Abstract
Oligosaccharyl transferase (OT) is a complex multisubunit enzyme that, in the case of Saccharomyces cerevisiae, contains nine different transmembrane proteins. One of our goals is to identify the OT subunit(s) responsible for recognizing the consensus sequence, -Asn-X-Thr/Ser-, and catalyzing the oligosaccharide transfer reaction. By using a substrate-based photoprobe, earlier we found that Ost1p was specifically linked to the radiolabeled photoprobe. We have now examined Ost1p in more detail. Deletion of the cytoplasmic tail of Ost1p caused no defects in growth and glycosylation. In addition, replacement of the transmembrane domain with other hydrophobic amino acids did not impair growth. In contrast, a construct containing only the luminal domain of Ost1p did not support cell growth. Given these observations, we concentrated on studying the luminal domain of Ost1p and localized the photoprobe attachment region within a sequence of nine amino acid residues. Because mutations in the photoprobe attachment region did not cause any severe growth or glycosylation defects, we conclude that this region is not involved in the recognition of the N-glycosylation site. By further mutagenesis of the conserved residues of Ost1p we conclude that the luminal domain mediates interactions with other subunits of OT and becomes labeled because of its proximity to the recognition and/or catalytic subunit in the OT complex, Stt3p.
The enzyme catalyzing N-linked glycosylation, called oligosaccharyl transferase (OT), is a remarkably complex multisubunit enzyme that, in the case of Saccharomyces cerevisiae, is composed of nine different transmembrane proteins (1–16). Even though genes encoding the nine subunits of yeast OT have been cloned, at present there is very limited information about the function of each of the subunits. We have undertaken the study of the mode of interaction of these transmembrane protein subunits within the endoplasmic reticulum to elucidate their function.
To identify the subunit(s) of yeast OT that recognizes -Asn-X-Thr/Ser- sites that can be glycosylated (17–19), we developed photoaffinity probes containing a photoreactive benzophenone derivative, p-benzoylphenylalanine (Bpa), as part of an 125I-labeled acceptor tripeptide, 125I-Bolton–Hunter (bh)-Asn-Bpa-Thr-Amide. By using this 125I-labeled Bpa containing tripeptide, which was shown to be a substrate of OT, we found that after photoactivation of yeast microsomes, Ost1p was specifically labeled (20).
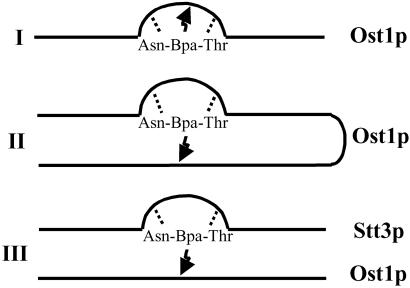
Our current objective was to localize the site of binding of the photoprobe and to determine whether Ost1p is the actual subunit that recognizes the N-glycosylation consensus sequence -Asn-X-Thr/Ser-. Three models to explain probe attachment to Ost1p were considered (Fig. 1). In model I, interaction of the Asn and Thr residues of the photolabeled tripeptide with the putative N-glycosylation site recognition domain occurs, and the photoreactive middle amino acid, Bpa, covalently binds to some nearby amino acid residue in the Ost1p backbone. Alternatively, in model II (Fig. 1) the probe may become attached in another region of Ost1p that does not encompass the glycosylation site recognition domain. However, this region must be in the vicinity in terms of its tertiary structure, because the photoreactive group (a diradical) can only react with carbon atoms within the vicinity of 3.1 Å (21). A third possibility, shown in model III, is that the N-glycosylation site recognition domain actually resides in another nearby subunit of OT, but the probe becomes attached to Ost1p. It is likely that the nine different subunits of OT are in reasonably close proximity because they do exist in one complex. Therefore, Ost1p could be located very close to the peptide glycosylation recognition subunit, and thereby become labeled by the photoreactive Bpa group. Our results are inconsistent with models I and II but support model III. Furthermore, our recent studies strongly suggest that the subunit containing the recognition and/or catalytic site is Stt3p, which is the most conserved subunit in the OT complex (22).
Fig 1.
Possible mechanisms for Ost1p labeling by the Asn-Bpa-Thr photoprobe. In model I, the probe attachment site (arrow) is postulated to be in close proximity to the N-glycosylation site recognition region (bend) in the primary sequence of Ost1p. In model II, the probe attachment site is not in the vicinity of the glycosylation site recognition region, but is elsewhere in the primary sequence of Ost1p. In model III, it is postulated that the glycosylation site recognition region and/or the catalytic site actually is in another OT subunit, but Ost1p is located in proximity to this other subunit and consequently is labeled by the photoprobe.
Materials and Methods
Strains.
W303 diploid strain (ade2 can1 his3 leu2 trp1 ura3) was used to generate QYY501 [ade2 can1 his3 leu2 trp1 ura3 OST1/Δost1:his 5+ (Schizosaccharomyces pombe)]. QYY501 was transformed with pRS316-OST1 and sporulated (5). The haploid strain QYY500 [MATa ade2 can1 his3 leu2 trp1 ura3 Δost1:his5+ (S. pombe) pRS316-OST1] was selected on -His -Ura (media) plates. All of the mutant strains were generated by using either QYY500 or QYY501 as the parental strain.
Plasmids.
pRS316-OST1 was obtained from Reid Gilmore's laboratory (5). A 2.1-kb fragment was generated by PCR using QYY101 (20) genomic DNA as template and 5′-CCACTTGAATTCCATTCATGTTAAC-3′ and 5′-CCGAGCTCCTGCAGCCCGGGGGATCCAC-3′ as primers. The resulting PCR product was subcloned into pRS314 and pRS306, which were digested with EcoRI and SacI to generate the plasmid pRS314-OST1HA and pRS306-OST1HA, respectively (23).
Procedures.
Photoprobes were synthesized as described (20). Yeast microsomes were prepared as described by Baker et al. (24).
Oligosaccharyl Transferase Activity Assay.
The enzyme assay for peptide glycosylation was performed as described (25).
PCR Mutagenesis for Block and Point Mutants.
PCR mutagenesis was performed according to the manufacturer's protocol (Stratagene). For glycosylation site mutations and Met mutations, pRS306-OST1HA was used as the PCR template. Mutagenized plasmids were sequenced and the correct plasmids were linearized by NcoI digestion and transformed into QYY501. The transformants were selected for Ura prototrophy. The diploid transformants were sporulated, dissected and HIS+ URA+ haploids were selected. For all of the other mutations mentioned in this paper, pRS314-OST1HA was used as the PCR template. Mutagenized plasmids were sequenced, and those with the expected sequence were transformed into QYY500. The transformants were selected for Trp and Ura prototrophy and then further for 5-fluoroorotic acid selection (26).
Conditions for Photoactivation and Immunoprecipitation.
Irradiation of crude yeast microsomes was conducted as described (20). After irradiation, the mixture reaction was centrifuged at 15,000 × g for 10 min, and the pellet was resuspended in a buffer containing 150 mM NaCl, 10 mM Hepes KOH (pH 7.4), 5 mM MgCl2, 1% Triton X-100, 0.2% SDS. After centrifugation of the solution at 15,000 × g for 10 min, the supernatant was immunoprecipitated with an anti-hemagglutinin (HA) antibody. After incubation with protein G agarose beads, the beads were recovered by centrifugation and washed three times with the same buffer and once with 50 mM Tris⋅Cl (pH 7.4), 150 mM NaCl and 5 mM EDTA. Samples were analyzed by SDS/10% PAGE and autoradiographed.
Coimmunoprecipitation Using Mild Detergent.
This method was modified from Karaoglu et al. (27). Yeast microsomes were collected by centrifugation and the membrane pellet was resuspended in 5% glycerol/20 mM Tris⋅Cl, pH 7.4/5 mM MgCl2/1 mM EDTA/1 mM EGTA/1 mM PMSF. The suspension was adjusted to contain 1.5% digitonin, 0.5 M NaCl, 20 mM Tris-Cl (pH 7.4), 3.5 mM MgCl2, and 1 mM MnCl2. The mixture was centrifuged for 20 min at 100,000 × g, and the clarified supernatant was used for immunoprecipitation. Immunoprecipitation conditions were the same as described earlier (27). Samples were separated on SDS/PAGE and transferred to nitrocellulose membrane. The membranes were probed with different antibodies.
Cyanogen Bromide (CNBr) Cleavage.
After photolabeling and immunoprecipitation, the proteins were eluted from protein G agarose beads with 1% SDS. CNBr was dissolved in 100% formic acid to 10 mg/ml and then added to the SDS eluant so that the final concentration of formic acid was 70%. Each tube was flushed with N2, capped, and stored at room temperature in the dark overnight. After CNBr cleavage, samples were dried in a vacuum centrifuge. Water was added to the tubes several times and the samples were evaporated to dryness to remove all of the formic acid. Dried samples were dissolved in SDS/PAGE sample buffer, analyzed by SDS/PAGE, and autoradiographed.
Quantitation of Immunoprecipitated OT Subunits and Radiolabeled Ost1p.
Quantitation of immunoprecipitated OT subunits and radiolabeled Ost1p was performed by scanning the films and analyzing the bands by using nih IMAGE 1.62 program. For the mutants that photolabeled Ost1p (D306A and R326A), we used same volume microsomes as the control for immunoprecipitation and photoactivation. We normalized the immunoprecipitated protein amounts and radiolabeled Ost1p to the wild-type control. Because the amount of protein varied, in some cases more Ost1p was immunoprecipitated in mutants than in the control. For the mutants that did not photolabel Ost1p (324-326AAA, 327-329AAA, 330-333AAAA, and D306K), the quantity of immunoprecipitated Ost1p was normalized to the control. Furthermore, we used an amount of microsomes for immunoprecipitation that contained the same amount of Ost1p.
Results and Discussion
Mutations Outside the Luminal Domain of Ost1p.
Based on hydropathy analysis, Ost1p is a type I membrane protein with a long luminal domain, one transmembrane span, and a short cytosolic tail containing only nine amino acid residues. We prepared a construct containing only the luminal domain of Ost1p [Ost1p(L)] to determine whether it could function as an essential component of OT. The mutated plasmid was introduced into an ost1 chromosomally deleted strain that contained a plasmid copy of the wild-type OST1 gene to support growth. The plasmid shuffling procedure revealed that the luminal protein was unable to support cell growth (26).
It is obvious that the transmembrane domain and/or the cytosolic tail could play an important role in the function of this protein. To differentiate between these possibilities, we prepared a construct without the C terminus, and found that this construct could support cell growth and showed no defect in glycosylation. This result indicated that the C-terminal nine amino acid residues are dispensable for Ost1p function, but based on the negative results with the luminal domain, it seemed that both the transmembrane and luminal domains were required. To determine whether there was a requirement for the specific transmembrane sequence of Ost1p, we divided the 17-aa peptide region into three portions, and each portion was mutated one at a time to polyleucine residues. After the plasmid shuffling procedure, all three mutant proteins were found to support cell growth. This result suggests that the transmembrane domain is only involved in anchoring the luminal domain of Ost1p to the lipid bilayer and has no role in the interaction with the transmembrane domains of other OT subunits.
Identification of the Site of 125I-Photoprobe Attachment.
We concluded from the mutational analysis discussed above that the luminal domain linked to the membrane was essential. Next we wanted to localize where the photoprobe attached to the primary sequence of Ost1p. Because Ost1p has six methionine residues, we used CNBr cleavage to determine to which fragment the photoprobe was attached (Fig. 2a).
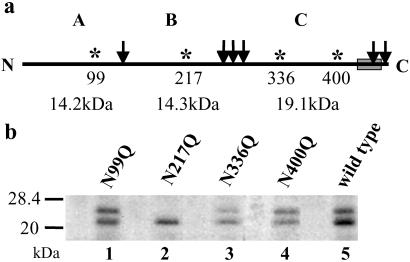
Fig 2.
Photoprobe binds to CNBr fragment B of Ost1p. (a) Diagram of Ost1p structure with the position of Met residues that are expected to be cleaved by CNBr indicated by the arrow. A, B, and C indicate the three large CNBr cleavage products with their calculated backbone molecular masses. In addition, potential N-glycosylation sites (asterisks) and their amino acid positions are shown. (b) Microsomes were prepared from each strain carrying an N to Q mutation and from the wild-type strain, and photolabeling was performed. Immunoprecipitation was carried out with the photolyzed microsomes and the eluants from the precipitants were subjected to CNBr cleavage, resolved on SDS/PAGE, and detected by autoradiography.
In initial experiments using microsomes from a strain carrying a HA epitope-tagged form of the OST1 gene integrated into the chromosome, we demonstrated that the tagged Ost1p (Ost1HAp) was labeled after photoactivation of the 125I-labeled probe. To localize the site of labeling, photolabeled membranes were solublized, and the HA-tagged protein was isolated by immunoprecipitation and subjected to CNBr cleavage. The results of SDS/PAGE of the cleavage products revealed two glycosylated, 125I-labeled CNBr fragments with apparent masses of 21 and 24 kDa (see Fig. 2b, lane 5). After protein glycanase F (PNGase F) treatment, these two bands shifted to one band at ≈18 kDa (data not shown). This result indicated that these two bands had the same peptide backbone, and differed only in the number of N-glycan chains.
As indicated in Fig. 2a, it was difficult to ascertain to which of these three large CNBr fragments (A, B, or C) the probe was attached because the masses of all three major fragments are not very different. It has been demonstrated that three of four sites of the four potential N-linked glycosylation sites of Ost1p can be glycosylated (5, 28), and therefore the protein often migrates as a doublet. Given this fact, we assumed that elimination of the glycosylation site(s) in the CNBr fragment that contained the covalently attached labeled probe would result in a mobility shift on SDS/PAGE. Consequently, to determine whether the 125I-labeled probe was located in CNBr fragment A, B, or C, mutants in which Asn was converted to Gln in each of the four glycosylation sites in Ost1HAp (Fig. 2a) were prepared. All four of these mutants exhibited 90% of the OT activity of the wild-type strain; clearly glycosylation is not essential for enzyme activity. Each of the four mutant constructs was integrated into the chromosome by homologous recombination, and microsomes were prepared from each strain. After photoactivation, immunoprecipitation, and then CNBr cleavage, the products were analyzed by SDS/PAGE to determine whether one of the CNBr fragments had decreased in size because a glycosylation site was removed. As shown in Fig. 2b, the results indicate that the photolabeled CNBr fragment was not A or C because the 125I-labeled fragment did not increase in mobility after mutations at the first, third or fourth glycosylation sites. However, when the glycosylation site located in fragment B was mutated, only a single 125I-labeled band was observed (Fig. 2b, lane 2), indicating that the probe was, in fact, bound to CNBr fragment B (amino acid residues 151–273). This result also indicated that the glycosylation site at N217 is partially glycosylated in the wild type. PNGase F treatment of this fragment resulted in a molecular mass shift to 18 kDa, indicating that the covalently attached probe itself had been glycosylated (data not shown). Our earlier results clearly demonstrated that the photolabeling of Ost1p was substrate specific and inhibited OT activity (20). Therefore, this tripeptide is probably positioned in the active site of the enzyme, and during photoactivation or after that, it was glycosylated as well. However, this glycosylation could only happen either during or after the photoactivation because glycosylated peptide is a product, not a substrate for OT.
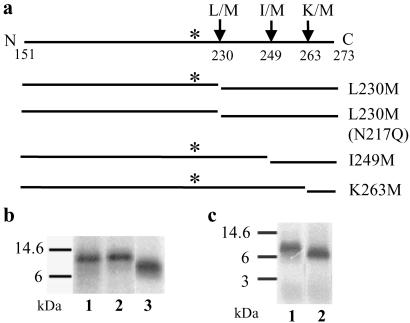
To further localize the site of probe binding, replacement of certain amino acids with Met in fragment B was performed (Fig. 3a). Only one residue was mutated to Met in each strain. The Met mutants used in this study affected neither growth nor OT activity of the strain (data not shown). After photoactivation with microsomes, the HA-tagged protein was immunoprecipitated, recovered, and subjected to CNBr cleavage. With the mutation of Leu-230 to Met, the CNBr fragment B would be expected to be cleaved to two fragments (Fig. 3a). One possible labeled fragment would encompass residues 151–230, and it should migrate as a doublet because the glycosylation site is at residue 217. The other possible labeled fragment is from residues 231–273. As shown in Fig. 3b, in lane 1, the 21/24-kDa doublet was absent, and instead a fragment migrating at ≈11 kDa appeared. This result indicated that the photoprobe was attached between amino acid residues 231 and 273.
Fig 3.
The photoprobe is attached between amino acid residues 264 and 273 in fragment B. Each of the indicated single Met mutations was generated in the region corresponding to fragment B, and microsomes were prepared from each of the mutant strains. After photolabeling was carried out, immunoprecipitation was performed with the photolyzed microsomes and the eluants were subjected to CNBr cleavage. The samples were analyzed by SDS/PAGE, and the peptide that contained the label was detected by autoradiography. (a) Diagram of Met mutation sites and the resultant CNBr cleavage products in fragment B. (b) Photoprobe was found to be located between residues 250 and 273. Lane 1, L230M; lane 2, L230M, N217Q; lane 3, I249M. (c) Photoprobe was found to be located between residues 264 and 273. Lane 1, I249M; lane 2, K263M.
To further confirm this result, we prepared a double mutant L230M, N217Q. If the photoprobe was attached between amino acid residues 151 and 230, photoactivation and subsequent CNBr cleavage with this mutant should yield one radiolabeled band instead of two because of the absence of the glycosylation. Conversely, if the photoprobe was attached between residues 231 and 273, the same radiolabeled band should be formed as shown above by using the L230M mutant. The data shown in Fig. 3b indicates that there is no difference in molecular mass between lane 1 and lane 2. This result clearly demonstrated that the photoprobe is attached between amino acid residues 231 and 273.
To further narrow down the site of attachment of the photoprobe, we mutated Ile-249 to Met (I249M). As shown in lane 3 of Fig. 3b, after CNBr cleavage only one radiolabeled band, migrating at ≈7 kDa, was observed. Based on this we concluded that the photoprobe was attached somewhere within amino acid residues 250 and 273.
A similar procedure was used to prepare another mutant, K263M (Fig. 3a). It is clear from the results in Fig. 3c that after CNBr cleavage, the radiolabeled band (lane 2) migrated faster than that of I249M (lane 1). Therefore, the probe was attached between residues 264 and 273. We also mutated Leu-264 to Met, and after photoactivation and CNBr cleavage, the resultant radiolabeled band migrated at ≈6 kDa. Therefore, the photoprobe was attached to the C-terminal side of Leu-264 (data not shown). These results clearly established that the photoprobe was attached in the luminal domain of Ost1p to one of the amino acid residues encompassing 265–273 (SKGFSRLEL).
Is the Probe Bound Within the Glycosylation Site Recognition Domain?
Next, we sought to distinguish between model I and model II (Fig. 1). If the probe attachment site is near the glycosylation site recognition domain in the primary sequence, mutations in this nine-residue region (residues 265–273) would be expected to result in severe growth defects or lethality and loss of OT activity. Block mutations of 3 aa replaced by three Ala residues within the 9-aa region were prepared and then tested to see whether these Ost1p mutants were functional. The results of the plasmid shuffling revealed that all of the mutants supported growth, though mutants 268-270AAA and 271-273AAA displayed slight growth defects at 37°C (data not shown).
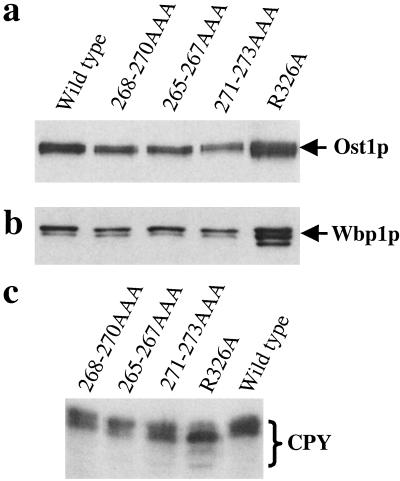
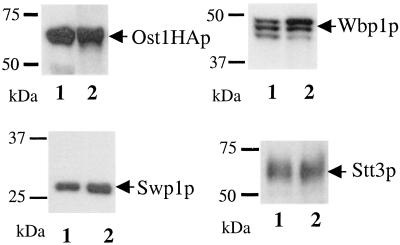
The glycosylation patterns of Ost1p, Wbp1p, and carboxypeptidase Y (CPY) were examined in these mutants because these proteins are known to be glycosylated. Ost1p and CPY contain four potential glycosylation sites, whereas Wbp1p contains two (1, 5, 28, 29). As shown in Fig. 4, Ost1p and Wbp1p were not underglycosylated in these three mutants when compared with the wild-type control. As for CPY, 271-273AAA showed slight underglycosylation, whereas the other two mutants were glycosylated normally. In comparison, the temperature sensitive mutant R326A (see below) displayed an underglycosylation pattern of all three proteins. These results clearly demonstrated that these nine amino acid residues are not required for protein glycosylation in vivo. Next, these mutants were assayed for their peptide binding affinity. Microsomes were prepared from each mutant strain, and the ability of the various mutants to be photolabeled was assessed. Mutant 268-270AAA bound less probe than mutants 265-267AAA, 271-273AAA or the wild-type control (data not shown). These three mutant proteins were expressed at wild-type levels in the cells. Two possibilities can be considered for reduced probe binding of the mutant protein. First, the mutant protein may not be integrated well into the OT complex. Therefore, we wanted to test these mutants for their ability to be incorporated into the OT complex. Previous observations demonstrated that, when using mild detergent to solubilize the microsomes followed by immunoprecipitation with an anti-HA antibody, all of the OT subunits can be precipitated even though the antibody tag is only attached to one of the subunits (27). We found these mutants were incorporated into the OT complex (part of the data are also shown in Fig. 4 a and b). The other reason for reduced probe binding could be because mutant 268-270AAA had undergone a modest protein conformational change that did not affect the peptide recognition site. But this change might increase the distance between the peptide recognition site and the probe attachment site. Because the Ala residues have a shorter side chain compared with the original three amino acids (FSR) in the Ost1p sequence, perhaps Bpa attached to the site with less efficiency. Based on the very modest effect on the growth phenotype and OT activity of these three mutants, we conclude that this nine-residue region probably is not the region in Ost1p that functions in recognizing -Asn-X-Thr/Ser- glycosylation sites. Therefore, these observations do not support model I.
Fig 4.
Mutations in the photoprobe attachment region do not cause glycosylation defects. (a and b) Microsomes were prepared from each mutant strain and the wild-type control. Nondenaturing immunoprecipitation was carried out. Samples eluted from protein G agarose beads were resolved by SDS/PAGE and followed by Western blot analysis using anti-Ost1p and anti-Wbp1p, respectively. (c) Spheroplasts were prepared from each mutant strain and the wild-type control. Samples were separated on SDS/PAGE and followed by Western blot analysis using anti-CPY antibody.
Studies on Conserved Residues of Ost1p.
If Ost1p is the peptide glycosylation recognition subunit, the residues involved in recognition should be conserved across different species. Therefore, after identification of the regions that are highly conserved across species, we prepared block mutations consisting of two to five residues replaced by Ala residues in an epitope-tagged OST1 construct. We found that 48 of the 52 block mutants displayed no growth phenotype and were not further studied. However, three of the block mutants were lethal, and one was temperature sensitive (Table 1).
Table 1.
Characteristics of mutants that display an impaired growth phenotype
| Mutant
|
Growth phenotype
|
Relative quantity of immunoprecipitated | Photolabeling of Ost1p
|
Relative labeling of Ost1p
|
OT activity
|
||
|---|---|---|---|---|---|---|---|
| Ost1p | Wbp1p | Swp1p | |||||
| 64-66AAA (30°C) | Normal | 0.761 | 0.773 | 0.498 | ND | ND | 1.051 |
| 64-66AAA (37°C) | t.s. | 0.617 | 0.508 | 0.161 | ND | ND | 0.305 |
| 324-326AAA | Lethal | 1.000 | 0 | 0 | 0 | 0 | ND |
| 327-329AAA | Lethal | 1.026 | 0 | 0 | 0 | 0 | ND |
| 330-333AAAA | Lethal | 0.923 | 0 | 0 | 0 | 0 | ND |
| Control | Normal | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| D306A (25°C) | t.s. | 1.431 | 0.251 | 0.317 | 1.264 | 0.883 | 0.649 |
| D306A (30°C) | t.s. | 1.196 | 0.198 | 0.238 | 1.043 | 0.872 | 0.505 |
| D306K | Lethal | 0.563 | 0 | 0 | 0 | 0 | ND |
| R326A (30°C) | Normal | 1.354 | 0.783 | 0.860 | 1.135 | 0.838 | 0.826 |
| R326A (37°C) | t.s. | 1.402 | 0.727 | 0.969 | 0.901 | 0.643 | 0.366 |
| Control | Normal | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
Growth phenotype was determined by 5-fluroorotic acid selection. t.s., temperature sensitive.
Relative quantity of immunoprecipitated Ost1p, Wbp1p, and Swp1p was determined by mild detergent immunoprecipitation followed by Western blot analysis. Quantitation was performed as described in Materials and Methods.
Photolabeling of Ost1p was performed as described in Materials and Methods and quantitation was based on densitometry of the autoradiograms. ND, not determined.
Relative labeling of Ost1p was determined by dividing the amount of photolabeled Ost1p by the relative quantity of immunoprecipitated ost1p. ND, not determined.
OT activity was carried out as described in Roos et al. (25). ND, not determined.
First we tested these mutants for their ability to be incorporated into the OT complex. Because each mutant Ost1p has a triple HA tag at its C terminus, we used a coimmunoprecipitation procedure followed by Western blot analysis to check for the presence of the other OT subunits. Because of the limitation of the antibodies available, we performed Western blots to check for the content of only certain OT subunits. It was found that mutants 324-326AAA, 327-329AAA, and 330-333AAAA did not coimmunoprecipitate other OT subunits, though similar amount of mutant Ost1p was immunoprecipitated. It is not surprising that these mutant proteins were not labeled by the photoprobe because they were not in the OT complex (Table 1). However, mutant 64-66AAA was incorporated into the complex at permissive temperature (Fig. 5 and Table 1). The protein conformation might be changed at the nonpermissive temperature and cause poor incorporation. As to the in vitro OT activity, 64-66AAA displayed only 30% activity compared with the wild-type control at 37°C.
Fig 5.
Ost1p block mutant 64-66AAA is incorporated into the OT complex. Microsomes were prepared from the block mutant strain and the wild-type control of Ost1p. Coimmunoprecipitation was carried out. Similar amounts of each sample that had been eluted from protein G agarose beads were analyzed by SDS/PAGE and followed by Western blot analysis using anti-Ost1p, anti-Wbp1p, anti-Swp1p, and anti-Stt3p antibodies as shown. The multiple bands of Wbp1p in lane 1 are believed to be caused by the underglycosylation of the mutant. Lane 1, 64-66AAA; lane 2, wild-type control.
Consequently, we generated single Ala mutations within these four block mutations to determine which of the individual amino acid residues are important for growth (13 mutants total). Surprisingly, only the R326A mutant showed a temperature sensitive phenotype at 37°C; the other 12 mutants showed no defects in growth. Therefore, the temperature sensitivity of TEY64-66 mutated to AAA was probably caused by an additive effect of the three single mutations. With respect to the 10-aa residue region (residues 324–333), we checked the glycosylation pattern of CPY and found no defects except in the case of R326A (Fig. 4). We speculate that these 10 residues might either comprise a segment that is essential for interacting with other OT subunits or form a loop region (as deduced by secondary structure predictions, unpublished observations) that is essential in positioning two functionally important helices. Perhaps when only one residue is mutated at a time, the effect is not sufficient enough to cause the cells to be impaired in growth except in the case of R326A. However, when three or four residues are mutated, the defect is pronounced enough that the mutant protein cannot be incorporated into the complex, and therefore these mutants are not viable.
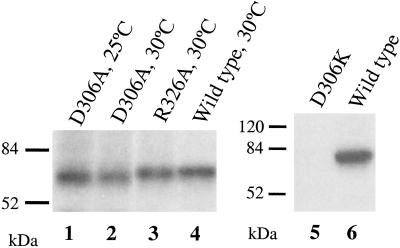
Subsequently, we checked the OT activity and the complex association of the R326A mutant. As shown in Fig. 4, Ost1p, Wbp1p and CPY were all underglycosylated in R326A mutant. It also displayed lowered in vitro OT activity at 37°C. It is incorporated into the OT complex (Table 1). We also investigated the photoprobe attachment of this R326A mutant protein and found it could be photolabeled well at 30°C (Fig. 6, lane 3). Therefore, it is unlikely that R326 residue is involved in peptide glycosylation site recognition.
Fig 6.
The binding of photoprobe to point mutants of Ost1p. Microsomes were prepared from each indicated mutant strain that had been grown at either 25 or 30°C, photolyzed, and immunoprecipitated. Samples were analyzed by SDS/PAGE and the labeled peptides were detected by autoradiography.
We also prepared a large number of single amino acid mutants of Ost1p and most of the single amino acid residue mutants had no growth phenotype. Besides R326A, a temperature sensitive mutation, only D306K was found to be a lethal mutation. When D306 was mutated to A instead of K, the cells grew very poorly. However, mutation of D306 to E did not cause any defect (data not shown). Based on its growth phenotype, D306 seems to be important only because it must be negatively charged. D306A had lower OT activity compared with the wild-type control (Table 1). These two mutants, D306K and D306A, were then checked for their ability to be integrated into the OT complex. The results indicated that in the case of D306A the mutant protein itself immunoprecipitated very well, but only low amounts of other OT subunits were coimmunoprecipitated with it compared with the wild-type control. Although the amount of D306K that immunoprecipitated was limited, none of the other OT subunits were present in the immunoprecipitated pellet (results are summarized in Table 1). These results imply that this charged residue D306 might be involved in interacting with other OT subunits by its negative charge; the slightly longer side chain of E was found to be able to mediate this interaction.
The photolabeling ability of these two mutants was also investigated. As shown in Fig. 6, mutant D306A was photolabeled quite well (lanes 1 and 2). We speculate that this mutant protein is in the OT complex, though its interaction with other OT subunits is weak. It could be radiolabeled by the photoprobe if it is in the OT complex. However, with mild detergent solublization, interactions with other subunits were perturbed and therefore limited amounts of other OT components were coimmunoprecipitated. But the antibody used for immunoprecipitation was against the HA tag on the C terminus of the mutant protein and therefore the labeled mutant protein was precipitated by the antibody very well. These findings indicated that D306 also could not be involved in the glycosylation recognition site. Furthermore, mutant D306K was not radiolabeled by the photoprobe (Fig. 6, lane 5). This mutant protein is not in the complex and therefore cannot be linked to the photoprobe. From this very extensive mutagenesis analysis of Ost1p, it is most likely the luminal domain of Ost1p that is involved in interacting with one or more OT subunits rather than serving as the peptide glycosylation recognition domain or the catalytic site. We believe model II is highly unlikely.
Conclusions
Initially we chose to place Bpa in the X position of Asn-X-Thr/Ser because we knew that, with the exception of proline, any amino acid could be located here and the tripeptide would still serve as a substrate (17, 19, 30, 31). This turned out to be the case with Bpa. However, this residue may not have been close enough to the glycosylation site recognition domain in the enzyme because residues with a great difference in the side chain length (i.e., Gly vs. Lys vs. Bpa) can be accommodated in the X position. Presumably this means that their side chains are extended away from the recognition site. This, of course, would explain why Bpa was not linked to the enzyme recognition site per se. Based on this study, we believe that Ost1p was photolabeled because of its close proximity to the OT subunit containing the glycosylation site recognition domain. Recently we have reported evidence (22) indicating that this subunit containing the glycosylation recognition site and/or catalytic site is Stt3p, as shown in model III in Fig. 1.
Acknowledgments
We thank Dr. Reid Gilmore for generous gifts of pRS316-OST1 plasmid, anti-Ost1p, and anti-Ost2p antibodies, Dr. Markus Aebi for anti-Wbp1p and anti-Swp1p antibodies, and Dr. Satoshi Yoshida for anti-Stt3p antibody. We give special thanks to Drs. Robert Haltiwanger, Ann Sutton, Hangil Park, and Tadashi Suzuki for helpful discussions. This work was supported by National Institutes of Health Grant GM33185.
Abbreviations
OT, oligosaccharyl transferase
bh, Bolton–Hunter reagent
Bpa, p-benzoylphenylalanine
CNBr, cyanogen bromide
HA, hemagglutinin
PNGase F, protein glycanase F
CPY, carboxypeptidase Y
References
- 1.te Heesen S., Rauhut, R., Aebersold, R., Abelson, J., Aebi, M. & Clark, M. W. (1991) Eur. J. Cell Biol. 56, 8-18. [PubMed] [Google Scholar]
- 2.te Heesen S., Janetzky, B., Lehle, L. & Aebi, M. (1992) EMBO J. 11, 2071-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.te Heesen S., Knauer, R., Lehle, L. & Aebi, M. (1993) EMBO J. 12, 279-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knauer R. & Lehle, L. (1994) FEBS Lett. 344, 83-86. [DOI] [PubMed] [Google Scholar]
- 5.Silberstein S., Collins, P. G., Kelleher, D. J., Rapiejko, P. J. & Gilmore, R. (1995) J. Cell Biol. 128, 525-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pathak R., Parker, C. S. & Imperiali, B. (1995) FEBS Lett. 362, 229-234. [DOI] [PubMed] [Google Scholar]
- 7.Silberstein S., Collins, P. G., Kelleher, D. J. & Gilmore, R. (1995) J. Cell Biol. 131, 371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaoglu D., Kelleher, D. J. & Gilmore, R. (1995) J. Cell Biol. 130, 567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida S., Ikeda, E., Uno, I. & Mitsuzawa, H. (1992) Mol. Gen. Genet. 231, 337-344. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida S., Ohya, Y., Nakano, A. & Anraku, Y. (1995) Gene 164, 167-172. [DOI] [PubMed] [Google Scholar]
- 11.Zufferey R., Knauer, R., Burda, P., Stagljar, I., te Heesen, S., Lehle, L. & Aebi, M. (1995) EMBO J. 14, 4949-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi J. H., Roos, J. & Dean, N. (1996) J. Biol. Chem. 271, 3132-3140. [DOI] [PubMed] [Google Scholar]
- 13.Reiss G., te Heesen, S., Gilmore, R., Zufferey, R. & Aebi, M. (1997) EMBO J. 16, 1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knauer R. & Lehle, L. (1999) J. Biol. Chem. 274, 17249-17256. [DOI] [PubMed] [Google Scholar]
- 15.Silberstein S. & Gilmore, R. (1996) FASEB J. 10, 849-858. [PubMed] [Google Scholar]
- 16.Knauer R. & Lehle, L. (1999) Biochim. Biophys. Acta 1426, 259-273. [DOI] [PubMed] [Google Scholar]
- 17.Marshall R. D. (1972) Ann. Rev. Biochem. 41, 673-702. [DOI] [PubMed] [Google Scholar]
- 18.Bause E. (1983) Biochem. J. 209, 331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavel Y. & von Heijne, G. (1990) Protein Eng. 3, 433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Q., Prestwich, G. D. & Lennarz, W. J. (1999) J. Biol. Chem. 274, 5021-5025. [DOI] [PubMed] [Google Scholar]
- 21.Dorman G. & Prestwich, G. D. (1994) Biochemistry 33, 5661-5673. [DOI] [PubMed] [Google Scholar]
- 22.Yan, Q. & Lennarz, W. J. (2002) J. Biol. Chem., in press.
- 23.Sikorski R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker D., Wuestehube, L., Schekman, R., Botstein, D. & Segev, N. (1990) Proc. Natl. Acad. Sci. USA 87, 355-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roos J., Sternglanz, R. & Lennarz, W. J. (1994) Proc. Natl. Acad. Sci. USA 91, 1485-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeke J. D., LaCroute, F. & Fink, G. R. (1984) Mol. Gen. Genet. 197, 345-346. [DOI] [PubMed] [Google Scholar]
- 27.Karaoglu D., Kelleher, D. J. & Gilmore, R. (1997) J. Biol. Chem. 272, 32513-32520. [DOI] [PubMed] [Google Scholar]
- 28.Kelleher D. J. & Gilmore, R. (1994) J. Biol. Chem. 269, 12908-12917. [PubMed] [Google Scholar]
- 29.Stevens T., Esmon, B. & Schekman, R. (1982) Cell 30, 439-448. [DOI] [PubMed] [Google Scholar]
- 30.Hart G. W., Brew, K., Grant, G. A., Bradshaw, R. A. & Lennarz, W. J. (1979) J. Biol. Chem. 254, 9747-9753. [PubMed] [Google Scholar]
- 31.Welply J. K., Shenbagamurthi, P., Lennarz, W. J. & Naider, F. (1983) J. Biol. Chem. 258, 11856-11863. [PubMed] [Google Scholar]