Abstract
The lymphatic microvasculature is uniquely adapted for the continuous removal of interstitial fluid and proteins and is an important entry point for leukocytes and tumor cells. Specialized functions of lymphatics suggest differences in the molecular composition of the lymphatic and blood vascular endothelium. However, the extent to which the two cell types differ is still unclear, and few molecules that are truly specific to lymphatic endothelial cells have been identified to date. We have isolated primary lymphatic and blood microvascular endothelial cells from human skin by immunoselection with the lymphatic marker LYVE-1 and demonstrate that the two cell lineages express distinct sets of vascular markers and respond differently to growth factors and extracellular matrix. Comparative microarray analysis of gene-expression profiles revealed a number of unique molecular properties that distinguish lymphatic and blood vascular endothelium. The molecular profile of lymphatic endothelium seems to reflect characteristic functional and structural features of the lymphatic capillaries. Classification of the differentially expressed genes into functional groups revealed particularly high levels of genes implicated in protein sorting and trafficking, indicating a more active role of lymphatic endothelium in uptake and transport of molecules than previously anticipated. The identification of a large number of genes selectively expressed by lymphatic endothelium should facilitate the discovery of hitherto unknown lymphatic vessel markers and provide a basis for the analysis of the molecular mechanisms accounting for the characteristic functions of lymphatic capillaries.
The lymphatic and blood vascular systems serve distinct yet complementary functions to maintain tissue homeostasis. The lymphatic system returns fluid and macromolecules from the tissues back to the blood circulation and, thus, plays a vital role in the regulation of fluid, protein, and pressure equilibrium in tissues (1, 2). The lymphatic vessels also play an important role in the immune response by directing antigen-presenting cells from tissues to the lymph nodes (3).
Lymphatic capillaries are responsible for the uptake of the components from the interstitium. Although endothelial cells (EC) of lymphatic capillaries have many properties in common with the endothelium of blood vessels, they also have distinct structural characteristics reflecting their specific functions (4–7). Lymphatic capillaries lack mural cells and are characterized by an incomplete or absent basement membrane. Lymphatic endothelium typically contains numerous invaginations and cytoplasmic vesicles as well as characteristic overlapping intercellular junctions. Whereas the junctions in blood vessels connect adjacent ECs over entire cell boundaries, the junctions in lymphatics are generally more sparse. Finally, one of the most striking characteristics of the lymphatic capillary is its integration within the interstitium; lymphatic ECs are connected to the extracellular matrix by fine strands of elastic fibers, i.e., anchoring filaments (5, 8–10).
The unique structural and functional characteristics of lymphatic capillaries suggest significant differences between the lymphatic and blood microvasculature at the molecular level. However, very few differentially expressed molecules have been identified to date, and most of these are either expressed at lower levels or absent from lymphatics (11). Recently, several positive markers of lymphatic vessels have been identified. These include VEGFR-3, the tyrosine kinase receptor for vascular endothelial growth factor (VEGF)-C and VEGF-D (12, 21, 48); podoplanin, a glomerular podocyte membrane mucoprotein (13, 14); Prox-1, the homeobox gene product that is involved in developmental regulation of the lymphatic system (15); and a hyaluronan receptor LYVE-1 (16, 17). Still, better discrimination between the two types of capillaries is crucial for addressing questions regarding the biology and pathology of the lymphatic system.
In the present study, we demonstrate the characteristic gene expression profile of human lymphatic microvascular ECs. The identification of distinct molecular characteristics of lymphatics should provide insight into the molecular basis of lymphatic vessel function and help identify hitherto unknown lymphatic vessel markers.
Materials and Methods
Isolation of Lymphatic and Blood Microvascular ECs.
Primary cultures consisting of a mixture of dermal cells were established from human neonatal foreskins according to a standard protocol (18). Cells were cultured on collagen-coated dishes in EC basal medium (Clonetics, San Diego) with 20% (vol/vol) FBS and supplements, as described (19). Magnetic beads (Dynal, Great Neck, NY) were used for immunomagnetic purification of cells, according to the manufacturers instructions. Rabbit IgG-conjugated Dynabeads were coated with the anti-human LYVE-1 antibody (16) and added to confluent primary cultures. Cells were incubated with beads for 15 min at 4°C, washed, and trypsinized as described (19). LECs attached to beads were separated with a magnetic particle concentrator and plated. Cells in the supernatant were repeatedly exposed to the magnet to ensure removal of any remaining LECs bound to beads, and BECs were subsequently purified by incubating cells in suspension with the CD31-conjugated beads. The second immunopurification step was performed at passage 2. LECs were first depleted of CD34+ cells and then purified by using CD31-coated beads. BECs were depleted of any remaining LYVE-1+ cells and purified by using CD34-coated beads. Beads were released from the cells with a DNase according to the manufacturer's instructions (Dynal).
Immunofluorescent Staining.
Cryosections of human foreskin tissue (6 or 50 μm thick) were stained as described (20) by using antibodies to human LYVE-1 (16) (1/300), PAL-E (1/50; Caltag, South San Francisco, CA), CD31 (1/50; DAKO), CD34 (1/50, PharMingen) or smooth muscle α-actin (1/100; DAKO) and corresponding secondary antibodies labeled with AlexaFluor488 or AlexaFluor594 (Molecular Probes). Cells were grown on coated tissue-culture slides (ICN) and fixed for 10 min in acetone before staining. Specimens were examined by using a Nikon E-600 microscope, and images were captured with a SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI).
Cloning, Expression, and Purification of Recombinant Human VEGF-C.
cDNA encoding mature VEGF-C (21, 22) was cloned from human umbilical vein ECs (HUVECs). Total RNA was isolated by using RNeasy kit (Qiagen, Chatsworth, CA); cDNA was generated by using SuperScript RT (Invitrogen); and human VEGF-C was amplified by PCR (nt 657–995; GenBank accession no. X94216). The purified PCR product was subcloned into EcoRI/BamHI-digested pSecTag2B expression vector containing C-terminal 6× His tag and c-myc epitope (Invitrogen). The sequence-verified pSecTag2B/VEGF-C ΔNΔC construct was transfected into 293T cells by using Fugene 6 Transfection Reagent (Roche Molecular Biochemicals), and stably transfected cells were selected in growth medium [DMEM with 10% (vol/vol) FBS] containing 100 μg/ml Zeocin. Conditioned medium was collected after a 48-h incubation and concentrated 10-fold by using Centriplus-10 filtration devices (Amicon). VEGF-C was purified by using Ni-NTA Agarose (Qiagen), according to the manufacturer's protocol. Concentration of the purified protein was determined with the Bio-Rad assay (Bio-Rad, Hercules, CA), and VEGF-C was analyzed by Western analysis by using antibodies to c-myc (Invitrogen) or VEGF-C (R & D Systems).
Collagen Gel Assay.
The ability of LECs and BECs to form capillary-like structures in vitro was assessed in a collagen gel “sandwich” assay, as described (23). ECs were seeded onto three-dimensional collagen gels at 2 × 104 cells per cm2 and allowed to attach. After 90–120 min, medium was aspirated, and the cells were overlaid with a second layer of collagen. Cultures were treated with 10 ng/ml rhFGF-2 (kindly provided by P. Sarmientos, Farmitalia Carlo Erba, Milan, Italy), 100 ng/ml rhVEGF (PeproTech, Rocky Hill, NJ) or 100 ng/ml rhVEGF-C. After 48 h, cells were analyzed by phase-contrast microscopy with a Nikon Diaphot TMD microscope. The total length of cell cords in each 1.0 × 1.4 mm field was measured. Results were expressed in μm as the mean total cell cord length ± SEM per field, from at least 15 measured fields per condition. Data were pooled from four experiments.
Northern and Western Analyses.
Northern and Western analyses were performed as described (24) by using confluent cells at passages 4–6. The LYVE-1 probe used for Northern analysis was a nt 90–1,092 human LYVE-1 cDNA fragment (GenBank accession no. AF118108) and was generated by RT-PCR from total RNA isolated from LECs by using an RNeasy kit (Qiagen). A single-tube RT-PCR was performed as indicated by the manufacturer (Stratagene). A human β-actin cDNA probe (CLONTECH) was used as a control for equal RNA loading. Western analyses were performed by using antibodies against human CD31, VEGFR-2, VEGFR-3 (all from Santa Cruz Biotechnology or R & D Systems), plakophilin 2 (Research Diagnostics, Flanders, NJ), or LYVE-1 (16).
Affymetrix Microarray Analysis.
Total RNA was isolated from LECs and BECs at passage 4, and DNA was removed with the DNase kit (Qiagen). Human GeneChips (HG-U95Av2) were purchased from Affymetrix (Santa Clara, CA). Array HG-U95Av2 is comprised of ≈12,000 sequences, most of which are previously characterized full-length genes; each gene is represented by ≈16 nonoverlapping oligonucleotide probes (25-mers). cDNA synthesis, hybridization, and signal intensity normalization were carried out at the Affymetrix facility of the Brigham and Women's Hospital (Boston). Data indicating presence or absence of gene expression (presence/absence call, determined by Affymetrix) were sorted, compared, and statistically analyzed by using SPOTFIRE software (Somerville, MA). Sequences whose presence call was designated A in both cell types or M in either cell type were filtered out. Genes were considered selectively expressed when present in one but absent in the other cell type; in this case, fold change designates the level of gene expression above an arbitrary threshold. Genes were considered differentially expressed when present in both cell types, with at least 2-fold difference in signal intensity; fold-change in this case indicates the relative difference in signal intensity for the gene between the two cell types. To search for the sequences encoding LYVE-1, Prox-1, and podoplanin (GenBank accession nos. AF118108, U44060, and U96449, respectively) among ESTs, because they were not present among known genes, we developed a custom algorithm that translated query sequences into all six reading frames and compared them with the six-reading-frame translation of the target-sequences (25). Genes were designated according to the annotations from Affymetrix and the Genecards databases (www.dkfz-heidelberg.de/GeneCards). Genecards were used to identify gene function, and the genes were classified into functional categories following the Gene Ontology Consortium guidelines (www.geneontology.org; ref. 26).
Results
Distinct Expression Patterns of Vascular Markers in Lymphatic and Blood Vasculature.
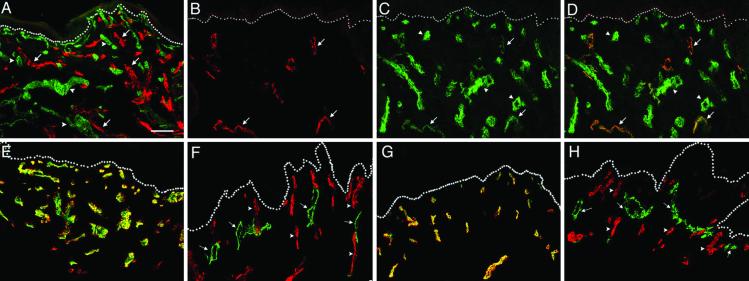
LYVE-1 is a hyaluronan receptor selectively expressed in lymphatic vessels in most tissues (16, 27), with the exception of spleen and liver, where it is expressed also by specialized sinusoids (16, 28). In human skin, LYVE-1 unequivocally distinguishes lymphatic from blood microvascular endothelium, as assessed by immunostaining with the anti-LYVE-1 antibody in combination with several antibodies against blood vascular endothelial antigens (Fig. 1). Expression of LYVE-1 was restricted to a subset of dermal vessels lacking expression of PAL-E, a specific blood vascular marker. In contrast, CD31 was detected in all vessels, although expression levels were lower in the LYVE-1+ endothelium (Fig. 1 B–D). Blood capillaries also stained strongly for CD34, which was completely absent from LYVE-1+ vessels (Fig. 1 E and F). Another important distinction between blood and lymphatic microvasculature is the lack of mural cells around lymphatics (4–6). Accordingly, expression of smooth muscle α-actin, a marker of mural cells, was restricted to blood capillaries and was not detected in association with LYVE-1+ vessels (Fig. 1 G and H). These results demonstrate differences in vascular marker expression between lymphatic and blood microvasculature and confirm the selectivity of LYVE-1 for lymphatics in the skin.
Fig. 1.
Selective expression of vascular markers in human skin vasculature. (A) Double immunofluorescent staining for LYVE-1 (red) and a PAL-E, a marker of blood vessels (green), in a 50-μm thick section of human foreskin. The stainings are mutually exclusive, indicating high specificity of LYVE-1 antibody for lymphatic vessels. Note high lymphatic vessel density. (B–D) Fluorescent staining for LYVE-1 (red), CD31 (green), or both together (merged), respectively, demonstrates that all LYVE-1+ vessels are also CD31+. (E and F) Double-staining for CD34 (red) and PAL-E (green) revealed identical expression pattern in blood vessels (E), whereas LYVE-1+ lymphatic vessels (green) do not express CD34 (F). (G and H) Smooth muscle α-actin (red) was colocalized with PAL-E (green) in blood vessels (G) but was absent from LYVE-1+ vessels (H). Arrows point to the lymphatics; arrowheads point to blood vessels; dots indicate dermal-epidermal junction. (Bar = 100 μm.)
Isolation of Lymphatic and Blood Microvascular ECs.
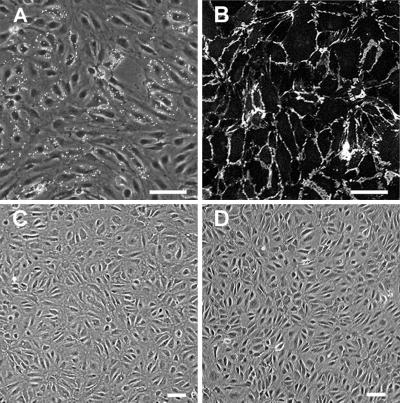
Pure populations of microvascular LECs and BECs were isolated from human neonatal foreskins by immunomagnetic purification using a combination of vascular markers LYVE-1, CD31, and CD34. First, primary cultures consisting of a mixture of dermal microvascular ECs, fibroblasts and some epidermal keratinocytes were established according to a standard protocol (18). Next, LECs were purified by using magnetic beads coupled to a LYVE-1 antibody. Within primary cultures, LYVE-1+ cells formed clusters that were segregated from the LYVE-1− endothelium (Fig. 2A). After the second purification step with the CD31 antibody, any remaining BECs were removed with the CD34 antibody. Immunofluorescent staining of LECs in culture demonstrated that all cells expressed CD31 at cell junctions (Fig. 2B). Corresponding BECs purified from the same pool of ECs were defined as LYVE-1−/CD31+/CD34+ cells. LECs and BECs exhibited similar morphology as monolayer cultures under standard growth conditions (Fig. 2 C and D) and were propagated for at least eight passages without their characteristics being altered. The procedure results in high yields; ≈109 LECs and BECs can be obtained from a single foreskin by passage five.
Fig. 2.
Purification of lymphatic and blood microvascular endothelium. (A) Primary culture consisting mainly of ECs, a few fibroblasts, and keratinocytes, with LYVE-1-coated magnetic beads attached to the subpopulation of ECs. (B) After the second purification step, all cells are stained with the CD31 antibody at cell junctions, indicating a pure endothelial population. (C and D) Confluent layers of lymphatic (C) and blood vascular (D) ECs at passage 7. (Bar = 25 μm.)
Selective Effects of VEGF-C on Cultured Lymphatic Endothelium.
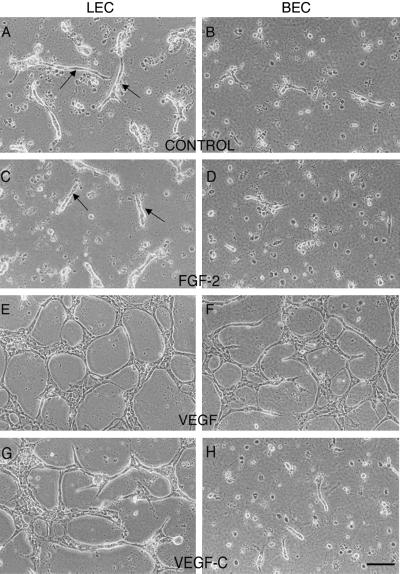
The ability of collagen type I and growth factors to induce capillary-like morphogenesis in LECs and BECs was assessed in a collagen gel “sandwich” assay (23). Within 48 h of exposure of the apical cell surface to collagen type I, the majority of BECs and LECs had undergone cell death. Interestingly, this effect was more prominent in BECs than in LECs (Fig. 3 A and B). LECs showed not only increased survival rates but also demonstrated the ability to form tubes without exogenously added growth factors. Addition of FGF-2 had no effect on survival of either cell type (Fig. 3 C and D). VEGF, however, was a potent survival factor for both blood and lymphatic vascular ECs and promoted the formation of an extensive network of tubes (Fig. 3 E and F). In contrast, VEGF-C selectively induced survival and tube formation of LECs (Fig. 3 G and H and Fig. 4). The effect of VEGF-C on LECs was comparable to that observed with VEGF treatment. The differential responsiveness of the two cell lineages to the extracellular matrix and to VEGF-C indicates that LECs and BECs retain their distinct phenotypes in culture.
Fig. 3.
Selective effects of VEGF-C on survival and tube formation by LECs in a collagen gel. Cells either received no treatment (A and B) or were exposed to exogenous FGF-2 (C and D), VEGF (E and F), or VEGF-C (G and H). (Bar = 25 μm.)
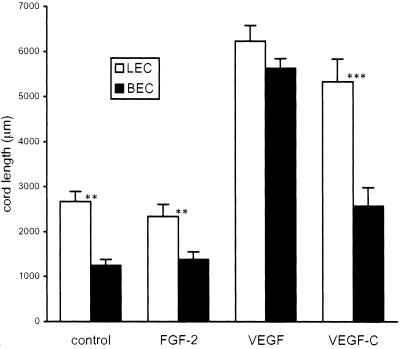
Fig. 4.
Quantitative assessment of LEC and BEC network formation in the collagen gel. Total length of cell cords formed after 48 h was measured. Data are expressed as total cord length (in μm ± SEM) per field from at least 15 fields. Data are pooled from four experiments. The unpaired Student's t test was used for statistical analyses. LEC control vs. FGF, P = 0.5; vs. VEGF, P < 0.001; vs. VEGF-C, P < 0.001. BEC control vs. FGF, P = 0.5; vs. VEGF, P < 0.001; vs. VEGF-C, P < 0.05.
Differential Expression of Specific Markers in Cultured Lymphatic and Blood Vessel ECs.
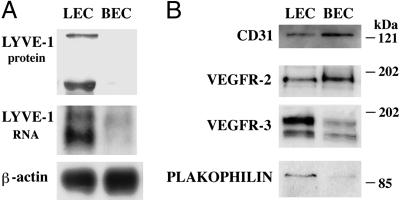
LECs and BECs were analyzed for expression of several lineage-specific genes by Western and/or Northern analysis (Fig. 5). LYVE-1 was selectively expressed in LECs, both at RNA and protein levels. Both cell lineages maintained expression of CD31 in culture, as determined by Western analysis (Fig. 5) and immunostaining (Fig. 2). Expression of CD31 was less pronounced in cultured LECs, recapitulating the expression pattern observed in vivo. Likewise, lower amounts of VEGFR-2 protein were detected in LECs than in BECs, whereas VEGFR-3 protein was predominantly expressed by LECs. Plakophilin 2, desmosomal protein found in nonclassical adherens junctions characteristic of lymphatic vessels (29, 30), was detected mainly in LECs. These results demonstrate that LECs and BECs stably maintain distinct patterns of gene expression in culture. Microarray analysis confirmed expression of CD31 and plakophilin 2 by LECs (Tables 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org). However, in contrast to the protein data, expression levels of VEGFR-2 and VEGFR-3 RNA were comparable between the two cell types, suggesting that differences in the amounts of the respective proteins result from posttranscriptional regulation. Sequences for lymphatic markers LYVE-1, podoplanin or Prox-1 were not present on the GeneChip among the known genes or ESTs, as determined by the custom algorithm (see Materials and Methods).
Fig. 5.
Differential expression of vascular markers by cultured LECs and BECs. (A) Western and Northern analyses of LYVE-1 expression. Two major bands of LYVE-1 protein (≈70 and 200 kDa) and RNA (2.0 and 2.6 kb) were expressed in LECs. Hybridization with a β-actin cDNA probe was performed as a loading control for the Northern analysis. For Western analyses, equal amounts of proteins were loaded. (B) Western analyses of CD31, VEGFR-2, VEGFR-3, and plakophilin expression. Positions of molecular mass markers are shown in kDa.
Molecular Profile of Lymphatic Vascular ECs.
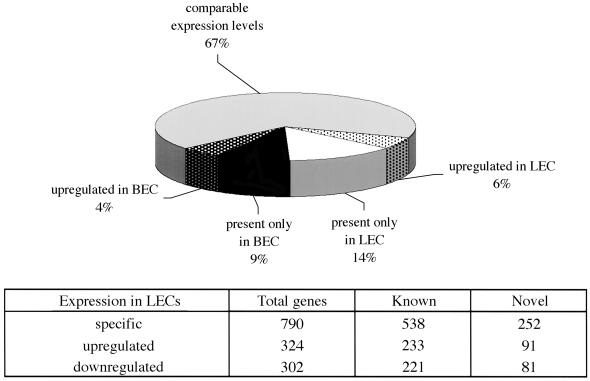
To further characterize differences between lymphatic and blood endothelium we carried out microarray analysis. Of the 12,626 genes represented on a GeneChip, 5,789 were expressed in one or other cell type (45%). Among the expressed sequences, 33% were differentially expressed. Of all genes expressed, 20% were either selectively present or significantly up-regulated in LECs, indicating notable quantitative as well as qualitative differences between the two cell lineages (Fig. 6 and Tables 1–3, which are published as supporting information on the PNAS web site).
Fig. 6.
Quantitative assessment of differential gene expression.
To investigate whether certain classes of genes were preferentially represented by the lymphatic endothelium, the differentially expressed genes were classified based on their function following the Gene Ontology consortium guidelines (26). The categories that were most highly represented in LECs comprised molecules involved in protein transport, secretion and metabolism (Table 4, which is published as supporting information on the PNAS web site). Synaptogyrin 3, the gene expressed at the highest level within this group, is a member of the family of proteins abundantly present in synaptic vesicles, with presumable function in exocytosis (31, 32). LECs selectively expressed high levels of various genes encoding proteins of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family such as syntaxins 1a, 5, 11 and 16, YKT6 protein that plays a central role in vesicular trafficking (32–34). Syntaxins are transmembrane proteins that regulate fusion of transport vesicles with target membranes. YKT6 and syntaxin 5, present on the vesicle and the target membrane, specifically interact to form complexes that catalyze lipid bilayer fusion (35). Besides SNAREs, LECs expressed transcripts encoding distinct members of other protein families that control the specificity of vesicle fusion, such as rab GTPases, sec-related proteins, vesicle-associated membrane proteins, and ATPases, as well as various genes that regulate vesicle docking (32, 34). Elevated levels of transcripts for certain enzymes whose activity is required for protein translocation within the cell were also typical for LECs.
Genes belonging to a number of other functional categories were differentially expressed. For example, LECs characteristically expressed several cell adhesion molecules that constitute adherens junctions, i.e., cadherin-13, plakophilin 2, and zona occludens 2 (ZO-2; refs. 30, 36, and 37), but did not express VCAM (vascular cell adhesion molecule) and N-cadherin, that were found selectively in BECs. As expected, BECs were characterized by prominent expression of several genes encoding the components of basement membrane, such as α1 type XV collagen, α3 laminin and nidogen. Several genes implicated in cell differentiation such as endothelial differentiation protein edg-1, ets-1, Id1, and Id2, were expressed in both cell types at the comparable levels. However, a transmembrane receptor implicated in endothelial differentiation, Notch4, was found expressed only by BECs. The differences also were found in the expression of chemokines and growth factors. Most prominently, BECs selectively expressed SDF-1 (stromal cell derived factor-1), whereas RANTES, a chemokine for T cells and monocytes, was primarily expressed by LECs. Several growth factors implicated in angiogenesis, such as bFGF, VEGF-B, and TGFβ, were expressed in both lineages. Interestingly, LECs expressed high levels of VEGF and Ang2, whereas PlGF was predominantly expressed by BECs. Differential expression of selected genes has been confirmed by Northern analysis (Fig. 7, which is published as supporting information on the PNAS web site). Taken together, these results reveal significant differences in the molecular make-up of the lymphatic and blood microvascular endothelium and provide insights into the molecular basis of the biological differences between the two cell lineages.
Discussion
The lymphatic microvasculature is uniquely adapted for the continuous removal of interstitial fluid and proteins and is an important point of entry for leukocytes and tumor cells (1–3). The exact mechanisms by which lymphatic capillaries accomplish these tasks, however, remain to be defined. Specialized functions of lymphatics suggest differences in the molecular composition of lymphatic and blood vascular endothelium, the understanding of which should provide valuable insight into the molecular basis of lymphatic function. We have compared the gene expression profiles of isolated primary lymphatic and blood microvascular ECs by using commercially available microarrays, and demonstrate unique differences between the two cell types at the molecular level.
LECs and BECs were isolated from human skin by immunomagnetic separation using a combination of positive and negative markers. The purification strategy was devised based on the specific expression of LYVE-1 and CD34 in the lymphatic and blood vasculature of the skin, respectively. LECs were identified as LYVE-1+/CD31+/CD34− cells, whereas BECs were defined as LYVE-1−/CD31+/CD34+ cells. Both cell lineages retained this characteristic expression pattern of markers in culture, as demonstrated by Northern and Western analysis. Moreover, the lymphatic vessel growth factor VEGF-C selectively induced tube formation of LECs but not BECs in an in vitro angiogenesis assay. In contrast, VEGF promoted survival and tube formation of both cell types, suggesting that it might play a role in the regulation of lymphatic vessel survival and/or formation in vivo. In agreement with the results of these functional studies, both cell lineages expressed VEGFR-2, whereas VEGFR-3 was predominantly expressed in LECs. Interestingly, in the absence of exogenous growth factors, LECs incorporated into collagen type I scaffolds exhibited a significantly higher survival rate than BECs. This difference in response to collagen type I may reflect differences in the type of the extracellular matrix that each vessel type is exposed to in its natural environment. Blood vascular endothelium is in immediate contact with components of the basal lamina, whereas in lymphatic capillaries basal lamina is largely absent and LECs form an intimate association with adjacent interstitial tissue (5, 9). In fact, one of the features that discriminate lymphatic capillaries from blood capillaries at the ultrastructural level are direct connections of LECs to the interstitial collagens by anchoring filaments (5, 9). Taken together, distinct expression patterns of vascular markers by cultured LECs and BECs and their differential responsiveness to the extracellular matrix and VEGF-C indicate that LECs and BECs represent distinct cell lineages which retain their differentiated phenotypes in culture.
Recently, the feasibility of isolating LECs by using two different lymphatic markers, podoplanin and VEGFR-3, has been reported (38, 39). In agreement with our results, these studies demonstrated that LECs maintained expression of their characteristic markers in culture. However, LECs isolated by the three different methods showed slightly different expression of some of the vascular markers examined, which may be because of the different isolation strategies selecting for distinct subpopulations of lymphatic ECs. Alternatively, the reason may be a different source of tissues used, i.e., adult vs. neonatal skin. LECs isolated from commercially available mixed cultures of ECs by employing VEGFR-3 antibodies (38) may be partly contaminated with BECs, because VEGFR-3 can also be expressed by blood vascular endothelium (40).
Comparative analysis of gene expression profiles revealed significant differences in the molecular signatures of LECs and BECs. The molecular profile of LECs indeed appears to reflect the characteristic functional and structural features of the lymphatic microvascular endothelium. Classification of the differentially expressed genes into functional groups revealed that LECs express remarkably high levels of genes implicated in protein metabolism, sorting, and trafficking. Particularly highly represented were genes encoding proteins that control specificity of vesicle targeting and fusion, such as proteins of the SNARE family, rab GTPases, AAA ATPases, and sec-related proteins (32, 34), indicating pronounced vesicular transport in LECs. Of interest, one of the typical features of lymphatic endothelial ultrastructure is the presence of membrane invaginations and cytoplasmic vesicles (10, 41, 42), whose functional significance has not been established. Intercellular clefts are considered to be a major passageway for fluid and proteins into the lymphatics, the entry of which is driven by pressure gradients across the endothelial wall (43). Some early studies, however, demonstrated the presence of interstitially injected molecular tracers within intracellular vesicles of lymphatic ECs (10, 41, 42). In agreement with these findings, our results strongly suggest that in addition to intercellular transport, transendothelial pathways also may be used as a mechanism for the entry of molecules into lymphatics. It is therefore tempting to speculate that lymphatics may have the capacity to remove selectively molecules from the interstitium and, therefore, actively control the composition of lymph and interstitial fluid.
Expression of several genes encoding proteins implicated in transport of solutes further suggests an active role of LECs in regulating interstitial homeostasis. The potassium/chloride cotransporter KCC1 for example, plays an important role in the control of extracellular fluid volume as well as in the control of membrane potential (44). Lymphatic endothelium is characterized by a high density of anionic sites on cell membranes, particularly along intercellular junctions, which have been suggested to facilitate movement of small solutes and molecules into the lymphatic lumen (45). Hence, the expression of specific ion transporters in lymphatic endothelium may directly or indirectly regulate transport of solutes and fluid into the vessel.
We have identified many other genes whose expression appears to be of relevance to the typical structure of lymphatic capillaries. Ang 2, for example, is implicated in destabilizing adhesion of mural cells to the endothelium of blood capillary (46). In lymphatic capillaries, constitutive expression of Ang2 by ECs may account for the characteristic lack of pericytes in these vessels. Lymphatic capillaries are further distinguished by the specific organization of its intercellular junctions, including the typical presence of a special type of adherens junctions (29). We have identified several genes encoding proteins that constitute adherens junctions, such as plakophillin 2, H-cadherin, and zona occludens 2 (ZO-2; refs. 30, 36, and 37). Finally, the lymphatic endothelial cytoskeleton is directly connected to the extracellular matrix by anchoring filaments composed mainly of elastin fiber microfibrils (9). Interestingly, an elastin microfibril protein (EMILIN) that is normally expressed in elastin-rich tissues (47) has been found to be selectively and abundantly expressed in LECs.
In conclusion, we reveal a number of unique properties by which lymphatic and blood microvascular endothelium can be distinguished. With the exception of the few newly identified positive lymphatic markers, most of the known vascular markers are present at lower levels or absent in lymphatics (11). Furthermore, none of the positive lymphatic markers known to date are exclusive for the lymphatic vasculature in all types of tissues. Therefore, the necessity for the discovery of markers that would more reliably discriminate the two types of the vasculature in physiological as well as in pathological conditions remains. Identification of a large number of genes selectively expressed by LECs should facilitate the discovery of such markers. Finally, lymphatics have traditionally been assigned a passive role in the uptake of fluid, proteins, and cells from the interstitium. Our results indicate a more active role of lymphatic endothelium than previously anticipated and should provide a basis for the future analysis of the molecular mechanisms that account for the characteristic functions of lymphatic capillaries.
Supplementary Material
Acknowledgments
We thank Dr. Tearina Chu at the Mount Sinai Microarray Facility (New York) for help with data analysis, Drs. Michael Detmar and Avrom Caplan for helpful discussions, Melanie Cassella for critical reading of the manuscript, and Lauren Janes and Mireille Quayzin for technical assistance. This work was supported in part by U.S. Department of Army Grant DAMD17-01-1-0552, Speaker's Fund for Biomedical Research, the Peter Sharp Foundation (to M.S.), Swiss National Science Foundation Grant 3100–064037.00 (to M.S.P.), and Association for International Cancer Research Project Grant 00-311 (to D.G.J.).
Abbreviations
EC, endothelial cell
LEC, lymphatic EC
BEC, blood EC
FGF, fibroblast growth factor
VEGF, vascular endothelial growth factor
VEGFR, VEGF receptor
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rusznyak I., Foldi, M. & Szabo, G., (1967) Lymphatics and Lymph Circulation (Pergamon, Oxford).
- 2.Swartz A. & Skobe, M. (2001) Microsc. Res. Tech. 55, 92-99. [DOI] [PubMed] [Google Scholar]
- 3.Yoffey J. M. & Courtice, F. C., (1970) Lymphatics, Lymph and the Lymphomyeloid Complex (Academic, London).
- 4.Casley-Smith J. R. & Florey, H. W. (1961) Q. J. Exp. Physiol. 46, 101-105. [DOI] [PubMed] [Google Scholar]
- 5.Leak L. V. (1970) Microvasc. Res. 2, 361-391. [DOI] [PubMed] [Google Scholar]
- 6.Daroczy J., (1988) The Dermal Lymphatic Capillaries (Springer, Berlin).
- 7.Skobe M. & Detmar, M. (2000) J. Invest. Dermatol. Symp. Proc. 5, 14-19. [DOI] [PubMed] [Google Scholar]
- 8.Pullinger B. D. & Florey, H. W. (1935) Br. J. Exp. Pathol. 16, 49. [Google Scholar]
- 9.Gerli R., Ibba, L. & Fruschelli, C. (1990) Anat. Embryol. 181, 281-286. [DOI] [PubMed] [Google Scholar]
- 10.Leak L. V. (1976) Fed. Proc. 35, 1863-1871. [PubMed] [Google Scholar]
- 11.Sleeman J. P., Krishnan, J., Kirkin, V. & Baumann, P. (2001) Microsc. Res. Tech. 55, 61-69. [DOI] [PubMed] [Google Scholar]
- 12.Kaipainen A., Korhonen, J., Mustonen, T., van Hinsbergh, V. W., Fang, G. H., Dumont, D., Breitman, M. & Alitalo, K. (1995) Proc. Natl. Acad. Sci. USA 92, 3566-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breiteneder-Geleff S., Soleiman, A., Kowalski, H., Horvat, R., Amann, G., Kriehuber, E., Diem, K., Weninger, W., Tschachler, E., Alitalo, K., et al. (1999) Am. J. Pathol. 154, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weninger W., Partanen, T. A., Breiteneder-Geleff, S., Mayer, C., Kowalski, H., Mildner, M., Pammer, J., Sturzl, M., Kerjaschki, D., Alitalo, K., et al. (1999) Lab. Invest. 79, 243-251. [PubMed] [Google Scholar]
- 15.Wigle J. T. & Oliver, G. (1999) Cell 98, 769-778. [DOI] [PubMed] [Google Scholar]
- 16.Banerji S., Ni, J., Wang, S. X., Clasper, S., Su, J., Tammi, R., Jones, M. & Jackson, D. G. (1999) J. Cell Biol. 144, 789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson D. G., Prevo, R., Clasper, S. & Banerji, S. (2001) Trends Immunol. 22, 317-321. [DOI] [PubMed] [Google Scholar]
- 18.Davison P. M., Bensch, K. & Karasek, M. A. (1980) J. Invest. Dermatol. 75, 316-321. [DOI] [PubMed] [Google Scholar]
- 19.Richard L., Velasco, P. & Detmar, M. (1998) Exp. Cell Res. 240, 1-6. [DOI] [PubMed] [Google Scholar]
- 20.Skobe M., Hawighorst, T., Jackson, D. G., Prevo, R., Janes, L., Velasco, P., Riccardi, L., Alitalo, K., Claffey, K. & Detmar, M. (2001) Nat. Med. 7, 192-198. [DOI] [PubMed] [Google Scholar]
- 21.Joukov V., Pajusola, K., Kaipainen, A., Chilov, D., Lahtinen, I., Kukk, E., Saksela, O., Kalkkinen, N. & Alitalo, K. (1996) EMBO J. 15, 290-298. [PMC free article] [PubMed] [Google Scholar]
- 22.Joukov V., Kumar, V., Sorsa, T., Arighi, E., Weich, H., Saksela, O. & Alitalo, K. (1998) J. Biol. Chem. 273, 6599-6602. [DOI] [PubMed] [Google Scholar]
- 23.Montesano R., Orci, L. & Vassalli, P. (1983) J. Cell Biol. 97, 1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skobe M., Hamberg, L. M., Hawighorst, T., Schirner, M., Wolf, G. L., Alitalo, K. & Detmar, M. (2001) Am. J. Pathol. 159, 893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevo R., Banerji, S., Ferguson, D. J., Clasper, S. & Jackson, D. G. (2001) J. Biol. Chem. 276, 19420-19430. [DOI] [PubMed] [Google Scholar]
- 28.Carreira C. M., Nasser, S. M., di Tomaso, E., Padera, T. P., Boucher, Y., Tomarev, S. I. & Jain, R. K. (2001) Cancer Res. 61, 8079-8084. [PubMed] [Google Scholar]
- 29.Schmelz M., Moll, R., Kuhn, C. & Franke, W. W. (1994) Differentiation 57, 97-117. [DOI] [PubMed] [Google Scholar]
- 30.Mertens C., Kuhn, C. & Franke, W. W. (1996) J. Cell Biol. 135, 1009-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugita S., Janz, R. & Sudhof, T. C. (1999) J. Biol. Chem. 274, 18893-18901. [DOI] [PubMed] [Google Scholar]
- 32.Hay J. C. (2001) Exp. Cell Res. 271, 10-21. [DOI] [PubMed] [Google Scholar]
- 33.Nelson W. J. & Yeaman, C. (2001) Trends Cell Biol. 11, 483-486. [DOI] [PubMed] [Google Scholar]
- 34.Mellman I. & Warren, G. (2000) Cell 100, 99-112. [DOI] [PubMed] [Google Scholar]
- 35.Zhang T. & Hong, W. (2001) J. Biol. Chem. 276, 27480-27487. [DOI] [PubMed] [Google Scholar]
- 36.Lee S. W. (1996) Nat. Med. 2, 776-782. [DOI] [PubMed] [Google Scholar]
- 37.Avila-Flores A., Rendon-Huerta, E., Moreno, J., Islas, S., Betanzos, A., Robles-Flores, M. & Gonzalez-Mariscal, L. (2001) Biochem. J. 360, 295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makinen T., Veikkola, T., Mustjoki, S., Karpanen, T., Catimel, B., Nice, E. C., Wise, L., Mercer, A., Kowalski, H., Kerjaschki, D., et al. (2001) EMBO J. 20, 4762-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kriehuber E., Breiteneder-Geleff, S., Groeger, M., Soleiman, A., Schoppmann, S. F., Stingl, G., Kerjaschki, D. & Maurer, D. (2001) J. Exp. Med. 194, 797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Partanen T. A., Alitalo, K. & Miettinen, M. (1999) Cancer 86, 2406-2412. [PubMed] [Google Scholar]
- 41.Dobbins W. O. & Rollins, E. L. (1970) J. Ultrastruc. Res. 33, 29-59. [DOI] [PubMed] [Google Scholar]
- 42.Leak L. V. (1972) J. Ultrastruc. Res. 39, 24-42. [DOI] [PubMed] [Google Scholar]
- 43.Schmid-Schönbein G. W. (1990) Physiol. Rev. 70, 987-1028. [DOI] [PubMed] [Google Scholar]
- 44.Delpire E. & Mount, D. B. (2002) Annu. Rev. Physiol. 64, 803-843. [DOI] [PubMed] [Google Scholar]
- 45.Leak L. V. (1986) Microvasc. Res. 31, 18-30. [DOI] [PubMed] [Google Scholar]
- 46.Maisonpierre P. C., Suri, C., Jones, P. F., Bartunkova, S., Wiegand, S. J., Radziejewski, C., Compton, D., McClain, J., Aldrich, T. H., Papadopoulos, N., et al. (1997) Science 277, 55-60. [DOI] [PubMed] [Google Scholar]
- 47.Doliana R., Mongiat, M., Bucciotti, F., Giacomello, E., Deutzmann, R., Volpin, D., Bressan, G. M. & Colombatti, A. (1999) J. Biol. Chem. 274, 16773-16781. [DOI] [PubMed] [Google Scholar]
- 48.Achen M. G., Jeltsch, M., Kukk, E., Makinen, T., Vitali, A., Wilks, A. F., Alitalo, K. & Stacker, S. A. (1998) Proc. Natl. Acad. Sci. USA 95, 548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.