 52.5 ± 9.27 | 101.77 ± 4.26 | 30.03 ± 1.63 | 36.33 ± 3.54 |
Creatine phosphate, ATP, ADP, AMP, ATP/AMP, and AMPK activity of white gastrocnemius muscles in WT and DN-AMPK mice treated with either saline or GPA (n = 5 in each group). Concentrations are expressed in μmol⋅g−1 wet weight, except for AMPK activity, which is expressed as pmol⋅mg−1⋅min−1.
, P < 0.01 vs. saline treated;
, P < 0.05 vs. saline treated.
Effect of GPA Treatment on CaMK II, CaMK IV, and PGC-1α Expression.
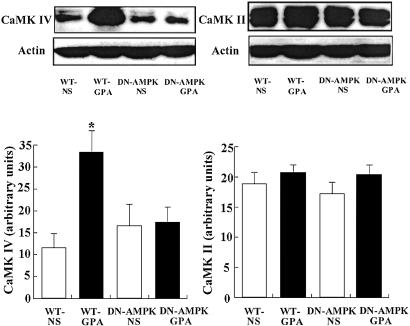
GPA treatment caused an ≈2-fold increase in the protein expression of CaMK IV, as assessed by Western blot analysis (Fig. 1), and PGC-1α mRNA expression, as assessed by Northern blot analysis (Fig. 2), from white gastrocnemius muscle of WT mice compared with untreated WT mice. In contrast GPA treatment had no effect on CaMK IV protein expression (Fig. 1) or PGC-1α mRNA (Fig. 2) expression in DN-AMPK transgenic mice. There was no effect of GPA treatment on the expression of CaMK II, as assessed by Western blot analysis, in either group (Fig. 1).
Fig 1.
Effect of GPA treatment on protein expression of CaMK IV and CaMK II in WT and DN-AMPK transgenic mice assessed by Western blot analysis (n = 5 in each group). *, P < 0.05 compared with other groups.
Fig 2.
Effect of GPA treatment on PGC-1α mRNA expression, as assessed by Northern blot analysis, in WT and DN-AMPK transgenic mice (n = 5 in each group). *, P < 0.01 compared with other groups.
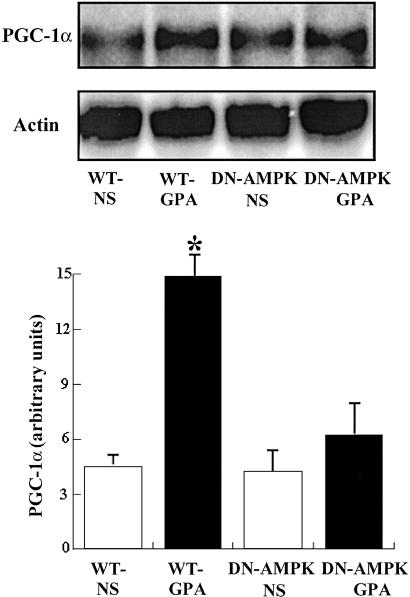
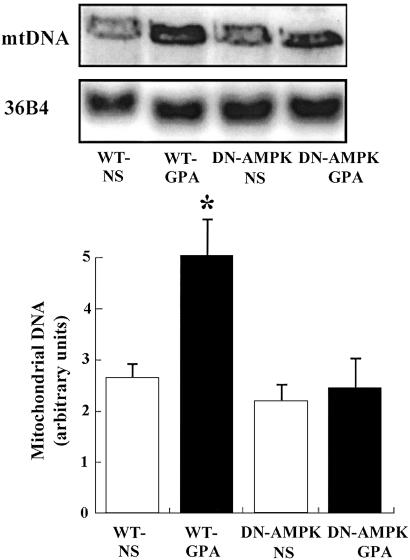
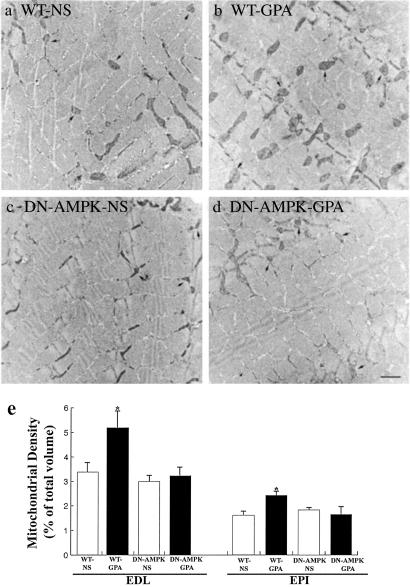
Effect of GPA on Mitochondrial Biogenesis and Mitochondrial Enzyme Content.
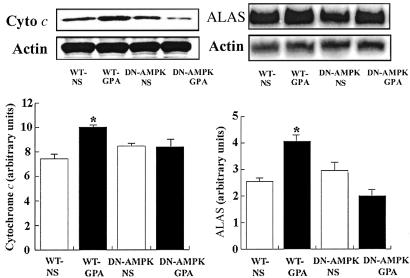
mtDNA content in white quadriceps muscle was assessed by Southern blot analysis. As shown in Fig. 3, mtDNA content in WT mice injected with GPA was increased ≈2-fold (P < 0.05). Mitochondrial densities of extensor digitorum longus (EDL) and the epitrochlearis (EPI) were also increased, as quantified by transmission electron microscopy, in WT mice injected with GPA (Fig. 4). Consistent with this finding, expression of cytochrome c protein content, as assessed by Western blot analysis, and ALAS mRNA content, as assessed by Northern blot analysis, were also increased in the muscle of GPA-treated WT mice (Fig. 5). In contrast, there were no changes in these parameters in GPA-treated DN-AMPK transgenic mice.
Fig 3.
Effect of GPA treatment on mtDNA in WT and DN-AMPK transgenic mice, as assessed by Southern blot analysis (n = 5 in each group). *, P < 0.05 compared with other groups.
Fig 4.
Effect of GPA treatment on mitochondrial content assessed by electron microscopy of the epitrochlearis (EPI) muscle of (a) WT mouse (saline treated), (b) WT mouse (GPA treated), (c) DN-AMPK transgenic mouse (saline treated), and (d) DN-AMPK transgenic mouse (GPA treated). Arrows point to mitochondria. (Bar, 1 μm.) (e) Mitochondria density (% of total volume) in the extensor digitorum longus (EDL; n = 10 in each group) and EPI muscles of each group mice (n = 5 in each group). *, P < 0.05 compared with other groups.
Fig 5.
Effect of GPA treatment on protein expression of cytochrome c, as assessed by Western blot analysis, and ALAS mRNA, as assessed by Northern blot analysis (n = 5 in each group). *, P < 0.05 compared with other groups.
Discussion
Endurance exercise training induces skeletal muscle mitochondrial biogenesis and improves performance by increasing oxidative capacity (2, 3). These adaptations also occur in nonexercising muscles chronically depleted of phosphocreatine and ATP following chronic GPA treatment (5). Under these conditions, muscle AMPK is activated and associated with increased expression of a number of transcription factors involved in mitochondrial biogenesis (5). The primary goal of this study was to directly assess the role of AMPK in skeletal muscle mitochondrial biogenesis. Using transgenic mice expressing dominant inhibitory catalytic subunit in muscle, we have demonstrated that there is no mitochondrial biogenesis or activation of important known signaling pathways involved in this process in response to chronic muscle energy deprivation. This constitutes direct evidence that muscle AMPK plays a critical role in mitochondrial biogenesis.
Prior studies in cultured C2C13 myoblasts transfected with a retroviral vector containing PGC-1α have shown that PGC-1α is a key regulator of mitochondrial biogenesis (19). Furthermore, transgenic mice overexpressing PGC-1α in the heart also show an induction of cardiac mitochondrial biogenesis (30). PGC-1α stimulates the expression of nuclear respiratory factors (NRF-1 and NRF-2) and mitochondrial transcriptional factor A (mtTFA), which in turn activates expression of nuclear and mitochondrial genes encoding mitochondrial proteins (19). Similarly, CaMK IV has recently been shown to be an important transcriptional regulation factor that regulates PGC-1α expression and mitochondrial biogenesis (21). Indeed in this study, we found that chronic GPA treatment caused a 2-fold increase in the expression of CaMK IV and PGC-1α in skeletal muscle, which did not occur in the DN-AMPK transgenic mice. Taken together, these data demonstrate that AMPK is required for increased expression of CaMK IV and PGC-1α in response to chronic energy deprivation. In contrast to CaMK IV, there was no effect of GPA on expression of CaMK II in the muscle of WT mice, suggesting that CaMK II does not play a major role in regulating mitochondrial biogenesis in response to chronic energy deprivation.
ALAS is the initial, and rate-limiting, enzyme in the synthesis of heme, an essential component for electron transfer and mitochondrial respiration. ALAS content depends on transcriptional and translational/posttranslational control (31, 32). The promoter of the ALAS gene has a functional binding site for NRF-1 (33). The expression of ALAS during physiological increases in energy demand is under the control of NRF-1 (34). This transcription factor is also involved in the up-regulation of cytochrome c expression (35), as observed in response to metabolic stressors such as electrical pacing of cardiac myocyte (36, 37). NRF-1 may also regulate mitochondrial transcription and replication through induction of mtTFA (38). In the present study, we measured ALAS mRNA content, cytochrome c protein expression, and mitochondrial density, and found all of these parameters to be increased in the muscles of GPA-treated WT mice. In contrast, GPA treatment had no effect on these parameters in the DN-AMPK transgenic mice. These results provide further evidence that activation of AMPK is a critical factor in the regulation of mitochondrial biogenesis.
In conclusion, the present study shows that activation of muscle AMPK is required for mitochondrial biogenesis in response to chronic energy deficiency in vivo. Furthermore, we found that AMPK is required for increased expression of CaMK IV and PGC-1α. Taken together, these data suggest that, by sensing the energy status of the cell, AMPK may be the initial signaling step that links the acute metabolic response to energy deprivation of increased glucose transport, and fat oxidation to the chronic adaptive response of increased mitochondrial biogenesis.
Acknowledgments
We are grateful for the technical assistance of Yan Chen, Yanlin Wang, Suzanne Neschen, Zhen-Xiang Liu, Christina Horensavitz, Gary Cline, and Varman Samuel. This work was supported by United States Public Health Service Grants R01 DK-40936, U24 DK-59635, P30 DK-45735, and R01 HL-6381 and a mentor-based fellowship award from the American Diabetes Association. M.J.B. and G.I.S. are investigators of the Howard Hughes Medical Institute.
Abbreviations
AMPK, AMP-activated protein kinase
CaMK, calcium/calmodulin-dependent protein kinase
DN-AMPK, dominant-negative mutant of AMPK expressed in muscle
GPA, β-guanidinopropionic acid
NS, normal saline
References
- 1.Holloszy J. O. (1975) Med. Sci. Sports 7, 155-164. [PubMed] [Google Scholar]
- 2.Hood D. A. (2001) J. Appl. Physiol. 90, 1137-1157. [DOI] [PubMed] [Google Scholar]
- 3.Williams R. S., Salmons, S., Newsholme, E. A., Kaufman, R. E. & Mellor, J. (1986) J. Biol. Chem. 261, 376-380. [PubMed] [Google Scholar]
- 4.Winder W. W., Holmes, B. F., Rubink, D. S., Jensen, E. B., Chen, M. & Holloszy, J. O. (2000) J. Appl. Physiol. 88, 2219-2226. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron R., Ren, J. M., Cadman, K. S., Moore, I. K., Perret, P., Pypaert, M., Young, L. H., Semenkovich, C. F. & Shulman, G. I. (2001) Am. J. Physiol. 281, E1340-E1346. [DOI] [PubMed] [Google Scholar]
- 6.Carling D., Aguan, K., Woods, A., Verhoeven, A. J., Beri, R. K., Brennan, C. H., Sidebottom, C., Davison, M. D. & Scott, J. (1994) J. Biol. Chem. 269, 11442-11448. [PubMed] [Google Scholar]
- 7.Woods A., Munday, M. R., Scott, J., Yang, X., Carlson, M. & Carling, D. (1994) J. Biol. Chem. 269, 19509-19515. [PubMed] [Google Scholar]
- 8.Hardie D. G., Carling, D. & Carlson, M. (1998) Annu. Rev. Biochem. 67, 821-855. [DOI] [PubMed] [Google Scholar]
- 9.Hardie D. G. (1999) Biochem. Soc. Symp. 64, 13-27. [PubMed] [Google Scholar]
- 10.Hardie D. G. & Carling, D. (1997) Eur. J. Biochem. 246, 259-273. [DOI] [PubMed] [Google Scholar]
- 11.Ihlemann J., Ploug, T., Hellsten, Y. & Galbo, H. (1999) Am. J. Physiol. 277, E208-E214. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi T., Hirshman, M. F., Fujii, N., Habinowski, S. A., Witters, L. A. & Goodyear, L. J. (2000) Diabetes 49, 527-531. [DOI] [PubMed] [Google Scholar]
- 13.Bergeron R., Previs, S. F., Cline, G. W., Perret, P., Russell, R. R., III, Young, L. H. & Shulman, G. I. (2001) Diabetes 50, 1076-1082. [DOI] [PubMed] [Google Scholar]
- 14.Kudo N., Barr, A. J., Barr, R. L., Desai, S. & Lopaschuk, G. D. (1995) J. Biol. Chem. 270, 17513-17520. [DOI] [PubMed] [Google Scholar]
- 15.Dagher Z., Ruderman, N., Tornheim, K. & Ido, Y. (2001) Circ. Res. 88, 1276-1282. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson L. L., Fischer, M. A. & Lopaschuk, G. D. (2002) J. Biol. Chem. 277, 29424-29430. [DOI] [PubMed] [Google Scholar]
- 17.Kemp B. E., Mitchelhill, K. I., Stapleton, D., Michell, B. J., Chen, Z. P. & Witters, L. A. (1999) Trends Biochem. Sci. 24, 22-25. [DOI] [PubMed] [Google Scholar]
- 18.Salt I., Celler, J. W., Hawley, S. A., Prescott, A., Woods, A., Carling, D. & Hardie, D. G. (1998) Biochem. J. 334, 177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C. & Spiegelman, B. M. (1999) Cell 98, 115-124. [DOI] [PubMed] [Google Scholar]
- 20.Lin J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418, 797-801. [DOI] [PubMed] [Google Scholar]
- 21.Wu H., Kanatous, S. B., Thurmond, F. A., Gallardo, T., Isotani, E., Bassel-Duby, R. & Williams, R. S. (2002) Science 296, 349-352. [DOI] [PubMed] [Google Scholar]
- 22.Mu J., Brozinick, J. T., Jr., Valladares, O., Bucan, M. & Birnbaum, M. J. (2001) Mol. Cell 7, 1085-1094. [DOI] [PubMed] [Google Scholar]
- 23.Sabina R. L., Kernstine, K. H., Boyd, R. L., Holmes, E. W. & Swain, J. L. (1982) J. Biol. Chem. 257, 10178-10183. [PubMed] [Google Scholar]
- 24.Heinz F. & Weiber, H. (1987) in Methods of Enzymatic Analysis, ed. Bergmeyer, H. U. (VCH, New York), Vol. 8, pp. 507–514. [Google Scholar]
- 25.Winder W. W. & Hardie, D. G. (1996) Am. J. Physiol. 270, E299-E304. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 1, pp. 6.3–6.62. [Google Scholar]
- 27.Weibel E. R., (1979) Stereological Methods (Academic, London), pp. 101–161.
- 28.Fitch C. D., Shields, R. P., Payne, W. F. & Dacus, J. M. (1968) J. Biol. Chem. 243, 2024-2027. [PubMed] [Google Scholar]
- 29.O'Gorman E., Beutner, G., Wallimann, T. & Brdiczka, D. (1996) Biochim. Biophys. Acta 1276, 161-170. [DOI] [PubMed] [Google Scholar]
- 30.Lehman J. J., Barger, P. M., Kovacs, A., Saffitz, J. E., Medeiros, D. M. & Kelly, D. P. (2000) J. Clin. Invest. 106, 847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holloszy J. O. & Winder, W. W. (1979) Am. J. Physiol. 236, R180-R183. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi M., McCurdy, D. T., Essig, D. A. & Hood, D. A. (1993) Biochem. J. 291, 219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braidotti G., Borthwick, I. A. & May, B. K. (1993) J. Biol. Chem. 268, 1109-1117. [PubMed] [Google Scholar]
- 34.Li B., Holloszy, J. O. & Semenkovich, C. F. (1999) J. Biol. Chem. 274, 17534-17540. [DOI] [PubMed] [Google Scholar]
- 35.Evans M. J. & Scarpulla, R. C. (1989) J. Biol. Chem. 264, 14361-14368. [PubMed] [Google Scholar]
- 36.Xia Y., Buja, L. M., Scarpulla, R. C. & McMillin, J. B. (1997) Proc. Natl. Acad. Sci. USA 94, 11399-11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Y., Buja, L. M. & McMillin, J. B. (1998) J. Biol. Chem. 273, 12593-12598. [DOI] [PubMed] [Google Scholar]
- 38.Virbasius J. V. & Scarpulla, R. C. (1994) Proc. Natl. Acad. Sci. USA 91, 1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]