Abstract
We have developed a method for simultaneous recording of high-resolution topography and cell surface fluorescence in a single scan which we call scanning surface confocal microscopy. The resolution of the system allows imaging of individual fluorescent particles in the nanometer range on fixed or live cells. We used this technique to record the interaction of single virus-like particles with the cell surface and demonstrated that single particles sink into the membrane in invaginations reminiscent of caveolae or pinocytic vesicles. This method provides a technique for elucidating the interaction of individual viruses and other nanoparticles, such as gene therapy vectors, with target cells. Furthermore, this technique should find widespread application for studying the relationship of fluorescently tagged molecules with components of the cell plasma membrane.
Crossing the cell membrane is the first critical event in achieving exogenous gene expression. Understanding the details of the passage of viruses, bacteria or other nanoparticles across this barrier is essential in developing drug or gene delivery systems. Most uptake is mediated by receptors that trigger internalization, where the cell membrane invaginates to form a vesicle around the particle. The membrane-associated events involved, whether fusion (as in the case of enveloped viruses) or endo/phagocytosis (for nonenveloped viruses, bacteria, and most drug and gene delivery vectors), have largely been studied by electron microscopy, confocal microscopy, and by using reagents that block uptake pathways (1–4). By using these techniques, the molecular mechanisms of endocytosis have been described in detail for viral infection, and attention is now focusing on the dynamics of the process (5). Real-time, single-particle tracing methods are being developed to characterize viral infection pathways in living cells; however, details of movement across cellular membranes cannot be visualized because the membrane is not detectable optically (6). As the dynamics of nanoparticle penetration of cellular membranes is a key element in their uptake and infectivity, there is a need to develop techniques that provide high-resolution measurements of this process as a function of time.
A number of methods have recently been developed to record surfaces at high resolution by using scanning probe microscopy. These methods scan a fine probe over a surface by using the local interaction between them to maintain constant probe-surface separation. By using scanning tunneling microscopy and atomic force microscopy (AFM), atomic resolution is possible; however, their application in the field of biological sciences is limited, particularly when applied to live cells. One method that has the potential to obtain both high resolution topography and fluorescence images of the cell surface is scanning near-field optical microscopy (SNOM). This technique uses the force between the probe and the surface to control the distance between them, combined with a light from a subwavelength aperture to obtain simultaneous subdiffraction limited optical or fluorescence images. Despite its potential, there have been few studies using SNOM on cells because of the difficulties in reliably regulating the distance control over the soft and responsive cell surface and operating in physiological buffers (7).
We have adopted an alternative scanning approach using scanning ion conductance microscopy (SICM; ref. 8). This method is based on scanning a nanopipette over the cell surface by using the ion current that flows to the pipette to control the sample distance. The pipette distance is maintained at 25–75 nm from the cell surface so that the pipette glides over the cell without touching it. This method enables us to image the surface of live cells with high topographic resolution both laterally (determined by the outer radius of the pipette) and vertically, typically of 75–150 nm and 10–20 nm, respectively. This method is very reliable, enabling distance control over a contracting heart cell (9) and imaging of live cells for 24 h. In addition, we have used the pipette to apply ions locally and measure their flow into the cell to detect the location of ion channels (10). The same scanning nanopipettes have also been used to perform patch clamp to localize and functionally characterize channels on the surface of a cardiac myocyte (11).
Here, we describe an extension of this technique, scanning surface confocal microscopy (SSCM), a combination of SICM and scanning confocal microscopy (SCM), to simultaneously record topographic and quantitative fluorescence images of the cell surface. We have chosen to use polyoma virus-like particles (VLPs) as a model system to validate our method and to gain insight into their mechanism of entry into cells. Polyoma VLPs are nanospheres composed of 360 molecules of a single viral coat protein, VP1, that have been shown to interact with and carry plasmid DNA into cells (12, 13). The protein nanospheres encapsidate 1–2 kb of the plasmid DNA whereas the remainder loosely associates with the outside of the VLP structure (14, 15). These particles enter cells by both receptor-dependent and -independent routes, but only the former is productive for gene transfer (16). Thus, the fate of the VLPs may be determined by their interaction with the cell membrane, making VLPs an ideal model to test our system.
The interaction of VLPs adsorbed to the cell surface was imaged by using SSCM and SCM. SSCM produced similar fluorescence images to SCM and could also be used to image live cells. The apparatus was sensitive enough to detect single VLPs and identify cell surface features corresponding to them from the topography scans. These experiments demonstrate that SSCM can provide high resolution topographical and fluorescence data for following nanoparticle uptake in live cells.
Materials and Methods
Reagents.
All chemicals were obtained from Sigma and BDH, unless otherwise stated.
Cell Culture and Plasmids.
Monkey COS 7 cells were routinely maintained at 37°C in 5% CO2, using DMEM (GIBCO/BRL) containing 5% (vol/vol) FCS (Helena Biosciences, Sunderland, Tyne and Wear, U.K.; ref. 17). The plasmid DNA used in all experiments was pCIKLuxOE, based on pCIKLux (18), and contained the simian virus 40 origin of replication and enhancer region from nucleotides 5,176 to 268 (N.K., unpublished data).
Preparation of Cy3-Labeled VLPs and Adsorption to Cells.
Capsid-like particles were purified by sucrose and CsCl gradient centrifugation from Hi5 insect cells infected with recombinant baculovirus, as described (12), and resuspended in sterile 20 mM Hepes, pH 7.5 at 3–6 mg/ml. Capsid-like particles were covalently labeled with Cy3 by using the FluoroLink Cy3 monofunctional dye (Amersham Pharmacia). Cy3-labeled capsid-like particles were incubated with Qiagen-purified, supercoiled plasmid DNA at a molar ratio of 5:1 to produce VLPs, as described (12).
COS 7 cells (5 × 104 cells per well) were plated on 13-mm glass coverslips in 24-well plates, incubated overnight at 37°C in DMEM containing 5% (vol/vol) FCS and then incubated for 30 min at 0°C with VLPs in 400 μl of DMEM containing 20 mM Hepes, pH 7.5, 0.5% BSA (0.25 μg of plasmid DNA per well). Cells then were washed with DMEM at 0°C, incubated at 37°C in DMEM containing 5% (vol/vol) FCS and harvested, 0 or 4 h posttransfection, by fixing for 15 min in PBS containing 3.7% (vol/vol) formaldehyde and 250 mM sucrose. Coverslips then were assembled for SSCM.
For live imaging, cells were plated and grown as above and then transferred to Leibowitz medium without phenol red (L15, GIBCO/BRL). Coverslips were mounted on a support Petri dish (see Fig. 1), and scanning was carried out at room temperature in L15. VLPs (0.25 μg of plasmid DNA) in 40 μl of L15 were added to the cells for 30 sec, the cells were washed twice in excess media, and scanning was resumed.
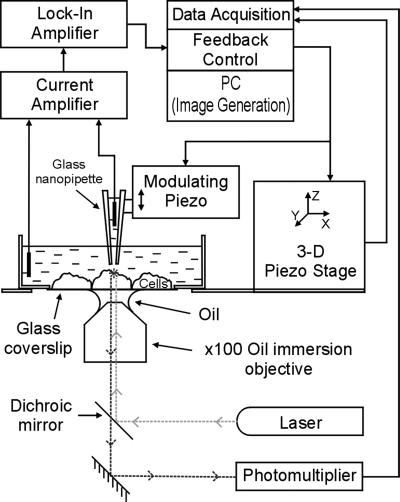
Fig 1.
Schematic diagram of the SSCM.
Analysis of VLPs by SCM and AFM.
Fluorescent Cy3-labeled VLPs (diluted to 0.15 mg/ml in water) were dropped onto glass and mounted in Vectorshield (Vector Laboratories). Samples were imaged by confocal microscopy and fluorescence detected by 0.2-μm sectioning with a Leica DM IRB SP1 SCM by using a plan-apochromat ×100/1.4 N.A. PH3 UV oil-immersion lens. Cy3 was excited with the 568-nm line (krypton laser), and emitted fluorescence was collected over 577–621 nm. Averaged digital images were captured by using the Sequential Scanning function (Leica Confocal Software; Leica Microsystems, Wetzlar, Germany), and maximum projections of six scans were used for fluorescence-intensity measurements.
AFM preparation and analysis was carried out as described (16).
Scanning Surface Confocal Microscope.
To perform SSCM, an apparatus was built (see Fig. 1) based on a combination of SICM and SCM. The SICM used here was described (9, 19, 20) and includes feedback control based on distance modulation, a technique developed independently by us (9) and others (21). The sensitive SICM probe consists of a glass nanopipette filled with electrolyte with an Ag/AgCl electrode plugged into it. The electrode is connected to a high impedance, head-stage current amplifier (Axopatch 200B, Axon Instruments, Foster City, CA). The probe detects the ion current passing from the liquid medium through the pipette opening. The probe is mounted on a piezo translation stage (Tritor, Piezosystem Jena, Germany) that is modulated vertically. In our experiments, vertical modulation was 40 nm at 200 Hz. As soon as the probe approaches the sample, the vertical movement of the pipette generates a modulated ion current. The modulated ion current is amplified and then fed into a lock-in amplifier (SR830 DSP, Stanford Research Systems, Sunnyvale, CA) tuned to the frequency of the modulation. The amplified and modulated current provided the signal for the feedback loop that was calculated by control/data acquisition software (East Coast Scientific, Cambridge, U.K.) for a DSP card (DSP32C PC, Loughborough Sound Images, Loughborough, U.K.). The feedback signal was used to control the probe position over the sample keeping constant probe-sample separation by means of a three-axis piezo-translation stage (Tritor, Piezosystem Jena, Germany) on which the sample was mounted. The control/data acquisition electronics drive the translation stage to scan the specimen under the nanopipette probe and record both the lateral and vertical positions, generating the topographical image.
To enable simultaneous SCM/SICM imaging of the same specimen, the following additional modifications were introduced to the system. The current control/data acquisition hardware and software were used to scan the specimen, to perform SSCM, or to acquire conventional confocal image sections. The excitation light source was provided by a GPNT-02 laser diode (532-nm wavelength, Lasertechnik Bremen, Germany). The optical recording system consisted of a Nikon Eclipse inverted microscope (Eclipse TE300, Nikon) equipped with a ×100 1.3 N.A. oil-immersion objective. The excitation light was fed through an epi-fluorescent filter block and emitted light collected by a photomultiplier with a pinhole (model D-104-814; Photon Technology International, Surbiton, U.K.).
The samples were imaged on glass coverslips, supported by a Petri dish, in the appropriate medium. The SICM nanopipettes were made from 1.00-mm outer diameter, 0.58-mm inner diameter glass microcapillaries (Intracel, Herts, U.K.) on a laser-based Brown-Flaming puller (model P-2000; Sutter Instruments, Novato, CA). The nanopipette tip outer radius estimated by scanning electron microscopy was ≈75 nm.
Results
SSCM.
SSCM is based on a combination of SCM and SICM. SICM is a scanning probe microscopy technique in which the ion current flowing into a nanopipette is used to control the vertical (z axis) position of the cell relative to the pipette tip. As shown diagrammatically in Fig. 2A, in SSCM the cell is moved up and down in the z direction while scanning in the x and y directions, so its surface is always the same distance from the nanopipette (typically, 25–75 nm). A laser is passed up a high numerical aperture objective so that it is focused just at the tip of the nanopipette, and a pinhole is positioned at the image plane so that the confocal volume is just below the pipette, as described (9). Thus, a fluorescence image of the cell surface is obtained in a single scan, as well as a simultaneously captured image of the cell topography. Any point on the cell surface is only exposed to the laser once during the scan, equalizing photobleaching across the sample.
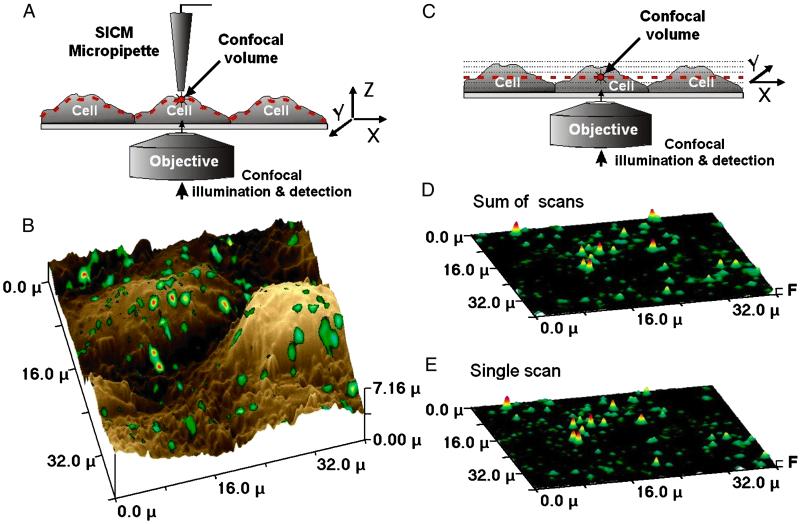
Fig 2.
Comparison of surface confocal and conventional confocal microscopy. (A) Principle of SSCM. Optical and topographical contouring with an SSCM. The dotted line indicates the position of the optical image of the cell surface obtained in a single scan. (B) Overlay of simultaneously obtained topographic and fluorescence images of fluorescent Cy3-labeled VLPs adsorbed to the surface of COS 7 cells. (C) Optical sectioning with a conventional scanning confocal microscope. Dotted lines indicate positions of multiple optical sections required to generate an image of the cell surface. (D) Sum of eight confocal sections of the same sample as shown in B. (E) Single surface confocal microscopy scan of the same sample as shown in B, projected on a flat surface.
Simultaneous Recording of Surface Fluorescence and Topography in a Single Scan.
To study the interaction of VLPs with the cell membrane, fluorescent Cy3-labeled VLPs were adsorbed to the surface of monkey COS 7 cells and imaged by using SSCM. The superimposed fluorescence and topographical images of the cell surface (Fig. 2B) show that the VLPs were scattered, apparently randomly, on or very close to the surface of the cell. By using conventional confocal microscopy, similar fluorescence data can be gathered, but because of the undulating surface of a cell, multiple scans are required to obtain the same amount of information (represented diagrammatically in Fig. 2C). To compare the data gathered by each method, scanning of the same area (as in Fig. 2B) by conventional confocal microscopy was carried out. The resulting image (a sum of eight scans, shown in Fig. 2D) displayed spots of a similar number and intensity to the SSCM scan (the SSCM image is shown flattened, for comparison, in Fig. 2E), demonstrating that a single SSCM scan generated data similar to the conventional confocal technique. Furthermore, by moving the confocal volume further from the pipette, it is possible to image inside the cell at different fixed distances from the cell surface, again in a single scan (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org).
Performing SSCM on Live Cells.
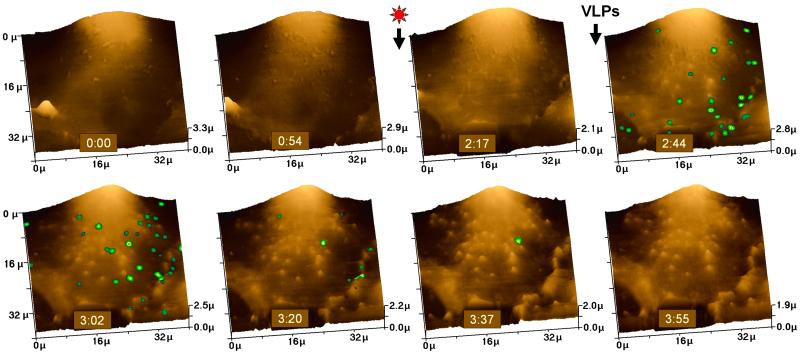
As one of our major goals is to apply SSCM to studying particle uptake in cells with time, we tested whether recordings of fluorescent VLPs on the surface of live cells could be made. Large-scale scans of 40 × 40 μm were carried out (≈18 min per scan) to gain a global impression of the cell with regard to morphology and distribution of VLPs. Selected images before and after adding VLPs are shown in Fig. 3, as three-dimensional (3D) images of the topography with superimposed fluorescence measurements. Eight scans were made over 2 h 17 min (scans at 0 and 54 min are shown). During this time, the cells were alive, as demonstrated by their morphology (flattened) and dynamic surface: microvilli (small projections scattered over the surface of the cell) formed and disappeared between scans, and a large feature (to the left of the images) could be seen migrating out of frame (first three panels). At 2 h 17 min, the laser was turned on. No significant autofluorescence was observed on the surface. Fluorescent VLPs then were added to the surface of the cells (see Materials and Methods) and scanning resumed (panel marked 2 h 44 min). VLPs bound to the cell surface, producing an image similar to that observed in fixed cells (compare with Fig. 2B). By the next scan (3 h 2 min), the distribution of surface fluorescence had altered dramatically. In the following image, much less surface fluorescence was observed (3 h 20 min), and by the last two scans, little remained (3 h 37 min and 3 h 55 min, respectively). At the end of the experiment, scanning by conventional confocal microscopy detected fluorescence within the cell (data not shown). A slowly moving fluorescent spot that was still visible on the surface at 3 h 37 min, when all of the other fluorescence had gone, indicated that the disappearance of fluorescence was not solely caused by photobleaching.
Fig 3.
Imaging live cells. Live COS 7 cells were sequentially scanned by SSCM approximately every 20 min for 4 h. Shown are topographical scans initiated at 0 min and 54 min and overlays of simultaneously obtained topographic and fluorescence scans at 2 h 17 min, 2 h 44 min, 3 h 2 min, 3 h 20 min, 3 h 37 min, and 3 h 55 min, respectively. Scanning for fluorescence was initiated at 2 h 17 min (red star); fluorescent VLPs were added at 2 h 40 min (VLPs). Fluorescence is represented as an intensity profile: black, lowest intensity; red, highest intensity. A slowly moving spot is clearly visible near the center of SSCM images at 3 h 2 min, 3 h 20 min, and 3 h 37 min (last remaining fluorescent spot in the image at 3 h 37 min).
Detection of Single VLP Interaction with the Cell Plasma Membrane.
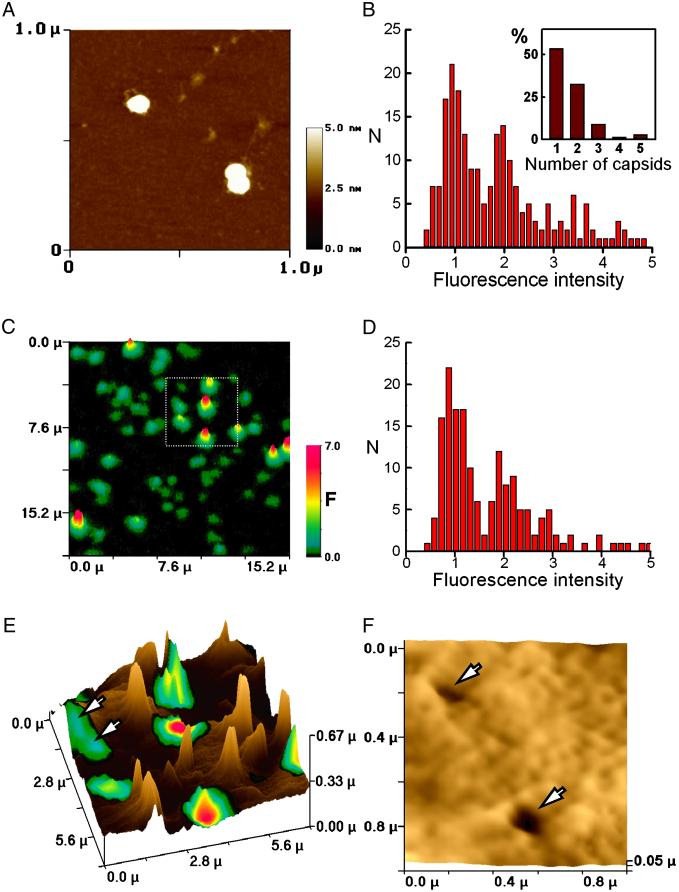
To determine whether SSCM would be sufficiently sensitive to track individual nanoparticles on the cell membrane, we first investigated whether single VLPs could be detected by fluorescence on a flat surface. Samples of fluorescent VLPs were analyzed in parallel by AFM (on mica) and conventional SCM (on glass). By AFM, the preparation consisted predominantly of single or pairs of VLPs, represented by 50-nm diameter spheres, loosely associated with strand-like structures presumed to be the plasmid DNA (examples are shown in Fig. 4A). Fluorescence intensities of the particles on glass, measured by confocal microscopy, were calculated per spot, and the distribution is shown in Fig. 4B. The fluorescence-intensity distribution fell into two major peaks, one having twice the intensity of the other, corresponding to the distribution of single and paired VLPs and occasional larger groupings, determined by AFM (Inset). Thus, the data are consistent with the smallest unit of fluorescence intensity corresponding to a single 50-nm VLP.
Fig 4.
Detection of single VLP entry. (A) AFM image of typical preparation of fluorescent Cy3-labeled VLPs on mica. (B) Fluorescence intensity distribution of VLPs on glass calculated from confocal images. (Inset) Distribution of VLPs in aggregates based on the AFM measurements of 33 aggregates of VLPs. (C) Surface confocal fluorescence image of VLPs on the cell membrane. (D) Fluorescence intensity distribution of VLPs on the cell membrane obtained from surface confocal images. (E) A 3D representation of a combination of surface confocal and SICM images of high resolution of the region marked with white box in C. Microvilli are visible as pale brown protrusions on the surface. (F) A zoomed topographic image acquired from the data gathered in the scan represented in E (a region containing two VLPs; arrows).
VLPs then were imaged on the surface of fixed cells, to prevent VLP movement, by using SSCM (Fig. 4C), and fluorescence intensity was measured for a random sample of ≈200 fluorescent spots. The majority of the fluorescent objects had similar fluorescence intensities (Fig. 4D, peak at 1 unit), a second smaller population had double the intensity, and there were a few larger aggregates, producing a histogram very similar to that obtained for VLPs on glass. Thus, we conclude that the smallest units of fluorescence we observe on the cell surface correspond to single VLPs.
To address the question of whether we could image structures of nanometer dimensions topographically, higher resolution scans (shown in 3D in Fig. 4E) of the area boxed in Fig. 4C were obtained. A zoomed topographic image of an area containing two single 50-nm VLPs (white arrows) is given in Fig. 4F. The topography of this area shows two hollows in the surface no larger than 100 nm, corresponding to each VLP. To estimate the size of the depressions, the images were compared with SSCM scans of 50- to 100-nm pores in a polymer (see Fig. 6, which is published as supporting information on the PNAS web site). The appearance of the holes recorded by SSCM for each structure was similar, confirming the size of the VLP-associated depressions to be within the 50–100 nm range.
Discussion
In this study, we developed SSCM, a combination of SICM and SCM, to permit analysis of single fluorescent nanoparticles associated with the cell surface. By using this technique, the interaction of single VLPs with the cell membrane was imaged. In two experiments (Figs. 2 and 4) the cells were fixed, allowing both reimaging of the same region at increasing resolutions and quantitative analysis of the fluorescence. However, imaging of live cells was also possible (Fig. 3).
The SSCM technique provides many advantages for studying the interaction of nanoparticles with the cell surface compared with other fluorescence microscopy methods. First, and most importantly, the position of fluorescently labeled particles can be related directly to the topography of the cell surface. Previously, this had only been possible in the context of markers such as plasma membrane proteins, cytoskeleton, or lipid dyes (16, 22, 23). Such studies can result in artifacts if the markers change during the uptake process and, as the surface of most cells is dynamic, determining whether particles have passed through the membrane or have moved with the membrane as it undulates is extremely difficult. Second, quantitative fluorescence measurements can be obtained because the laser beam is focused at the cell membrane throughout the whole scan; thus, photobleaching is equivalent at each point. Finally, less background from intracellular autofluorescence is collected compared with similar data obtained by conventional confocal microscopy, as only the surface is scanned. These factors have allowed accurate measurements of fluorescence to be made, as demonstrated by our ability to count single VLPs on the undulating cell surface as accurately as if they were spread on a flat surface (Fig. 4 A–D).
By using ion conductance to control the distance from the sample, we have been able to obtain much greater resolution than has previously been possible and have been able to image fluorescent VLPs on the surface of live cells. We found that cells could be scanned repeatedly at high resolution over 4 h (see Fig. 3). The SSCM imaging did not prevent internalization of the VLPs, and uptake occurred over a time scale reported for this virus family (24, 25).
To refine this technique for studying particle interactions with live cells, factors such as time resolution and photobleaching need to be considered. The scan rate used in these preliminary experiments was relatively slow (averaging 18 min), but it could be reduced to 30–50 sec by using a faster piezo (2–3 kHz resonant frequency) and by scanning a smaller area (7 × 7 μm, unpublished results). Thus, it would be feasible to use this technique for studying viral entry, where average internalization rates are 5–30 min (26–28). Although it is possible to perform continuous topographic imaging, this is not possible for fluorescence imaging as, in common with most fluorescence methods, especially in live-cell fluorescence microscopy, photobleaching and photodamage limit the observation time. In our case, minimizing the cell exposure to the laser by scanning at a faster rate should extend the observation time. Furthermore, to follow a particle on the cell surface, it is not necessary to obtain a fluorescence image with every topographic image, allowing the cell exposure to the laser light to be reduced further. Once a particle has passed through the membrane, its physical interaction with the cells can no longer be observed by topographic imaging. However, by lowering the confocal volume in relation to the membrane, it is possible to obtain fluorescence images under the cell surface to follow subsequent steps. In this way, we were able to image VLPs internalized after 4 h (see Fig. 5).
By using SSCM, we observed single or small aggregates of VLPs associated with the surface of the cell in an apparently random distribution. No tendency to associate with surface features such as microvilli was found (Fig. 4E). Larger aggregates were not associated with the surface with any significant affinity: no topological structures or large patches of fluorescence were detected on the surface of the cells at low resolution, but some large fluorescent objects were dislodged by the scanning pipette. The live cell experiment demonstrated the dynamic nature of the interaction of VLPs with the cell surface. We observed large changes in the distribution of particles in the first 30 min after addition and, subsequently, loss of surface fluorescence, consistent with the particles being internalized. Although in the fixed-cell experiments (Figs. 2 and 4) adsorption was carried out at 0°C, single particles were found to have sunk into the surface, creating depressions of a size consistent with caveolae or pinocytic vesicles (Fig. 4 E and F), routes described as being used by polyoma virus or VLPs to enter cells (25, 29). The fact that apparent invagination of VLPs has occurred under these conditions suggests that it is possible that some internalization may occur at the lower temperature. Similar internalization events have been observed both by electron (30) and confocal microscopy (24). Some small aggregates stood proud of the surface (data not shown), but it is currently not known whether these represent partially invaginated or superficially bound material. Because uptake of VLPs seems to be rapid or difficult to arrest with temperature, to assist real-time measurements, particles could be locally delivered to the cell surface with the same, or a second, pipette, and then the entry process followed in a small area of the cell at high resolution. These studies should provide real-time structural information only previously imaged statically by electron microscopy.
By using this new surface-orientated approach, a number of cell plasma membrane phenomena can be addressed. For instance, the initial stages in virus, bacteria, gene delivery vehicle, or drug uptake may now be observed in relation to the actual membrane surface, allowing characterization of lateral movements and penetration of the membrane. Elucidation of their roles in uptake could be carried out by relating their position with fluorescently tagged cellular functions, such as receptors, lipids, or cytoskeleton. Further, apart from characterizing the basic process, it would be possible to identify new drug targets, test the ability of drugs to modulate uptake, or characterize uptake topography in a semiquantitative manner.
The data in this paper demonstrate that, with SSCM, sensitive and quantitative imaging of fluorescence can be obtained in conjunction with topographic data gathering. This method will allow specific study of nanoparticles on the surface of cells and detailed characterization of their interactions with the plasma membrane. Thus, SSCM represents a valuable tool for understanding the earliest stages of particle uptake and will contribute to the development of new drugs and gene therapy vehicles in a range of clinical applications.
Supplementary Material
Acknowledgments
We are grateful to L. Ying for some initial fluorescence characterization of the VLPs and C. Williamson and A. Minson for critical reading of the manuscript. This work was funded by the Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council, and the Medical Research Council.
Abbreviations
SICM, scanning ion conductance microscopy
VLP, virus-like particle
SSCM, scanning surface confocal microscopy
SCM, scanning confocal microscopy
AFM, atomic force microscopy
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chimini G. & Chavrier, P. (2000) Nat. Cell Biol. 2, E191-E196. [DOI] [PubMed] [Google Scholar]
- 2.Godbey W. T., Wu, K. K. & Mikos, A. G. (1999) Proc. Natl. Acad. Sci. USA 96, 5177-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sieczkarski S. B. & Whittaker, G. R. (2002) J. Gen. Virol. 83, 1535-1545. [DOI] [PubMed] [Google Scholar]
- 4.Zabner J., Fasbender, A. J., Moninger, T., Poellinger, K. A. & Welsh, M. J. (1995) J. Biol. Chem. 270, 18997-19007. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett J. S. & Samulski, R. J. (1998) Nat. Med. 4, 635-637. [DOI] [PubMed] [Google Scholar]
- 6.Seisenberger G., Ried, M. U., Endress, T., Büning, H., Hallek, M. & Bräuchle, C. (2001) Science 294, 1929-1932. [DOI] [PubMed] [Google Scholar]
- 7.de Lange F., Cambi, A., Huijbens, R., de Bakker, B., Rensen, W., Garcia-Parajo, M., van Hulst, N. & Figdor, C. G. (2001) J. Cell Sci. 114, 4153-4160. [DOI] [PubMed] [Google Scholar]
- 8.Hansma P. K., Drake, B., Marti, O., Gould, S. A. & Prater, C. B. (1989) Science 243, 641-643. [DOI] [PubMed] [Google Scholar]
- 9.Shevchuk A. I., Gorelik, J., Harding, S. E., Lab, M. J., Klenerman, D. & Korchev, Y. E. (2001) Biophys. J. 81, 1759-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korchev Y. E., Negulyaev, Y. A., Edwards, C. R. W, Vodyanoy, I. & Lab, M. J. (2000) Nat. Cell Biol. 2, 616-619. [DOI] [PubMed] [Google Scholar]
- 11.Gu Y., Gorelik, J., Spohr, H. A., Shevchuk, A., Lab, M. J., Harding, S. E., Vodyanoy, I., Klenerman, D. & Korchev, Y. E. (2002) FASEB J. 16, 748-750. [DOI] [PubMed] [Google Scholar]
- 12.Krauzewicz N., Cox, C., Soeda, E., Clark, B., Rayner, S. & Griffin, B. E. (2000) Gene Ther. 7, 1094-1102. [DOI] [PubMed] [Google Scholar]
- 13.Forstová J., Krauzewicz, N., Sandig, V., Elliott, J., Palková, Z., Strauss, M. & Griffin, B. E. (1995) Hum. Gene Ther. 6, 297-306. [DOI] [PubMed] [Google Scholar]
- 14.Soeda E., Krauzewicz, N., Cox, C., Štokrová, J., Forstová, J. & Griffin, B. E. (1998) Gene Ther. 5, 1410-1419. [DOI] [PubMed] [Google Scholar]
- 15.Štokrová J., Palková, Z., Fischer, L., Richterová, Z., Korb, J., Griffin, B. E. & Forstová, J. (1999) FEBS Lett. 445, 119-125. [DOI] [PubMed] [Google Scholar]
- 16.Krauzewicz N., Štokrová, J., Jenkins, C., Elliott, M., Higgins, C. F. & Griffin, B. E. (2000) Gene Ther. 7, 2122-2131. [DOI] [PubMed] [Google Scholar]
- 17.Gluzman Y. (1981) Cell 23, 175-182. [DOI] [PubMed] [Google Scholar]
- 18.Gill D. R., Smyth, S. E., Goddard, C. A., Pringle, I. A., Higgins, C. F., Colledge, W. H. & Hyde, S. C. (2001) Gene Ther. 8, 1539-1546. [DOI] [PubMed] [Google Scholar]
- 19.Korchev Y. E., Milovanovic, M., Bashford, C. L., Bennett, D. C., Sviderskaya, E. V., Vodyanoy, I. & Lab, M. J. (1997) J. Microsc. 188, 17-23. [DOI] [PubMed] [Google Scholar]
- 20.Korchev Y. E., Bashford, C. L., Milovanovic, M., Vodyanoy, I. & Lab, M. J. (1997) Biophys. J. 73, 653-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pastre D., Iwamoto, H., Liu, J., Szabo, G. & Shao, Z. (2001) Ultramicroscopy 90, 13-19. [DOI] [PubMed] [Google Scholar]
- 22.Bkaily G., Jacques, D. & Pothier, P. (1999) Methods Enzymol. 307, 119-135. [DOI] [PubMed] [Google Scholar]
- 23.Vanderplasschen A. & Smith, G. L. (1999) Methods Enzymol. 307, 591-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelkmans L., Kartenbeck, J. & Helenius, A. (2001) Nat. Cell Biol. 3, 473-483. [DOI] [PubMed] [Google Scholar]
- 25.Richterová Z., Liebl, D., Horák, M., Palková, Z., Štokrová, J., Hozák, P., Korb, J. & Forstová, J. (2001) J. Virol. 75, 10880-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greber U. F., Willetts, M., Webster, P. & Helenius, A. (1993) Cell 75, 477-486. [DOI] [PubMed] [Google Scholar]
- 27.Gromeier M. & Wetz, K. (1990) J. Virol. 64, 3590-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sodeik B., Ebersold, M. W. & Helenius, A. (1997) J. Cell Biol. 136, 1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffith G. R. & Consigli, R. A. (1984) J. Virol. 50, 77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stang E., Kartenbeck, J. & Parton, R. G. (1997) Mol. Biol. Cell 8, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.