Abstract
We have devised a high-resolution protein footprinting methodology to dissect HIV-1 reverse transcriptase (RT) contacts to the viral RNA:tRNA complex. The experimental strategy included modification of surface-exposed lysines in RT and RT-viral RNA:tRNA complexes by the primary amine selective reagent NHS-biotin, SDS/PAGE separation of p66 and p51 polypeptides, in gel proteolysis, and comparative mass spectrometric analysis of peptide fragments. The lysines modified in free RT but protected from biotinylation in the nucleoprotein complex were readily revealed by this approach. Results of a control experiment examining the RT-DNA:DNA complex were in excellent agreement with the crystal structure data on the identical complex. Probing the RT-viral RNA:tRNA complex revealed that a majority of protein contacts are located in the primer-template binding cleft in common with the RT-DNA:DNA and RT-RNA:DNA species. However, our footprinting data indicate that the p66 fingers subdomain makes additional contacts to the viral RNA:tRNA specific for this complex and not detected with DNA:DNA. The protein footprinting method described herein has a generic application for high-resolution solution structural studies of multiprotein-nucleic acid contacts.
Initiation of HIV-1 reverse transcription requires formation of a specific nucleoprotein complex comprising the viral RNA (vRNA) template, tRNA primer, and viral reverse transcriptase (RT) (1). Eighteen nucleotides at the 3′ end of the cellular tRNALys,3 hybridize to a specific site on the RNA genome termed the primer binding site (PBS; see ref. 2 and references therein). RT uses tRNALys,3 to prime synthesis of a single-stranded DNA product termed minus-strand strong-stop DNA. Alterations in both primer and HIV-1 proviral DNA by deletion and site-directed mutagenesis, as well as in vitro chemical and nuclease footprinting studies on the vRNA:tRNA complex, revealed additional critical intermolecular interactions in addition to that involving the PBS (3–12). Most prominently, an A-rich loop in the U5 inverted repeat of HIV-1 RNA upstream of the PBS interacts with the U-rich anticodon loop of tRNALys,3 (4, 6, 7, 9–11, 13). In addition, Berkhout and coworkers have hypothesized that base pairing between an 8-nt motif in the U5 region of vRNA and the TΨC loop of tRNALys,3 favors initiation of reverse transcription (14). Additional interactions between the tRNA primer and the viral genomic RNA have also been described for HIV-2 (15), feline immunodeficiency virus (16), avian retroviruses (17), and the yeast retrotransposons Ty1 and Ty3 (18–20).
Relatively limited information is available regarding HIV-1 RT contacts to its cognate vRNA:tRNA. Using site-specific cross-linking, Mishima and Steitz revealed two contacts between tRNALys,3 and RT (21). However, these contacts were localized to very large protein segments of 124 and 203 aa. In later site-directed mutagenesis studies, R307 of p66 was implicated in RT binding to vRNA:tRNA, whereas mutations of K249 or K311 of p66 did not affect initiation of DNA synthesis but significantly impaired completion of minus-strand strong-stop species (22). Isel et al. used RNA-specific nucleases to probe the vRNA-tRNA structure complexed with RT (9). Although this approach was useful in revealing structural reorganizations in several unliganded parts of vRNA and tRNA on RT binding, accurate identification of direct protein-nucleic acid contacts was hampered by high background protection in the vRNA and tRNA footprints derived from the steric conflict between bulky nucleases and RT (9, 23). Such limited information on protein contacts to the template-primer prompted us to design a high-resolution protein footprinting methodology allowing comprehensive and fine mapping of direct RT contacts to vRNA:tRNA in the initiation complex. Using a primary amine selective reagent and a mass spectrometric approach, we identified nine lysine contacts to vRNA:tRNA. The majority of these contacts are located in the template-primer binding cleft, whereas the p66 fingers subdomain makes additional specific contacts to vRNA:tRNA.
Materials and Methods
Enzymes and Nucleic Acid Duplexes.
p66/p51 HIV-1 RT was prepared as described previously (24). A model DNA duplex containing a 5-nt template overhang at the 3′ primer end was obtained by annealing the following two oligos: 5′-ATGCATCGGCGCTCGAACAGGGACTGTG-3′ and 5′-CACAGTCCCTGTTCGAGCGCCGA-3′. pShortPBS with a 99-nt region surrounding the HIV-1 (HXB2) PBS was a gift from K. Musier-Forsyth (Department of Chemistry, University of Minnesota, Minneapolis; ref. 25). HIV-1 (HXB2) PBS RNA was prepared by in vitro transcription of RNA nucleotides 125–223 with the trinucleotide sequences GGG and CCC added at the 5′ and 3′ ends, respectively. Because both a fully modified tRNALys,3 primer and its synthetic counterpart are specifically recognized and used by RT to initiate minus-strand DNA synthesis in vitro (11), we used the T7 transcribed synthetic tRNA in this study.
RNA-Dependent DNA Synthesis Reactions.
Equimolar concentrations of the vRNA template and tRNA primer were annealed in 10 mM Hepes (pH 7.5) and 25 mM NaCl, and the homogeneous preparation of the complex was confirmed by nondenaturing gel electrophoresis. DNA synthesis reaction mixtures contained the following: 200 nM RT, 1 μM template, primer, 200 μM dNTPs, 42.9 mM Tris⋅HCl, pH 7.8, 6 mM MgCl2, 60 mM KCl, 1 mM DTT. The reactions were incubated for 30 min at 37°C and analyzed by high voltage denaturing polyacrylamide gel electrophoresis. Quantitation was performed by using imagequant software (Bio-Rad).
Biotinylation and in Gel Proteolysis of RT.
N-hydroxysuccinimidobiotin (NHS-biotin) reacts specifically with primary amines on proteins, resulting in covalent addition of a biotin molecule (226.30 daltons) to Lys and the concomitant release of N-hydroxysuccinimide. In our experiments, protein–nucleic acid complexes were preformed by mixing 10 μM DNA:DNA or vRNA:tRNA with 2 μM RT in a 10-μl reaction mix of 50 mM Hepes (pH 7.5), 50 mM NaCl and then modified by adding 1 μl NHS-biotin (Pierce). After incubation at 25°C for 30 min, the reactions were quenched with 10 mM lysine and the mixture was subjected to SDS/PAGE. p66 and p51 were visualized by Coomassie blue stain, excised, and processed separately. The gel slices were extensively destained in 50% methanol/10% acetic acid. SDS was removed by two 10-min washes with 50 mM NH4HCO3. The gel pieces were then dehydrated with 100% acetonitrile, vacuum desiccated, and exposed to 1 μg of trypsin or Lys-C in 50 mM NH4HCO3 buffer at 37°C for 16 h. The supernatant was recovered and subjected to MS and MS/MS analysis.
Mass Spectrometric Analysis.
MS analysis reveals the precise molecular weight of a peptide ion, whereas MS/MS analysis yields amino acid sequence information of the parent peptide ion based on internal fragmentation of peptide bonds. MS spectra were recorded by using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) or quadrupole–time of flight (Q-TOF) techniques. MALDI-TOF experiments were performed with the Kratos Axima-CFR instrument (Kratos Analytical Instruments) using α-cyano-4-hydroxy-cinnamic acid as a matrix. MS and MS/MS analyses were carried out by using a Micromass (Manchester, U.K.) Q-TOF-II instrument equipped with an electrospray source and a Micromass cap-LC. Peptides were separated with the Thermo Hypersil Keystone 72105-030315 precolumn (Thermo Environmental, Franklin, MA) and the Micro-Tech Scientific (Vista, CA) ZC-10-C18SBWX-150 column using two sequential linear gradients of 5–40% acetonitrile (ACN) for 35 min and 40–90% ACN for 10 min. MS/MS analysis data and the mascot search engine (www.matrixscience.com) were used to identify RT peptide peaks from the NCBInr primary sequence database. The matched peptides were located in the MS spectra. For accurate quantitative analysis of the biotinylated peptides peaks, the intensities of all MS peptide peaks during the entire run, as well as at least two adjacent unmodified RT peptide peaks, were considered as controls.
Results
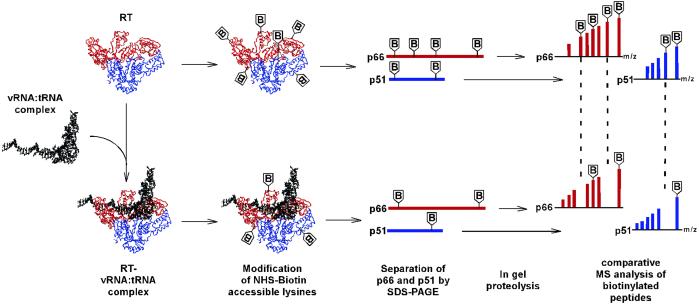
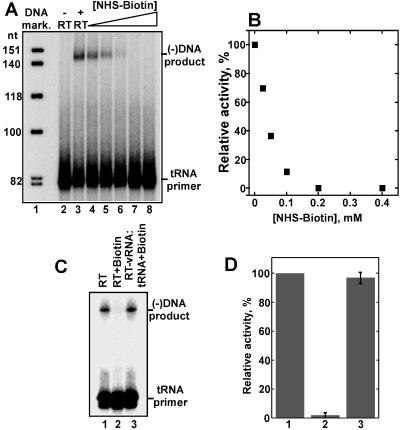
Our protein footprinting strategy is depicted in Fig. 1. We chose to dissect solvent accessible lysines in RT and the RT-vRNA:tRNA complex by using the primary amine-specific reagent NHS-biotin for several reasons. Most importantly, lysine is the most abundant residue in RT, with a total of 112 lysines present in both p66 and p51. In addition, crystallographic studies of RT-DNA:DNA and RT-RNA:DNA showed that lysine-nucleic acid contacts are abundant in both complexes. Finally, the powerful search engine mascot offers reliable identification of biotinylated lysines by using MS/MS data. To guarantee successful structural analysis, the modification reactions should be carried out under conditions in which the integrity of the nucleoprotein complex is fully preserved. Therefore, we examined the effect of increasing concentrations of NHS-biotin on the DNA polymerase activity of RT. The enzyme was first modified with the reagent, and then preannealed vRNA:tRNA was provided as a template:primer for the DNA synthesis reaction. The data in Fig. 2 A and B show that treatment with 200 μM NHS-biotin resulted in full inactivation of the enzyme. To ensure that the activity loss was due to blocking the active site lysines rather than disruption of RT structure, we repeated the modification reaction. But first we allowed the RT-vRNA:tRNA complex to form and then exposed it to 200 μM NHS-biotin. The result was remarkable protection by the vRNA:tRNA complex, yielding >97% recovery of the original enzymatic activity (compare lane 3 with lane 1, Fig. 2 C and D). Thus, these experiments helped us to define optimal reaction conditions for subsequent structural studies.
Fig 1.
Protein footprinting strategy. Heterodimeric RT comprises two subunits: p66 (red) and p51 (blue). The vRNA:tRNA complex is depicted by the black helical structure. Biotinylation reactions of free RT and the preformed RT-vRNA:tRNA complex were carried out in parallel. Surface exposed lysines were modified by NHS-biotin in free RT, whereas those coordinating vRNA:tRNA become protected from modification in the nucleoprotein complex. p66 and p51 were first separated by SDS/PAGE and then subjected to in gel proteolysis. Comparative MS analysis of the peptide fragments enabled us to dissect lysines shielded by the nucleic acid contacts from those remaining susceptible to modification in the complex.
Fig 2.
Effect of biotinylation on the RNA-dependent DNA polymerase activity of RT. (A) Denaturing gel electrophoresis analysis of minus-strand strong-stop DNA synthesis by using a 5′ end-labeled tRNA primer annealed to the vRNA template. Lane 1, DNA markers; lane 2, negative control with no RT; lane 3, DNA synthesis performed with unmodified RT. In lanes 4–8, the enzyme was first subjected to biotinylation, and then primer-template and dNTPs were provided for the DNA synthesis reaction. The following concentrations of NHS-biotin were used: lane 4, 25 μM; lane 5, 50 μM; lane 6, 100 μM; lane 7, 200 μM; lane 8, 400 μM. (B) Quantitation of the results depicted in A. (C) Preservation of RT activity by vRNA-tRNA. Lane 1, DNA synthesis performed with unmodified RT. Lane 2, RT was first treated with 200 μM NHS-biotin, and then vRNA:tRNA and dNTPs were provided. Lane 3, The RT-vRNA:tRNA complex was preformed and then treated with 200 μM NHS-biotin. dNTPs were subsequently added to allow DNA synthesis. (D) Quantitation of the data illustrated in C. The bars represent the mean of three independent experiments.
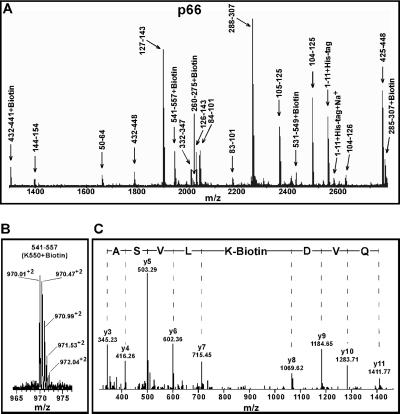
RT is a heterodimer comprising two polypeptides termed p66 and p51. The latter lacks the C-terminal RNase H domain, whereas the 440-aa N-terminal stretch is identical in both subunits. To differentiate between the two subunits in MS analysis, we first subjected the modified protein to SDS/PAGE. This step not only enabled us to separate p66 and p51, but was also essential for effective proteolysis. Polypeptides present in a gel slice have an unfolded “linear” structure, with all proteolytic sites being equally exposed for Trypsin or Lys-C cleavage. As a result, we achieved complete and reproducible hydrolysis. MALDI-TOF analyses of the proteolytic peptide mixtures yielded singly charged peptide peaks (Fig. 3A), whereas Q-TOF analysis of the same samples generated doubly and triply charged peptide peaks (Fig. 3B). We unequivocally assigned 59 peptide peaks to p66 and 47 peaks to p51. Naturally, p51 lacked peptides representing the RNase H domain. The majority of the peptide peaks were detected with both MALDI-TOF and Q-TOF. However, some peptide peaks were detected by only one of the two techniques, indicating that these two analyses were complementary.
Fig 3.
Mass spectrometric analysis of biotinylated RT peptide fragments. (A) MALDI-TOF spectrum of tryptic digested fragments of p66. Monoisotopic resolution was obtained throughout, allowing us to unequivocally assign singly charged unmodified and biotinylated peptides to the RT sequence. Conventional amino acid sequence numbering for p66 is used to indicate the peptide fragments. (B) A typical Q-TOF spectrum of a doubly charged biotinylated RT peptide fragment. The molecular mass of this ion corresponds to RT peptide 541–557 plus biotin. (C) MS/MS analysis of the parent biotinylated peptide ion shown in B confirms that K550 is biotinylated. The “y” ion peaks derived from internal fragmentation of peptide bonds provide amino acid sequence information read from the peptide C terminus (left) toward its N terminus (right). The mass increment between y7 and y8 ions corresponds to a biotinylated lysine, whereas the masses of preceding and following y ions assign perfectly to the RT sequence.
Q-TOF analysis of the proteolytic peptides also generated MS/MS data. This information was used in a mascot database search that revealed 15 biotinylated lysines in p66 and 12 biotinylated residues in p51. A typical MS/MS spectrum of a biotinylated RT peptide is shown in Fig. 3C. Importantly, treatment of RT with 50, 100, or 200 μM NHS-biotin each resulted in a modification pattern that differed only in that the biotinylated peptide peak intensities increased with increasing concentrations of reagent (data not shown). These results correlate well with the activity data (Fig. 2), indicating that, under our experimental conditions, the structural integrity of RT and/or the RT-vRNA:tRNA complex was preserved. Interestingly, this technique allowed us to detect structural differences between p66 and p51 in free RT (Table 1). For example, K30 of p66 and K390 of p51 were readily biotinylated, whereas K30 of p51 and K390 of p66 were not amenable to modification. These data correlate well with that of the RT crystal structure (26–28), which show that K30 of p66 is fully surface exposed whereas K30 of p51 is sterically hindered (PDB ID codes and ). Both K390 of p51 and that of p66 are solvent accessible, but their surrounding environments differ. K390 of p51, which is susceptible to NHS-biotin modification, is located in the primer-template binding cleft, whereas K390 of p66 is positioned on the opposite surface of the protein (PDB ID codes and ). Importantly, the subsequent experiments with RT-vRNA:tRNA reveal that K30 of p66 and K390 of p51 are involved in coordination of the nucleic acid.
Table 1.
Biotinylation pattern of RT, RT-DNA:DNA, and RT-vRNA:tRNA
|
|
p66 | p51 | ||||
|---|---|---|---|---|---|---|
| RT + biotin | RT-DNA:DNA + biotin | RT-vRNA:tRNA + biotin | RT + biotin | RT-DNA:DNA + biotin | RT-vRNA:tRNA + biotin | |
| K13 | + | + | + | + | + | + |
| K22 | + | + | + | + | − | − |
| K30 | + | + | − | NA | NA | NA |
| K64 | + | + | − | + | + | + |
| K101 | + | + | + | + | + | + |
| K218 | + | + | + | + | + | + |
| K263 | + | − | − | + | + | + |
| K287 | + | − | − | + | + | + |
| K353 | + | − | − | + | + | + |
| K366 | + | − | − | + | + | + |
| K374 | + | − | − | + | + | + |
| K390 | NA | NA | NA | + | − | − |
| K450 | + | + | + | NP | NP | NP |
| K540 | + | + | + | NP | NP | NP |
| K550 | + | + | + | NP | NP | NP |
+, Lysines readily modified by NHS-biotin; −, lysines significantly protected from biotinylation in the presence of nucleic acid; ND, lysines not amenable to biotinylation in free RT or either nucleoprotein complex; NP, lysines representing the RNase H domain are not present in p51 sequence. Key lysines are in bold.
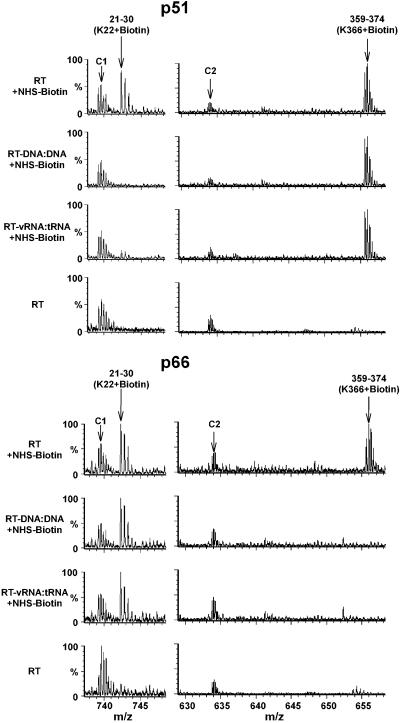
Our next goal was to identify lysines protected from modification in the nucleoprotein complex. In parallel with examining the RT-vRNA:tRNA complex, we also performed control experiments using duplex DNA as an alternative ligand to directly compare our footprinting results with those of the crystal structure studies (26–28). Comparative analysis of the MS data enabled us to clearly dissect the lysines protected by nucleic acid contacts from those remaining freely accessible to NHS-biotin in the complex. In addition, differences between the ability of either p66 or p51 to coordinate the cognate nucleic acid were immediately revealed. More specifically, treatment of free RT with NHS-biotin resulted in modification of K22 in both subunits. In RT complexed with nucleic acids, only K22 of p51 was significantly protected from biotinylation whereas K22 of p66 remained freely accessible to the reagent (Fig. 4). A converse picture was observed with K366. This p66 residue was shielded from NHS-biotin by either nucleic acid ligand, whereas no detectable protection was observed for K366 of p51 (Fig. 4). Quantitative analysis of all biotinylated peptide peaks indicated that five lysines (K263, K287, K353, K366, and K374) of p66 and two lysines (K22 and K390) of p51 were significantly protected in the RT-DNA:DNA complex (Table 1). Importantly, these residues are located in the primer-template binding cleft identified previously by crystallography (26–28). All other lysines that remained freely modified by NHS-biotin in the RT:DNA-DNA complex are located at least 10 Å from the nucleic acid in the crystal structure (PDB ID codes and ). Taken together, our data are in excellent agreement with crystallographic studies.
Fig 4.
Identification of lysines protected from biotinylation by nucleic acid contacts. The Q-TOF data illustrate differences between ability of p51 (Upper) and p66 (Lower) to coordinate DNA:DNA and/or vRNA:tRNA. Treatment of free RT with NHS-biotin results in modification of K22 in both p51 and p66, yielding peaks corresponding to the 21–30 peptide containing biotinylated K22. In RT-DNA:DNA or RT-vRNA:tRNA complexes, this peak is significantly diminished in p51 but remains unchanged in p66. A converse picture is observed with K366. The 359–374(K366 + biotin) peak in p51 persists in RT-DNA:DNA or RT-vRNA:tRNA, whereas the same peak in p66 is significantly reduced in both complexes. These data indicate that K22 of p51 and K366 of p66 are coordinating cognate nucleic acids, whereas K22 of p66 and K366 of p51 remain surface exposed in the nucleoprotein complexes. Unmodified RT peptide peaks C1 and C2 serve as controls. Each multiply charged peptide ion resulted in a clearly resolved peak cluster, indicating monoisotopic resolution in our Q-TOF analysis.
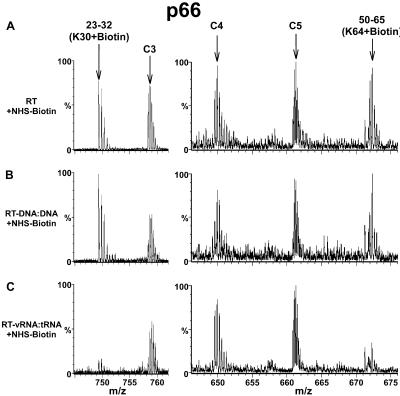
We next examined the biotinylation pattern for RT-vRNA:tRNA. Whereas the results in Fig. 4 show similarity between RT-DNA:DNA and RT-vRNA:tRNA, the data in Fig. 5 highlight differences between these two complexes. Seven lysines in the primer-template binding cleft were equally protected from biotinylation in both RT-DNA:DNA and RT-vRNA:tRNA (Table 1). In contrast, K30 and K64 of p66 were significantly protected in RT-vRNA:tRNA (Fig. 5C) and not in RT-DNA:DNA (Fig. 5B). The extent of biotinylation of K30 and K64 of p66 in RT complexed with a blunt ended or a 5 nt overhang containing DNA:DNA hybrid equaled that of free RT. In contrast, vRNA:tRNA specifically shielded the access of NHS-biotin to these sites (Fig. 5C).
Fig 5.
A comparison of RT-DNA:DNA and RT-vRNA:tRNA complexes. The peptide peaks containing biotinylated K30 and K64 (A) persist in RT-DNA:DNA (B) but diminish significantly in RT-vRNA:tRNA (C). RT peptide peaks C3, C4, and C5 serve as controls.
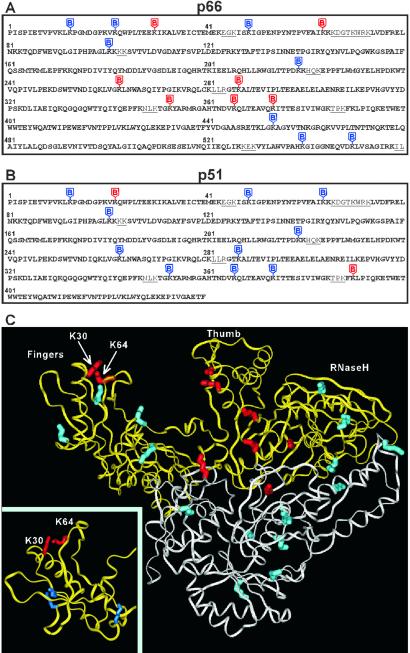
A summary of our footprinting results in the context of RT structure is depicted in Fig. 6. Assigning MS peptide peaks to p66 and p51 has yielded >95% coverage of the entire RT sequence (Fig. 6 A and B), which is important for high-resolution protein footprinting. Modified lysines (highlighted in red or blue) are evenly distributed throughout the protein, providing comprehensive structural analysis. Only lysines in red are significantly protected in RT-vRNA:tRNA. Such protection seems to be sufficient for full recovery of the DNA polymerase activity of RT (Fig. 2). The primer-template binding channel consisting of five lysines of p66 and two lysines of p51 can clearly be seen in the main panel of Fig. 6C. In addition, K30 and K64 of p66 are protected in RT-vRNA:tRNA and not in RT-DNA:DNA. Interestingly these two lysines are very closely (≈4 Å) positioned in the 3D structure of RT. In contrast, K13, K22, and K49 of the p66 fingers subdomain remain susceptible to NHS-biotin modification in RT-vRNA:tRNA, and are at least 20 Å from K30 or K64 of p66 (PDB ID codes and ). This spatial separation is better illustrated in Fig. 6C Inset.
Fig 6.
Summary of the footprinting results in the context of RT structure. Mapping biotinylation sites in p66 (A) and p51 (B). RT sequences to which we assigned MS peptide peaks are in bold (A and B). The segments that could not be detected by MS are in plain text and underlined. Lysines modified by NHS-biotin in free RT are highlighted throughout in blue or red. However, only those highlighted in red are significantly protected from modification in RT-vRNA:tRNA. (C) HIV-1 RT structure. p66 is in yellow and p51 is in white. The Inset in C provides a magnified view of the p66 fingers subdomain. This figure was generated with insight ii (Accelrys, San Diego) and PDB ID code .
Discussion
Structure of the RT-vRNA:tRNA Complex.
We present here a previously undocumented approach to high-resolution protein footprinting analysis of the RT-template-primer complex. Our data reveal nine lysine contacts to the vRNA:tRNA complex. Where are these protein contacts located with respect to the cognate nucleic acid? Because minus-strand DNA synthesis commences from the 3′ end of the tRNA primer (1), RT should bind the 18-bp PBS vRNA:tRNA duplex structure with the polymerase active site positioned at this 3′ end. Indeed, our results indicate that RT contacts to this nucleic acid segment resemble those with DNA:DNA or RNA:DNA duplexes. We show that five lysines of p66 and two of p51 are equally protected from modification in the presence of DNA:DNA or vRNA:tRNA. However, we have also found that K30 and K64 of the p66 fingers subdomain, which are positioned outside the template-primer cleft, make additional contacts to vRNA:tRNA. We suggest that these contacts may be located on the duplex structure just ahead of the primer [nucleotides 132–139 of HIV-1 RNA paired with nucleotides 168–175 (numbering according to the Mal isolate) and denoted helix 2 in ref. 23]. It is this structure that RT must disrupt for effective elongation to occur. It has been recently shown that deletion or extension of the 3-nt single stranded RNA junction that separates helix 2 from the PBS duplex markedly decreased minus-strand strong-stop DNA synthesis (23). Thus, the relative orientation of helix 2 and the PBS duplex, together with the specific RT contacts reported herein, may be important for correct initiation of minus-strand DNA synthesis.
It is now intriguing to apply our methodology for structural studies of other transient species formed during reverse transcription. RT pauses after addition of 3 to 5 nts to the tRNA primer, whereas the polymerization rate increases about 3,000-fold after the addition of the 6th nt (13). Arts et al. have shown that mutations of K249 and K311 in p66 do not significantly alter synthesis of the +3 and +5 initiation products but result in remarkable reduction of full length minus-strand strong-stop DNA synthesis (22). Thus, RT contacts to vRNA:tRNA during the initiation and elongation phases may substantially differ. The high-resolution protein-footprinting approach we describe here should prove to be useful in revealing these differences. In addition, group-specific reagents are also available for a large number of amino acids (reviewed by ref. 29). These reagents should help to dissect more detailed contacts between RT and the structurally distinct nucleic acid species encountered during reverse transcription.
Protein Footprinting Methodology.
The protein footprinting strategy we have devised has a generic application for high-resolution solution structural studies of multiprotein-nucleic acid complexes. Previous attempts to map protein-nucleic acid contacts by using primary amine-specific reagents have yielded only partial information due to the intrinsic limits of the analysis methods used (30, 31). For example, only a single lysine (K263) contact was identified in the RT-DNA:DNA complex by analyzing pyridoxal 5′-phosphate modifications of the protein with HPLC and Edman degradation sequencing (30). In another work with vaccinia virus topoisomerase-DNA:DNA complex, the peptides modified by citraconic anhydride and N-hydroxysuccinimide acetate were analyzed by SDS/PAGE, which restricted structural mapping to large peptides rather than single amino acids (31). On the other hand, probing protein surface topology studies with NHS-biotin and mass spectrometry have been reported (32–34), but this method, to our knowledge, has not been extended for fine mapping protein-nucleic acid contacts.
Here, we report a combination of the protein-footprinting steps comprising nucleoprotein modification by NHS-biotin, SDS/PAGE fractionation of different protein subunits, in gel proteolysis, and comparative MS analysis of peptide fragments. Our data demonstrate that this methodology represents a powerful tool for high-resolution solution structural studies of protein-nucleic acid complexes. We have obtained greater than 95% coverage of RT, successfully dissected multiple p66 and p51 contacts to nucleic acids, and revealed differences between RT-vRNA:tRNA and RT-DNA:DNA. One can envisage application of this strategy for other multiprotein-nucleic acid complexes, which are ubiquitous throughout transcription, translation, recombination, and DNA repair processes. Thus, after modification of the nucleoprotein species, different subunits of the complex can be separated by SDS/PAGE, and the role of each polypeptide can be assessed separately by mass spectrometry.
Our data also open a doorway for accurate mapping of drug-protein complexes. Drugs that bind irreversibly can be evaluated directly, whereas reversible drugs must be converted by attaching a covalent modifier. For this purpose, photoactivable reagents can be used, or, alternatively, a reaction mechanism mimicking the protein modification with NHS-biotin can be exploited.
Finally, the present work provides a platform for structural analysis of drug-resistant RT variants. Here, we used 0.5 μg RT per footprinting experiment. The ability to perform high-resolution structural analysis with such small protein quantities makes the mass spectrometric approach very attractive.
Abbreviations
RT, reverse transcriptase
vRNA, viral RNA
PBS, primer binding site
MALDI-TOF, matrix-assisted laser desorption ionization–time of flight
Q-TOF, quadrupole–time of flight
NHS-biotin, N-hydroxysuccinimidobiotin
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Telesnitsky A. & Goff, S. P. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 121–160.
- 2.Kleiman L. (2002) IUBMB Life 53, 107-114. [DOI] [PubMed] [Google Scholar]
- 3.Yu Q. & Morrow, C. D. (2000) Nucleic Acids Res. 28, 4783-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z., Kang, S. M., Li, Y. & Morrow, C. D. (1998) RNA 4, 394-406. [PMC free article] [PubMed] [Google Scholar]
- 5.Lund A. H., Duch, M., Lovmand, J., Jorgensen, P. & Pedersen, F. S. (1997) J. Virol. 71, 1191-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Zhang, Z., Wakefield, J. K., Kang, S. M. & Morrow, C. D. (1997) J. Virol. 71, 6315-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanchy J. M., Isel, C., Keith, G., Le Grice, S. F. J., Ehresmann, C., Ehresmann, B. & Marquet, R. (2000) J. Biol. Chem. 275, 12306-12312. [DOI] [PubMed] [Google Scholar]
- 8.Isel C., Lanchy, J. M., Le Grice, S. F. J., Ehresmann, C., Ehresmann, B. & Marquet, R. (1996) EMBO J. 15, 917-924. [PMC free article] [PubMed] [Google Scholar]
- 9.Isel C., Westhof, E., Massire, C., Le Grice, S. F. J., Ehresmann, B., Ehresmann, C. & Marquet, R. (1999) EMBO J. 18, 1038-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang S. M. & Morrow, C. D. (1999) J. Virol. 73, 1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arts E. J., Stetor, S. R., Li, X., Rausch, J. W., Howard, K. J., Ehresmann, B., North, T. W., Wohrl, B. M., Goody, R. S., Wainberg, M. A. & Grice, S. F. J. (1996) Proc. Natl. Acad. Sci. USA 93, 10063-10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Shalom, A., Li, Z., Wang, J., Mak, J., Wainberg, M. A. & Kleiman, L. (1996) J. Virol. 70, 4700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanchy J. M., Keith, G., Le Grice, S. F. J., Ehresmann, B., Ehresmann, C. & Marquet, R. (1998) J. Biol. Chem. 273, 24425-24432. [DOI] [PubMed] [Google Scholar]
- 14.Beerens N. & Berkhout, B. (2002) J. Virol. 76, 2329-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freund F., Boulme, F., Litvak, S. & Tarrago-Litvak, L. (2001) Nucleic Acids Res. 29, 2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J. T., Ehresmann, B., Hubscher, U. & Le Grice, S. F. J. (2001) J. Biol. Chem. 276, 27721-27730. [DOI] [PubMed] [Google Scholar]
- 17.Morris S., Johnson, M., Stavnezer, E. & Leis, J. (2002) J. Virol. 76, 7571-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friant S., Heyman, T., Bystrom, A. S., Wilhelm, M. & Wilhelm, F. X. (1998) Mol. Cell. Biol. 18, 799-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabus C., Ficheux, D., Rau, M., Keith, G., Sandmeyer, S. & Darlix, J. L. (1998) EMBO J. 17, 4873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeney J. B., Chapman, K. B., Lauermann, V., Voytas, D. F., Astrom, S. U., von Pawel-Rammingen, U., Bystrom, A. & Boeke, J. D. (1995) Mol. Cell. Biol. 15, 217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishima Y. & Steitz, J. A. (1995) EMBO J. 14, 2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arts E. J., Miller, J. T., Ehresmann, B. & Le Grice, S. F. J. (1998) J. Biol. Chem. 273, 14523-14532. [DOI] [PubMed] [Google Scholar]
- 23.Goldschmidt V., Rigourd, M., Ehresmann, C., Le Grice, S. F. J., Ehresmann, B. & Marquet, R. (2002) J. Biol. Chem. 277, 43233-43242. [DOI] [PubMed] [Google Scholar]
- 24.Le Grice S. F. J. & Gruninger-Leitch, F. (1990) Eur. J. Biochem. 187, 307-314. [DOI] [PubMed] [Google Scholar]
- 25.Hargittai M. R., Mangla, A. T., Gorelick, R. J. & Musier-Forsyth, K. (2001) J. Mol. Biol. 312, 985-997. [DOI] [PubMed] [Google Scholar]
- 26.Sarafianos S. G., Das, K., Tantillo, C., Clark, A. D., Jr., Ding, J., Whitcomb, J. M., Boyer, P. L., Hughes, S. H. & Arnold, E. (2001) EMBO J. 20, 1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobo-Molina A., Clark, A. D., Jr., Williams, R. L., Nanni, R. G., Clark, P., Ferris, A. L., Hughes, S. H. & Arnold, E. (1991) Proc. Natl. Acad. Sci. USA 88, 10895-10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H., Chopra, R., Verdine, G. L. & Harrison, S. C. (1998) Science 282, 1669-1675. [DOI] [PubMed] [Google Scholar]
- 29.Lundblad R. L., (1995) Techniques in Protein Modification (CRC, New York).
- 30.Basu A., Tirumalai, R. S. & Modak, M. J. (1989) J. Biol. Chem. 264, 8746-8752. [PubMed] [Google Scholar]
- 31.Hanai R. & Wang, J. C. (1994) Proc. Natl. Acad. Sci. USA 91, 11904-11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zappacosta F., Ingallinella, P., Scaloni, A., Pessi, A., Bianchi, E., Sollazzo, M., Tramontano, A., Marino, G. & Pucci, P. (1997) Protein Sci. 6, 1901-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett K. L., Matthiesen, T. & Roepstorff, P. (2000) in Mass Spectrometry of Proteins and Peptides, ed. Chapman, J. R. (Humana, Totowa, NJ), pp. 113–132.
- 34.Suckau D., Mak, M. & Przybylski, M. (1992) Proc. Natl. Acad. Sci. USA 89, 5630-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]