Abstract
Force production by type IV pilus retraction is critical for infectivity of Neisseria gonorrhoeae and DNA transfer. We investigated the roles of pilus number and the retraction motor, PilT, in force generation in vivo at the single-molecule level and found that individual retraction events are generated by a single pilus fiber, and only one PilT complex powers retraction. Retraction velocity is constant at low forces but decreases at forces greater than 40 pN, giving a remarkably high average stall force of 110 ± 30 pN. Further insights into the molecular mechanism of force generation are gained from the effect of ATP-depletion, which reduces the rate of retraction but not the stall force. Energetic considerations suggest that more than one ATP is involved in the removal of a single pilin subunit from a pilus. The results are most consistent with a model in which the ATPase PilT forms an oligomer that disassembles the pilus by a cooperative conformational change.
Type IV pili are polymeric filaments found on many Gram-negative bacteria. They play a crucial role in human pathogenesis, because they mediate adhesion to host mammalian cells evoking downstream cellular responses (1), biofilm formation (2), twitching motility (3, 4), and DNA uptake (5). Recently, type IV pili have been shown to generate considerable force by retraction (3). For some pilus-dependent functions, the amount of force generated by pilus retraction is critical, e.g., in host-cell responses (6) and movement of the bacteria through viscous mucous layers. Thus, it is of major interest to determine the level of force generated by a single pilus retraction and the factors that regulate force development.
Molecular motors normally move objects by repetitive, ATP-powered motions on polymer filaments. Type IV pilus polymers are several micrometers in length and 6 nm in diameter with 1,300 PilE (pilin) subunits/μm (7). It has been shown that PilT, an ATPase associated with various cellular activities (AAA), is required for pilus retraction (3, 8) and force generation. However, it is unknown whether the pilus retraction process, which most likely involves depolymerization into a membrane, is functionally similar to other motors or fundamentally different because of membrane depolymerization. PilT may act as a molecular motor (9), as a chaperone in a ratchet-like process (10), or as a decapping protein.
We address these issues by inducing the expression of pili (11) and the presumed motor molecule PilT in the human pathogen Neisseria gonorrhoeae at varying levels. The force–velocity dependence of a motor provides information on the molecular mechanism of force generation. Our data suggest that PilT is indeed an active motor molecule and that a single motor complex, presumably a hexamer, powers the retraction of a single pilus. Of particular note is the fact that this system generates force in excess of 100 pN, making PilT the strongest molecular motor reported so far.
Materials and Methods
Bacterial Strains and Media.
N. gonorrhoeae were maintained on GCB agar with supplements (Difco) and 0–10 mM isopropyl β-d-thiogalactoside (IPTG). Retraction assays were carried out in phenol red-free DMEM (GIBCO), supplemented with 2 mM L-glutamine/4 mM Na-pyruvate/5 mM ascorbic acid/1 mg/ml BSA/15 mM Hepes/0–10 mM IPTG (pH 7.9) at 33°C, unless stated otherwise. N. gonorrhoeae MS11 (WT) and its isogenic strain MS11C9.10 (pilE) are genetically described by Long et al. (11). Transformation was used to transfer the IPTG-inducible pilT gene in strain MW4 into MS11 to yield strain MS11–600 (pilT; ref. 5).
Electron Microscopy.
Bacteria were streaked into experiment buffer without BSA and adsorbed to a poly(L-lysine)-coated carbon grid for 10 min. After washing with PBS, cells were stained with 2% uranyl acetate for 1 min and washed three times with water.
Retraction Assay.
For retraction experiments, 3-μm silica beads (Polysciences) were coated with poly(L-lysine) and adsorbed to glass cover slides by centrifugation. Subsequently, 1.5-μm silica beads or 2-μm carboxylated latex beads (Polysciences) were added without further treatment to a suspension of gonococci, mounted on a microscope slide, and sealed.
The optical tweezers system consisted of a neodimium doped yttrium aluminum-garnet (Nd:YAG) laser (2 W), an inverted Zeiss microscope, and a movable mirror system that allowed computer-controlled deflection of the laser beam. Acquisition of positional data was performed as described by Simmons et al. (12) at 1 kHz time resolution by using a quadrant photodiode (Hamamatsu Photonics, Hamamatsu City, Japan) and LABVIEW software (National Instruments, Austin, TX). Before each acquisition, the detector signal was calibrated by recording the response bead movement caused by triangular movement of the optical trap by the mirrors. Displacement was linear in a range of 400 nm. To check for linearity and record bead movements larger than 400 nm, we used an image acquisition system that contained a video camera (nuvicon), an S-VHS video recorder, and an SGI workstation running ISEE particle-tracking software (Inovision, Raleigh, NC). We calibrated the trap stiffness by the viscous drag method as described in ref. 13 and verified the calibration with a spectrum analysis of the bead's Brownian motion (14).
Determination of Retraction Rate.
To determine the local rate of retraction, one has to average over the spatial fluctuations. Brownian motion, as well as an instability of the mirrors controlling the position of the laser beam, generates an SD of σ ≈ 7–10 nm. The oscillations of the mirror occur at multiples of 60 Hz; removing the peaks in the spectrum of the bead's motion yields an SD of σ ≈ 4 nm at a trap stiffness of ktrap = 0.3 pN/nm. The amplitude of fluctuations is decreased when the pilus binds and retracts. To average over at least two oscillations, we fitted a polygon to the data obtained from the photodetector with an average step length of 30 ms. The fitting procedure has been described in detail in ref. 15. Because of Brownian motion, we estimate an error on the velocity of 2%. (For details of this estimation, see ref. 15.) To check whether increasing the step length significantly influences the value obtained for the retraction rate, we compared the velocity vs. forces curves obtained from 20 retraction events averaged over 30 and 100 ms, respectively. The data overlapped within the statistical error bars for F > 20 pN (at lower forces, the start of a retraction is smeared out when the step length becomes large, resulting in a decreased velocity). Because pauses were rare events, we did not remove them.
We assume that the major error in the measurement of retraction rates and stall force arises from the finite size of the bacterium and thus from a velocity and force component in the vertical direction. The vertical direction is defined as perpendicular to the cover slide. With a total distance of 3 μm between the center of the bead and the bacterium and an uncertainty of δh ≈ 0.5 μm in the height of the point of attachment of the pilus, we estimate an error of 15% in the retraction velocity and the stall force in the horizontal direction.
To assess the effect of pilus/bacterium elasticity on the velocity vs. force behavior, we attached pilT mutant cells that produce pili but cannot retract to the surface-bound poly(L-lysine) beads. Then we moved a bead attached to a pilus at a velocity of 5 μm/sec. The elasticity, i.e., the force vs. elongation curve, shows almost linear behavior. Because it is likely that the orientation of the bacterium with respect to the bead and the location of pilus attachment influences the elastic behavior, we averaged nine force vs. deflection curves and calculated an approximate stiffness kbact ≈ 1.3 ± 0.2 pN/nm by a linear fit (data not shown). Then, we corrected the retraction velocity according to vretract = (l + ktrap/kbact)vbead (16).
The error on the retraction velocity Δv was calculated as a sum of the statistical error Δvs = σs/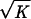 , geometrical error Δvg = 0.15 v, and elastic error Δve = 0.05 v; i.e., Δv = Δvs + Δvg + Δve, where σs is the SD and K is the number of measurements.
, geometrical error Δvg = 0.15 v, and elastic error Δve = 0.05 v; i.e., Δv = Δvs + Δvg + Δve, where σs is the SD and K is the number of measurements.
Results
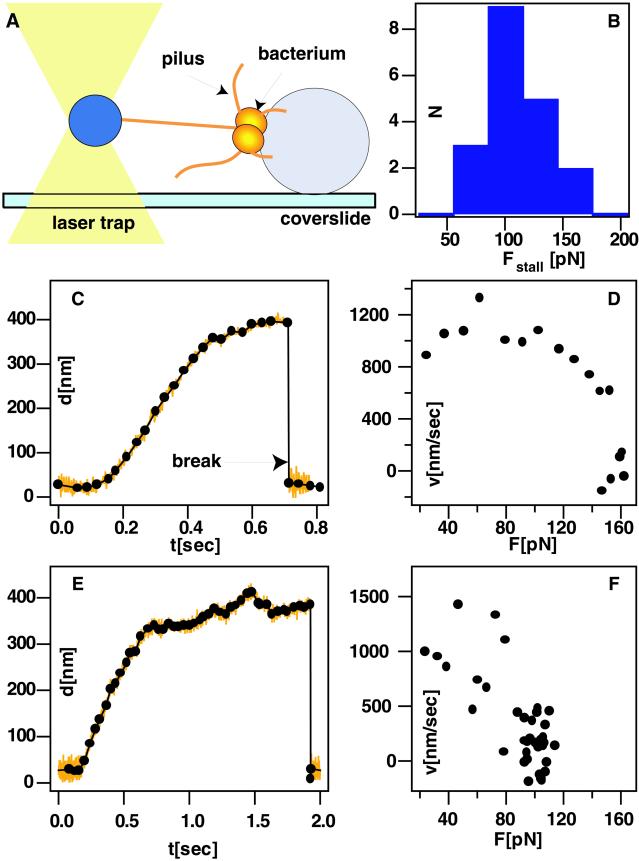
We measured the force–velocity relationship for individual type IV pilus retraction events in N. gonorrhoeae. A quantitative bead-based retraction assay was used (Fig. 1A); single gonococci were immobilized by attachment to a poly(L-lysine)-coated silica bead. By using a laser trap, a small bead was positioned 1–2 μm from the bacterium. Eventually, a pilus bound to the bead, and upon retraction, the bead was pulled out of the center of the laser trap. For small deflection, the force acting on the small bead is proportional to its deflection and allowed to determine the force acting on the pilus after calibration of the trap stiffness (see Materials and Methods). Likewise, the pilus-bound bead served as a probe to determine the retraction rate by measuring its deflection as a function of time.
Fig 1.
(A) Sketch of the setup. For retraction experiments, 3-μm silica beads (Polysciences) were coated with poly(l-lysine) and adsorbed to glass cover slides by centrifugation. Subsequently, 1.5-μm silica beads or 2-μm carboxylated latex beads (Polysciences) were added without further treatment to a suspension of gonococci, mounted on a microscope slide, and sealed. (B) Histogram of stalling force for MS11. (C and E) Deflection of the bead as a function of time (orange line). The fit (•) is a polygon fit with an average over 30 ms, as described in Materials and Methods. (D and F) Velocity vs. force for the event shown in C and E, respectively. (C and D) MS11C9.10 at 0.1 mM IPTG. (E and F) MS11.
Determination of the Stall Force of Pilus Retraction.
Retraction events were rapid (Fig. 1 C–F) but were easily quantified to determine the retraction rate. Pili retracted at an initial velocity of 〈v0〉 = 1,200 ± 200 nm/sec, and the velocity dropped dramatically at forces greater than 40 pN. Because we used a stationary trap, the external force increased as the pilus shortened. Thus, force and/or subunit concentration in the membrane could have slowed retraction. To test whether a local accumulation of pilin subunits affected the retraction rate, we performed experiments at very low trap stiffness, i.e., k = 0.08 pN/nm, where the maximum force on the bead was far below the stalling force. We found that the retraction rate did not depend on the retracted length of the pilus.
At high trap stiffness, i.e., k > 0.3 pN/nm, pilus-generated forces were not high enough to overcome the maximum force of the optical trap, and the pilus retraction stalled. Usually, a retraction event was terminated by release of the bead into the laser trap. We interpreted this release as a breakage event, although the location of the break was not known; the pilus, the attachment between pilus and bacterium, or the bond between the tip of the pilus and the bead may break. Most breakage events occurred at low forces (3). A stall was operationally defined as a retraction where the rate dropped below 40 nm/sec for at least 100 ms. A broad distribution of stalling forces was observed with an average stalling force of 110 ± 30 pN and a maximum of 140 pN (Fig. 1B).
Kinetics of a Single Retracting Pilus.
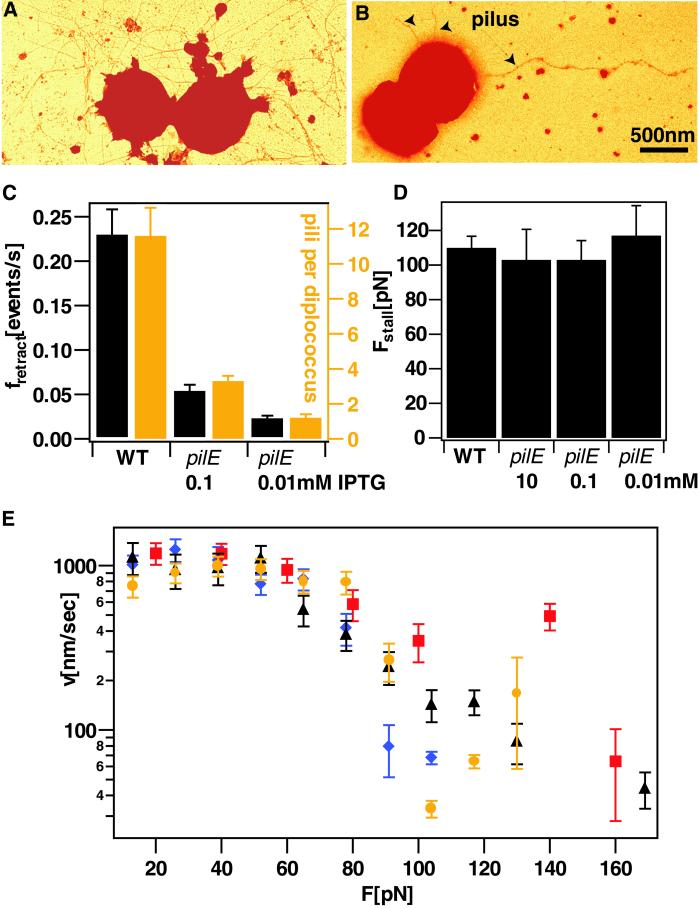
It was possible that the retraction of multiple pili was responsible for the generation of the high forces in excess of 80 pN (3). To assess whether forces as high as 140 pN were generated by a single pilus, we used a pilE strain (MS11C9.10) in which pilin expression was inducible by IPTG (11). Western blots were performed to verify that the concentration of pilin increased with increasing concentrations of IPTG (data not shown). At very low IPTG induction levels, electron micrographs showed that 80% of the diplococci had no pili, and the remainder had predominantly a single pilus with a maximum of three well separated pili (Fig. 2B). With increasing pilin concentration, cells expressed a higher number of pili (Fig. 2 A and C), and we found that the frequency of retraction events increased linearly with the number of expressed pili. However, neither the velocity at low forces (F < 80 pN) nor stall force changed significantly with an increasing number of pili (Fig. 2 D and E). Although we cannot exclude that minor modulations may be concealed by the scatter of the data near the stall force, the force–velocity profile revealed no strong dependence on the level of pilus expression (Fig. 2E). These observations suggest that single pili are responsible for each retraction event in the inducible PilE strain.
Fig 2.
Retraction kinetics of pilE mutant under load. Electron micrograph of WT MS11 (A) and pilE mutant MS11C9.10 (B) at 0.01 mM IPTG. (C) Frequency of retraction events and number of pili visible by electron microscopy. Only those cells were counted that retracted/showed pili. (D) Average stall force of WT compared with pilE mutant at varying IPTG concentrations. (E) Velocity as a function of force for varying expression level of pilin subunits. WT, black triangle; pilE, 10 mM IPTG, blue diamond; pilE, 0.1 mM IPTG, red square; pilE, 0.01 mM IPTG, yellow circle; averaged over K > 90 retraction events for each IPTG concentration. Note that many retraction events were terminated by breakage before the stalling force was reached.
For WT cells, we cannot exclude the possibility that multiple pili or even a bundle of pili are attached to the bead in the laser trap. However, pilE mutants expressing on average only one pilus and retracting only once in 50 sec are stalled by the same external force as the WT. Thus, we conclude that in a WT gonococcus, retraction events are temporally well separated and generated by a single pilus. Direct visualization of pilus retraction in Pseudomonas aeruginosa (4) showed that single retraction events were not correlated in time in good agreement with our findings.
Effect of PilT Concentration on Pilus Retraction.
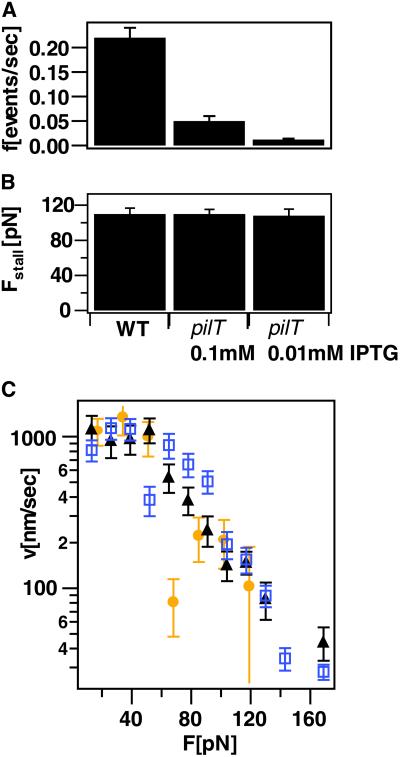
If single retraction events are driven by a single pilus, then a single motor molecule (PilT) may be driving the retraction. Therefore, we next investigated whether the retraction rate or the stall force depended upon the concentration of PilT. Recent experiments showed that the speed of flagella rotation in Escherichia coli increases stepwise with the concentration of force-generating subunits (17). By using a pilT strain (MS11–600) in which PilT is regulated by IPTG, we varied the number of PilT molecules per cell (5). On a glass surface, the pilT strain exhibited a significantly different twitching phenotype from the WT strain: whereas WT MS11 was continuously motile, the pilT strain made sporadic steps of 0.2–2 μm at low levels of IPTG. In the absence of IPTG, no significant movement was observed. The frequency of retraction events increased with increasing concentration of IPTG (Fig. 3A), indicating that PilT concentration was the limiting factor in motility. At low level of IPTG and low force, i.e., F < 80 pN, only 3% of the retractions paused with a maximum pause length of 47 msec, as compared with a pausing frequency of 2% for the WT, with a maximum pause length of 80 msec. (A pause was defined as a drop in velocity below 40 nm/sec for at least 30 msec.) Although the retraction frequency strongly depends on the level of PilT expression, the pausing frequency is largely independent on PilT concentration, indicating that PilT is highly processive.
Fig 3.
Retraction kinetics of pilT mutant. (A) Frequency of retraction events. (B) Average stall force of WT compared with pilT mutant at varying IPTG concentration. (C) Retraction rate for varying IPTG concentrations. WT, black triangle; pilE, 0.01 mM IPTG, yellow circle; pilE, 0.1 mM IPTG, blue square; averaged over K > 34 retraction events for each IPTG concentration.
Even at the lowest IPTG level when retraction events were very rare, the retraction events that did occur had the same stall force and unloaded velocity as the WT strain and the inducible PilE strain (Fig. 3 B and C). Likewise, the force–velocity profiles are similar for varying levels of PilT induction, although the data does not allow analyzing details of the velocity decay for forces larger than 50 pN. Thus, whether we decrease the number of pili or the number of PilT molecules, we observe the same stall force and similar force–velocity dependence. Taken together, these results indicate that the retractions represent the movement of a single pilus by a single PilT functional unit.
ATP Depletion.
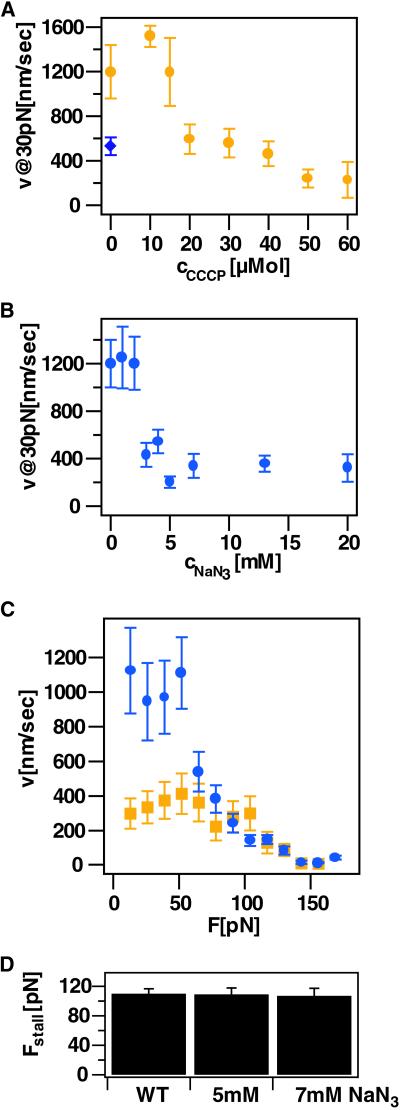
To determine whether ATP is required for retraction, ATP was depleted by exposing cells to ionophores carbonylcynanide m-chlorophenylhydrazone (CCCP), 2,4-dinitrophenol (DNP), and sodium azide (NaN3), an inhibitor of electron transport, for 30–90 min. Note that we cannot quantify the concentration of ATP available for PilT because the efficiency of ATP depletion may not be proportional to the amount of inhibitor added; furthermore, proteins with a higher binding affinity for ATP than PilT may compete for ATP-binding. Qualitatively, these reagents decreased the retraction rate but not the stall force, as shown in Fig. 4 A–D. This finding suggests that pilus retraction is powered by ATP, consistent with the hypothesis that PilT is an ATPase (8).
Fig 4.
Depletion of ATP. (A) Retraction rate at varying concentrations of CCCP (yellow circle) and 1 mM DNP (blue diamond) at 30 pN. (B) Rate at varying concentrations of NaN3 at 30 pN. (C) Comparison of the velocity vs. force curve at 0 mM (blue circle) and 5 mM NaN3 (yellow square); averaged over K > 10 retraction events for each concentration. (D) Average stall force for 5 and 7 mM NaN3.
Discussion
All of the evidence that we have obtained indicates that single retraction events are powered by the action of a single PilT complex on a single pilus. Decreasing either the number of pili or the number of PilT molecules per cell decreases the frequency of retraction events but does not alter the force–velocity relationship. The force produced by a single pilus is unexpectedly high. A variety of molecular motors like kinesin, polymerases, and a bacteriophage portal motor have been studied in vitro (15, 18–20). These motors stall at considerably lower forces, i.e., between 5 and 57 pN. Interestingly, the strongest motor reported so far, the portal motor of phage Φ29, comprises five ATPases (21).
Molecular Models for Pilus Retraction.
Genetic studies and structural data support the following molecular model. The cytoplasmic membrane has a reservoir of the prepilin subunits, which are proteolyzed by the prepilin peptidase PilD and then polymerized by the AAA PilF into pili. PilT acts as an antagonist of PilF and is responsible for the depolymerization of the pilus into the inner membrane. It is unknown, however, how PilT induces the process of pilus retraction.
One possible mechanism is a translocation ratchet, where the pilus diffuses forward and backward through the membrane. In this model, PilT acts as a chaperone that binds either the pilus or the pilin subunits on the cytoplasmic side and inhibits repolymerization. A similar mechanism has been discussed to explain posttranslational protein translocation through a membrane pore (22). In this scenario, we would expect that the retraction rate depended on the concentration of PilT molecules and possibly on the concentration of PilE. However, our data show no effect of PilT or PilE concentration on the retraction kinetics, and, therefore, we rule out this model.
We found that pilus retraction is highly processive; this is consistent with a model in which pilin is in an unstable state in the pilus that is stabilized by a cap, and PilT is an uncapping protein. In this scenario, the energy stored in the polymerized form (possibly provided by PilF) is released during pilus retraction and used to develop an external force. This model seems unlikely for the following reasons: we observed that depletion of ATP decreases the rate of retraction, which argues against an initial uncapping process. Furthermore, reducing the PilT concentration neither decreases the retraction rate nor introduces a significant number of pausing events, which would be expected if PilT action on a capping protein were ATP-dependent.
Hence, we favor a model in which PilT is actively involved in the dissociation of a pilus. PilT is a member of the GspE family of hexameric AAAs. The latter are known to be involved in the dissociation of stable protein interactions (23). A single PilT complex could bind to the cytoplasmic end of the pilus. ATP binding or hydrolysis could then induce a conformational change in the PilT complex, which would exert strain on a pilin subunit and thereby dissociate it from the pilus.
How can a single motor molecule generate large retraction forces? In a cell, hydrolysis of a single ATP commonly yields energy of ≈80 pN/nm (24). The type IV pilus is a single-stranded helix with five pilin subunits per turn and a pitch of 4 nm (7). Assuming that the relevant step length is set by the translocation of a single subunit into the cytoplasmic membrane, i.e., 0.8 nm, the energy involved in removing a single subunit is, on average, 88 pN/nm, with a maximum of 112 pN/nm. This energetic consideration suggests that hydrolysis of more than a single ATP is involved in the removal of one pilin subunit near the stall force. Because PilT belongs to the GspE family of hexameric AAAs (25), one PilT functional unit could hydrolyze several (up to six) ATP molecules in the process of dissociating one pilin subunit. The shape of the velocity–force curve shows a velocity plateau at low force and a steep decrease in velocity for F > 40 pN (Fig. 4). Thus, the mechanical translocation step is not rate-limiting at low force. A logical model to explain these observations is that ATP binding is rate-limiting at low force, but when the process is slowed by mechanical inhibition of the translocation step, a second ATP could bind to another subunit of the hexamer and thereby alter the kinetics. We have shown that pilus disassembly is highly processive, and we suppose that PilT and PilF alternately bind to the cytoplasmic end of the pilus. Because PilT does not induce pilus assembly, it is unlikely that PilT can relax force by reversal of pilus disassembly or microslippage, and thus, retraction is effectively irreversible. Further, the proteolysis of the pilin subunits also may be coupled with pilus assembly, making it impossible for the dissociated pilin subunits to contribute to reversal of disassembly. On the other hand, PilF, another AAA, is likely to hydrolyze ATP during pilus formation. The energy stored in the polymerized pilus could be released during pilus retraction. Thus, energy from PilT and/or PilF could contribute to the generation of large forces, and further experiments will be required to assess these possibilities.
How Do High Retraction Forces Affect Bacterial Function?
Neisserial infection of epithelial cells triggers the formation of cortical plaques containing high concentrations of signaling proteins (26). A pilT mutant adheres to cells as well as the WT parent strain but fails to stimulate plaque formation and is reduced in its ability to invade cells (A. J. Merz and M.S., unpublished results). These results suggest that pilus retraction during attachment promotes subsequent steps in infection. Cortical plaque formation is similar to formation of focal adhesion complexes, which respond to force. Focal complexes on the upper cell surface were generated when force was applied to beads acting as extracellular attachment sites (27). It is, therefore, reasonable to hypothesize that epithelial cells will sense the amount of force generated by pilus retraction and respond in a similar manner. High forces may also be necessary for bacterial movement through the viscous mucous.
WT Neisseria express multiple pili and eventually form bundles as shown in Fig. 2A. Our observations indicate that bundle formation is not involved in generation of high forces. However, we found that most retraction events were terminated by breakage of the link between the bead and bacterium. Furthermore, the pilE mutants at low level of pilus expression detached more frequently from a glass surface than the WT. These observations indicate that bundle formation is important for bacterial attachment, rather than force generation.
Acknowledgments
We thank M. Koomey and C. Freitag for MW4 and K. Brown for the opportunity to use the electron microscopy facilities. We are grateful for discussions with N. Heckenberg, A. Merz, and S. Clary. This work was funded by Columbia University Funds (to M.P.S.) and National Institutes of Health Grant AI49973. B.M. acknowledges support by the Deutsche Forschungsgemeinschaft (Emmy Noether Program).
Abbreviations
IPTG, isopropyl β-d-thiogalactoside
AAA, ATPase associated with various cellular activities
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lee S. L., Bonnah, R. A., Hugashi, D. L., Atkinson, J. P., Milgram, S. L. & So, M. (2002) J. Cell Biol. 156, 951-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh P. K., Parsek, M. R., Greenberg, E. P. & Welsh, M. J. (2002) Nature 417, 552-555. [DOI] [PubMed] [Google Scholar]
- 3.Merz A. J., So, M. & Sheetz, M. P. (2000) Nature 407, 98-101. [DOI] [PubMed] [Google Scholar]
- 4.Skerker J. M. & Berg, H. C. (2001) Proc. Natl. Acad. Sci. USA 98, 6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfgang M., Lauer, P., Park, H.-S., Brossay, L., Hébert, J. & Koomey, M. (1998) Mol. Microbiol. 29, 321-330. [DOI] [PubMed] [Google Scholar]
- 6.Geiger B., Bershadsky, A., Pankov, R. & Yamada, K. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 793-805. [DOI] [PubMed] [Google Scholar]
- 7.Parge H. E., Forest, K. T., Hickey, M. J., Christensen, D. A., Getzoff, E. D. & Tainer, J. A. (1995) Nature 378, 32-38. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser D. (2000) Curr. Biol. 10, R777-R780. [DOI] [PubMed] [Google Scholar]
- 9.Wall D. & Kaiser, D. (1999) Mol. Microbiol. 32, 1-10. [DOI] [PubMed] [Google Scholar]
- 10.Merz A. J. & Forest, K. T. (2002) Curr. Biol. 12, R297-R303. [DOI] [PubMed] [Google Scholar]
- 11.Long C. D., Hayes, S. F., Putten, J. P., Harvey, H. A., Apicella, M. A. & Seifert, H. S. (2001) J. Bacteriol. 183, 1600-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons R. M., Finer, J. T., Chu, S. & Spudich, J. A. (1996) Biophys. J. 70, 1813-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai J. & Sheetz, M. P., (1998) Cell Membrane Mechanics (Academic, New York). [PubMed]
- 14.Gittes F. & Schmidt, C. F., (1998) Signals and Noise in Micromechanical Measurements (Academic, New York). [DOI] [PubMed]
- 15.Maier B., Bensimon, D. & Croquette, V. (2000) Proc. Natl. Acad. Sci. USA 97, 12002-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visscher K. & Block, S. M., (1998) Versatile Optical Traps with Feedback Control (Academic, San Diego). [DOI] [PubMed]
- 17.Ryu W. S., Berry, R. M. & Berg, H. C. (2000) Nature 403, 444-447. [DOI] [PubMed] [Google Scholar]
- 18.Visscher K., Schnitzer, M. J. & Block, S. M. (1999) Nature 400, 184-189. [DOI] [PubMed] [Google Scholar]
- 19.Wang M. D., Schnitzer, M. J., Yin, H., Landick, R., Gelles, J. & Block, S. M. (1998) Science 282, 902-907. [DOI] [PubMed] [Google Scholar]
- 20.Smith D. E., Tans, S. J., Smith, S. B., Grimes, S., Anderson, D. L. & Bustamante, C. (2001) Nature 413, 748-752. [DOI] [PubMed] [Google Scholar]
- 21.Simpson A. A., Tao, Y., Leiman, P. G., Badasso, M. O., He, Y., Jardine, P. L., Olson, N. H., Morais, M. C., Grimes, S., Anderson, D. L., et al. (2000) Nature 408, 745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peskin C. S., Odell, G. M. & Oster, G. F. (1993) Biophys. J. 65, 316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vale R. D. (2000) J. Cell Biol. 150, F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan S. & Sheetz, M. P. (1997) Annu. Rev. Biochem. 66, 785-805. [DOI] [PubMed] [Google Scholar]
- 25.Krause S., Bárcena, M., Pansegrau, W., Lurz, R., Carazo, J. M. & Lanka, E. (2000) Proc. Natl. Acad. Sci. USA 97, 3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merz A. J., Enns, C. A. & So, M. (1999) Mol. Microbiol. 32, 1316-1332. [DOI] [PubMed] [Google Scholar]
- 27.Choquet D., Felsenfeld, D. P. & Sheetz, M. P. (1997) Cell 88, 39-48. [DOI] [PubMed] [Google Scholar]