Abstract
Steroid receptor RNA activator (SRA) is an RNA transcript that functions as a eukaryotic transcriptional coactivator for steroid hormone receptors. We report here the isolation and functional characterization of distinct RNA substructures within the SRA molecule that constitute its coactivation function. We used comparative sequence analysis and free energy calculations to systematically study SRA RNA subdomains for identification of structured regions and base pairings, and we used site-directed mutagenesis to assess their functional consequences. Together with genetic deletion analysis, this approach identified six RNA motifs in SRA important for coactivation. Because all nucleotide changes in the mutants that disrupted SRA function were silent mutations presumed not to alter deduced encoded amino acid sequence, our analysis provides strong evidence that SRA-mediated coactivation is executed by distinct RNA motifs and not by an encoded protein.
Nuclear receptors play critical roles in eukaryotic development, metabolism, reproduction, and disease through regulation of gene expression (1, 2). Recent advances in transcription have suggested that nuclear receptors form large, multicomponent complexes with coactivators and corepressors at the enhancer and promoter regions of target genes (3, 4). These complexes include factors exerting different enzymatic functions such as chromatin remodeling activities, and a variety of combinations of coregulators are believed to be essential for regulated gene expression.
We have previously reported the isolation and functional characterization of a novel transcriptional coactivator termed SRA (steroid receptor RNA activator; ref. 5). SRA acts as a catalytic RNA transcript by regulating eukaryotic gene expression mediated by the steroid receptors (SRs). When overexpressed in mammalian cells, recombinant SRA showed potent coactivation activity with the receptors for androgens, estrogens, glucocorticoids, and progestins (PR). We showed that several isoforms of SRA exist, and we grouped them into three splice classes based on their sequences outside a common core region. The SRA sequences of all of our cDNA clones contain an ORF but are devoid of an initiation ATG. Although the existence of a translation product of SRA cannot be categorically excluded, we have provided evidence to indicate that SRA functions as an RNA transcript (5).
Because the function of an RNA can be best understood in terms of its secondary or tertiary structure, we wished to further define the coactivation function we previously observed in SRA by using low-resolution RNA structure modeling. RNA secondary structure is a composite of hydrogen bonds between bases allowing certain noncanonical pairings, forming structures with double-helical motifs, bulges, bubbles, and loops that then spatially arrange to assemble specific intra- and intermolecular interactions. By using comparative sequence analysis and low-resolution RNA modeling, we set out to predict functional substructures of SRA and to obtain a framework within which multiple aspects of SRA could be viewed simultaneously. We reasoned that site-directed mutagenesis targeted exclusively to the wobble positions of the presumptive SRA amino acid sequence would not only identify functional substructures and possible docking motifs for interacting proteins, but also provide additional strong evidence that it is the RNA that mediates SR coactivation.
We report here the identification and partial characterization of six distinct secondary RNA structures within SRA that impaired transcription activation upon silent mutagenesis. We also show that multiple RNA substructures work together to contribute to the overall coactivation by SRA. In addition, because of the close genomic proximity of two other genes and ambiguous computer-predicted SRA genes, we present expression analyses strongly supporting an autonomous SRA gene locus.
Materials and Methods
Plasmids.
The human cytomegalovirus-driven mammalian expression vectors pSCT-1 (−), pSCT-SRA, AbΔBbs, Mfe, and the hormone-responsive luciferase reporter plasmid MMTV-Luc have been described (5). SRA deletion mutants were constructed by using generic restriction sites of the SRA cDNA and subsequent ligation of the fragments into fittingly restricted pSCT-1 or pSCT-SRA vectors. Construct names indicate the endonuclease(s) used for the mutation, whereby BaPs is BamHI–PstI, BaPa is BamHI–BanII, NB is NaeI–BamHI, and MS is MfeI–SgrAI.
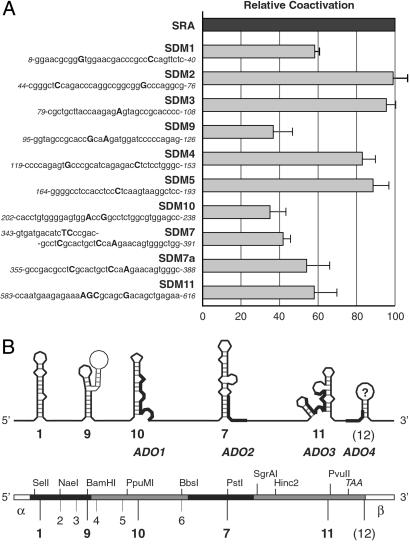
Site-directed SRA mutants (SDMs) were generated by using synthetic oligonucleotides (sequences are shown in Fig. 4A) and either the GeneEditor in vitro Site-Directed Mutagenesis System (Promega) or the QuikChange Site-Directed Mutagenesis Kit (Stratagene) following the manufacturers' protocols. Oligonucleotide 5′-CAGACTCACCGaACtCCAagcCCTATGCCTTGGAGACC-3′ was used to generate SDM12. Mutations were verified by dideoxy-sequencing. To authenticate remaining SRA and vector sequence, cDNA fragments enclosing the desired mutations were excised by endonuclease digestion (SDM1–3, AvrII–BamHI; SDM4–10, BamHI–SgrA1; SDM11, Hinc2–PvuII; SDM12; PvuII–NsiI) and subcloned into the parental pSCT-SRA vector.
Fig 4.
Composite SRA coactivation function. (A) Multiple secondary RNA structure motifs are involved in SRA coactivation. Transactivation properties of different SRA variants are shown relative to wild-type SRA. Site-directed SRA mutants (SDM1–11) and corresponding cDNA sequences near the sites of mutations are shown on the left; nucleotide changes are shown in uppercase, bold letters; numbers indicate nucleotide positions of the sequence relative to the SRA core region. SDM7a differs from SDM7 by omitting mutations at the serine codon. Transient transfection experiments were as described in Fig. 3. (B) Deduced map of functional substructures of SRA. At the top is a schematic representation of SRA and the identified secondary substructures, their assigned motif numbers (underneath), and location of the ADOs (and thickened lines in the illustrations of the substructures). At the bottom is a schematic illustration of the SRA cDNA showing selected restriction sites, the exon structure (different shadings), and position of inferred substructures. α and β indicate different SRA isoform-specific sequences outside the SRA core region.
Genomic Sequence and Expression Analysis.
We frequently used NIX (www.hgmp.mrc.ac.uk/NIX/) that runs the following programs on submitted genomic DNA sequences: grail, fex, hexon, mzef, genemark, genefinder, fgene, blast (against many databases) polyah, repeatmasker and trnascan, and algorithms freely available at the National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov/Database/index.html). NCI60 array data were retrieved through the University of California, Santa Cruz, Genomic Bioinformatic site (http://genome.ucsc.edu/) and the Stanford NCI60 Microarray homepage (http://genome-www.stanford.edu/nci60/index.shtml). For Northern analysis, total RNA was extracted and purified from T-47D and LNCaP-derived C4-2 cells by using TRIzol Reagent (Life Technologies, Carlsbad, CA). Total RNA from each cell line (30, 15, and 5 μg) was loaded on 6.6% formaldehyde/1.4% agarose gel, electro-separated in Mops buffer, and transferred by capillary force and 20× SSC to Zetaprobe GT membrane (Bio-Rad). The transferred RNA were immobilized by UV-crosslinking at 125mJ and first probed with a [32P]dCTP-labeled EIF4EBP3 cDNA probe under high stringency conditions over night at 65°C in hybridization buffer (0.5 M NaHPO4, pH7.2/7% SDS/1 mM EDTA/100 μg/ml salmon sperm DNA). EIF4EBP3 cDNA was obtained by RT-PCR of total human brain RNA (CLONTECH) by using the primers 5′-TAAGCAGTCAGAAGGTGACTTACTCC-3′ and 5′-AGGATCATCTACGACCGAAAGTTCCTG-3′. The radiolabeled blot was stripped in 0.5% SDS at 100°C and reprobed with a [32P]dCTP-labeled SRA core cDNA.
Low-Resolution Structure Modeling.
Pairwise sequence comparisons on orthologous SRA sequences of human, mouse, rat, and goat were carried out by using public-domain databases running blast and fasta algorithms and a proprietary database (Pangaea at Ibis Therapeutics, Carlsbad, CA). Michael Zuker's mfold program (http://bioinfo.math.rpi.edu/≈zukerm/) was used for low-resolution RNA structure modeling. Homologous SRA sequences were first computed by using default parameters (ΔG = −9.8 kcal/mol at 37°C, P = 5%), then resubmitted for folding with changed sequence window size (40–200 bp), and/or altered level of promiscuity of pairing with other bases. The energy dot blot of mfold was used as a first-level evaluation for the quality of the inferred structures.
Tissue Culture and Transient Transfection Assay.
Human cervical carcinoma (HeLa) cells were routinely maintained in DMEM/10% FBS. Twenty-four hours before transfection, 3 × 105 cells per well of a six-well plate or 105 cells per 12-well dish were plated in DMEM containing 5% dextran-coated, charcoal-stripped FBS. Cells were transfected with Superfect Transfection Reagent (Qiagen, Valencia, CA) according to the manufacturer's protocol. The transfection assay was described previously (5).
Results
SRA Is an Autonomous Gene Product.
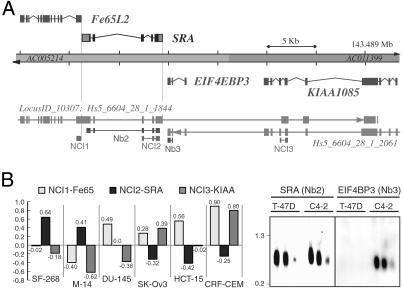
To confirm our initial identification of multiple isoforms of SRA, we used National Center for Biotechnology Information-based resources (6) and integrated gene assembly suites such as NIX to place isolated SRA sequences in perspective with available genomic DNA sequences. Fig. 1 shows a basic genomic characterization of SRA and illustrates the fact that SRA is flanked by two other genes: on its 5′ terminus by Fe65L2, and on its 3′ terminus by EIF4EBP3 on the reverse strand. Its proximity to these two genes, together with our previous demonstration that it functions as an RNA and exists as different splice forms, suggested the possibility that SRA may be a viable 3′ UTR of either flanking gene. In support of this notion, several gene-finding algorithms indicated that sequences we initially attributed to SRA may in fact be part of the Fe65L2 gene (Fig. 1A).
Fig 1.
Autonomous SRA gene locus. (A) Genomic illustration of the SRA locus. Two DNA contigs (AC005214 and AC011399), together comprising the Homo sapiens loci for Fe65L2, SRA, EIF4EBP3, and KIAA1085, are shown at the top. SRA is located on the direct strand of human chromosome 5, band q31 (≈143,460.7–143,469.3 kb), and is juxtaposed by genes encoding Fe65-LIKE2 (Fe64L2) and on the reverse strand by eukaryotic translation initiation factor 4E binding protein 3 (EIF4EBP3). Integrated results of gene predictions as alignments of mRNAs including alternative splice models against the genome are shown at the bottom. Gene assembly algorithms indicate that adjacent genes encroach upon SRA sequence. On the direct strand, the H. sapiens locus LocusID_10307: Hs5_6604_28_1_1844 is defined by several hundred cDNAs encoding FE65-LIKE 2. Some splice variants also include the SRA locus, and others extend to sequences that are also used on the reverse strand by the KIAA1085 gene. On the reverse strand, the locus Hs5_6604_28_1_2061 includes at least 16 different cDNA clones and produces eight different transcripts. Alternative spliced mRNAs overlap the genes KIAA1085 and EIF4EBP3, and some variants are predicted to also include SRA sequence. Annotated lines with small boxes underneath the integrated gene predictions indicate the exon probes used for expression analysis shown in B. (B) Expression analyses suggest an autonomous SRA locus. (Left) A subset of expression data of the NCI60 microarray experiment for the genes Fe65L2 (NCI1), SRA (NCI2), and KIAA1085 (NCI3) is shown. The values (y axis, log base 2) represent the variation in transcript levels relative to reference samples (8): positive numbers indicate a relative increase and negative numbers indicate a relative decrease in transcript levels. Selected cell lines are: SF-268, central nerve system; M14, melanoma; DU-145, prostate; SK-Ov-3, ovarian; HCT-15, colon; CRF-CEM, leukemia. (Right) Northern blots for SRA (Nb2) and EIF4EBP3 (Nb3) are shown for human breast cancer cell line T-47D and the LNCaP-derivative prostate cancer cell line C4-2. Both cell lines express SRA, but EIF4EBP3 was not detected in T-47D cells. A membrane containing three different concentrations (30, 15, and 5 μg) of total RNA was first analyzed for EIF4EBP3 expression, then stripped and subsequently reprobed with labeled SRA cDNA. Size markers are in kb.
To investigate this possibility, we next studied the expression pattern of the genes in question. A literature search identified a comprehensive expression analysis for Fe65L2, SRA, and KIAA1085 in the NCI60 study of Ross et al. (8), in which DNA microarrays were used to explore variations in expression of 8,000 genes among 67 human cancer cell lines. In this study, 38 (57%) of the 67 tested cell lines showed a difference in expression levels for SRA and Fe65L2 greater than a standard deviation, and 24 (34%) cell lines displayed inversely proportional expression levels for SRA and Fe65L2. The difference in expression pattern was even more apparent when KIAA1085 was compared with SRA: 41 (61%) cell lines showed significantly different expression levels, and 45% of cell lines exhibited an opposing expression pattern for SRA and KIAA1085. Fig. 1B contains the data of a subset of cell lines tested by the NCI60 project. These results suggested that the SRA expression unit is independent and distinct from Fe65L2 and KIAA1085, contradicting certain in silico predictions that incorporate SRA sequences in Fe65L2 transcripts. We next addressed the possibility that splice variants of EIF4EBP3 might include SRA sequences by probing tissue culture RNA membranes with cDNA probes of both genes. Whereas SRA was expressed at relatively high levels in human breast cancer T-47D cells and in prostate LNCaP-derived C4-2 cells, the EIF4EBP3 transcripts were found in LNCap/C4-2 cells but were not detected in T-47D cells in our assays (Fig. 1B). These results strongly suggest that the SRA gene is also distinct from the EIF4EBP3 gene. Together, our gene expression analyses strongly indicate that SRA is not part of either flanking gene but rather constitutes an autonomous genetic unit.
The SRA transcripts identified by our initial bacteriophage screen contained an ORF but lacked an initial ATG. Subsequent attempts to extend the SRA sequences by 5′ RACE failed to yield transcripts that included the presumptive SRA protein start codon. Moreover, RNase protection and primer extension assays pointed to possible different splice variants of SRA but did not indicate a 5′ extension beyond the sequence of SRA I (data not shown). In addition, the GRAIL/CpG algorithm predicted a CpG island and a putative TATA box sequence upstream of the SRA core, and ESTs covering the 5′ end of SRA at the time of our analysis did not indicate the presence of transcripts with 5′-extended exon I sequence. Given these data, we therefore decided not to further pursue a presumptive full-length SRA protein-encoding transcript and focused instead on its characterization as an RNA-based coactivator for SRs.
Composite Coactivation Function.
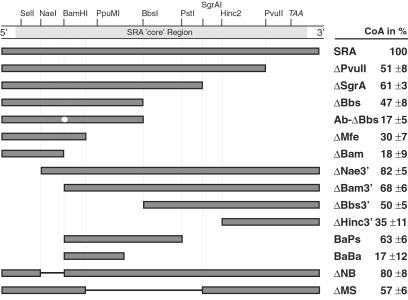
To further define the SR coactivation function attributed to the SRA RNA (5), we first generated a series of SRA mutants containing a variety of truncations and internal deletions, and tested these mutants in mammalian cells for the coactivation of PR-mediated transcription (Fig. 2). We found that the coactivation function of SRA cannot be attributed to one distinct portion of the transcript but is rather distributed over the SRA molecule. Relatively small truncations at both ends of the core sequence individually reduced PR coactivation (compare the constructs ΔPvuII and ΔNae3′ with full-length SRA). Further removal of terminal sequences also identified the center portion of SRA as important for coactivation. Because regions containing deduced coactivation functions (ΔBam, ΔMfe, ΔHinc3′, and BaBa) were not sufficient to significantly coactivate PR transactivation, we concluded that the coactivation function of SRA is not restricted to a single, discrete domain.
Fig 2.
SRA deletion analysis. Genetic deletion analyses indicate complex SRA coactivation function(s). SRA cDNA core region with selected restriction sites and presumptive ORF termination codon (TAA) is shown at the top. Shown underneath are the schematic representation of wild-type SRA and SRA deletion mutants and the corresponding relative coactivation of PR-mediated transactivation (CoA) as averaged from three separate transfection experiments (right). HeLa cells were transfected with 0.2–1 μg of pSCT-SRA expression plasmid or 200 nM ADOs along with 20 ng of PR expression vector and 2.5 μg of MMTV-Luciferase reporter. Ab-ΔBbs differs from ΔBbs by containing a point mutation at the BamHI site.
Structure Modeling.
Having established the existence of a number of distinct sequences involved in SRA coactivation function, we next decided to use low-resolution RNA structure modeling to predict functional substructures of SRA. Low-resolution RNA modeling is the analysis of the canonical Watson–Crick and wobble base-pairing patterns of short, double-helical RNA motifs. We used a combination of phylogenetic and thermodynamic approaches to determine topological conservations in SRA, and we tested their functional significance in tissue culture cells for coactivation of PR-mediated transcription. Given the small number of SRA orthologs available at the time of the study, the phylogenetic comparative analysis was limited to a sequence comparison between the human and mouse SRA cDNAs we identified in our initial bacteriophage screening and the analysis of deposited ESTs from human, mouse, rat, and goat.
Discrete portions of the related sequences were analyzed in silico by using the mfold program, which predicts optimal and suboptimal RNA secondary structures based on the free energy minimization method (9). The energy dot blot of mfold, a superimposition of various possible foldings (10), was used as a first-level, crude measurement for the quality of the inferred structures. Homologous SRA sequences were first computed by using default parameters and then resubmitted for folding with changed sequence window size and/or altered level of promiscuity of pairing with other bases. The persistent appearance of a particular prediction indicated a motif, which was assumed, at a high confidence level, to represent a functionally relevant structure of SRA.
Altogether, this approach identified 11 topological substructures contained in the core-sequence of human and mouse SRA (Fig. 4B shows a schematic representation of SRA and the identified secondary RNA motifs). We subjected all 11 predicted substructures to targeted mutagenesis and tested the resulting SRA variants for PR coactivation in tissue culture cells. Most importantly, we attempted to uncouple the biological function of the RNA while simultaneously preserving a putative SRA protein function by directing nucleotide changes to the wobble position of the presumptive amino acid sequence predicted by the ORF in SRA, or used alternative genetic codons if possible.
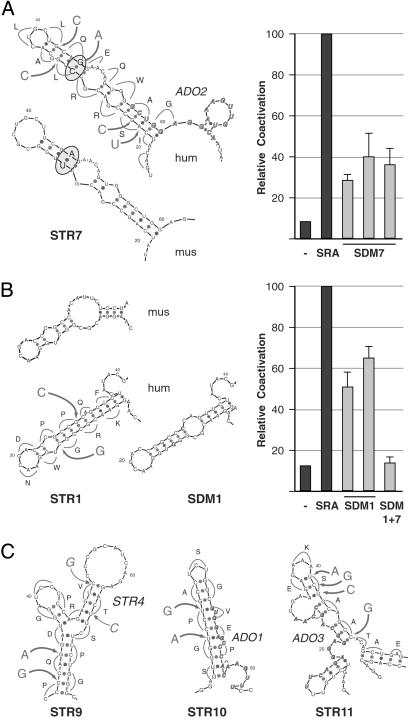
An example illustrating our silent mutagenesis strategy is shown in Fig. 3A by the plotted representation of the inferred structure for the sequence comprising exon IV of SRA. In this case, the SRA wild-type sequence is predicted to form a compact stem-loop with a large bulge and additional noncanonical pairings. Topological conservation of this motif between mouse and human SRA is supported by covariation of the underlying nucleotide sequence, which, following the bulge, shows a U-A pairing in mouse SRA and a C-G pairing in the human ortholog. To disable the function potentially mediated by this structure, we applied five nucleotide changes, none of which altered the presumptive amino acid sequence, but all of which abolished the indicated RNA motif. Three nucleotide changes were targeted to the wobble positions of codons located between the internal and end loops, and an alternative codon for serine was used at the stem of the structure. When tested in transient transfection assays, recombinant human SRA containing these mutations [called SDM7 for site-directed mutation in structure (STR) 7] consistently exhibited a significantly reduced capacity for coactivation of PR-mediated transcription (Fig. 3A). RT-PCR analysis of these cells indicated that SDM7 was expressed at similar levels as SRA was produced in transfected control cells (not shown). Because the presumptive SRA peptide sequence was not altered by the silent mutations, this result further indicated to us that the coactivation function of SRA is directly mediated by the RNA transcript.
Fig 3.
Discrete RNA structure modeling and silent, site-directed mutagenesis. Representation of comparative low-resolution RNA modeling (STR) and silent mutagenesis strategy of discrete portions of SRA sequences from human (hum) and mouse (mus) as predicted by the mfold program (Left), and relative coactivation of corresponding human site-directed SRA mutants (SDMs) tested in tissue culture experiments (Right). The single-letter code of the presumptive amino acid sequence given by the ORF in SRA, nucleotide changes (large letters), and sites of mutations (arrows) are shown alongside the plotted representation of the inferred wild-type structure of the human SRA sequence. Bold, italic sequences in STR7, STR10, and STR11 annotate the complement sequences of ADOs (see Discussion). Relative transactivation of wild-type SRA (SRA) and site-directed mutants (SDM) were obtained by transient cotransfection of recombinant cDNAs (1–2 μg) or empty vector (−) along with PR expression vector (20 ng) and MMTV-Luciferase reporter (2.5 μg) into HeLa cells. Luciferase activities are shown as the mean (±SE) values of at least four separate experiments. (A) Site-directed mutations in predicted substructure 7. Covariation is indicated by the U-A pairing in mouse and C-G pairing in the human ortholog (circled). Three independent cDNA constructs of mutated STR7 (SDM7) were assayed in transient transfection experiments as described above. (B) Inferred STR1 located at the 5′ end of the SRA core sequence (mus, hum), targeting strategy (hum), and predicted structure after site-directed mutagenesis (SDM1). Two independent silent mutants were generated and tested. SDM1 + 7 is a SRA variant with silent mutations in both structure 1 and structure 7. (C) Predicted substructures 9–11, silent mutagenesis and target sequences for ADOs as indicated. Transactivation data are shown in Fig. 4.
Similar results were obtained for another SRA variant containing two nucleotide changes in a deduced motif located at the 5′-end of the SRA core sequence (STR1). Free energy calculation suggested an extended Watson–Crick helix in human SRA whose distal stem-loop structure region was also predicted for the mouse sequence (Fig. 3B). Silent mutagenesis resulted in a new inferred stem-loop structure (SDM1) which significantly lowered, but did not eradicate, SRA function. This result identified another RNA motif involved in SRA coactivation, and is in agreement with our previous genomic deletion analysis (ΔNae3′). An SRA variant containing both silent mutations (SDM1 and SDM7), however, nullified SRA coactivation, indicating a functional relationship between the two distinct RNA motifs (Fig. 3B).
In addition to these regions of SRA, our initial genomic deletion and truncation analysis implied other sequences of SRA as important for coactivation. We therefore applied silent mutagenesis to the remaining inferred structures and tested the resulting SRA variants (SDMs) for PR coactivation. Fig. 4A shows the transactivation pattern of all SDMs obtained by averaging multiple transfection experiments. Not all deduced secondary structures were effective in significantly changing SRA coactivation after the introduction of motif-disrupting silent mutations. For example, mutations in predicted structures (SDM2, SDM3, SDM4, and SDM5) showed a stimulation of PR transactivation similar to that of wild-type SRA.
Comparative sequence analysis and in silico energy calculations suggested three other RNA motifs whose functional consequences we tested by silent mutation and subsequent transactivation analysis. Both SDM9 and SDM10 contain two nucleotide substitutions at wobble positions in predicted stem-loop structures (Fig. 3C); SDM9 targets a sequence containing the BamHI restriction site that we have identified to be sensitive to mutations (Ab-ΔBbs in Fig. 2A), whereas SDM10 marks a central sequence of exon III. When tested in tissue culture cells for PR coactivation, both SRA variants showed significantly impaired activity (Fig. 4A). With SDM11 we targeted a deduced secondary structure located at the 3′ end of the core sequence by using an alternative codon for serine and substituting a guanine for a cytosine at a wobble position (Fig. 3C). SDM11 also resulted in reduced SRA function (Fig. 4A). These results further support the notion that SRA coactivation is mediated at the RNA level by different structural motifs and that functional relationships exist between diverse substructures in the RNA molecule.
In summary, 5 of the 11 in silico predicted secondary structures exhibited impaired transcription coactivation after mutagenesis. Because all nucleotide changes were silent mutations presumed not to alter deduced encoded amino acid sequence, this analysis provides strong evidence that SRA-mediated coactivation is executed by distinct RNA motifs and not by an SRA protein. In addition, because mutations of individual substructures were not sufficient to completely abrogate SRA function in our assays, we concluded that multiple motifs in SRA contribute in a concerted manner to the overall coactivation function of the molecule.
Discussion
The knowledge of relatively stable motifs within an RNA molecule allows for experimental verification of the postulated structures by genetic analysis to assess their functional consequences. In the case of SRA, this not only would precisely identify the coactivation functions, but also, if site-directed mutagenesis were targeted to the wobble positions of the presumptive SRA amino acid sequence, would provide additional strong evidence that it is the RNA that mediates SR coactivation. Moreover, knowledge of secondary structure, together with additional information on structural constraints or tertiary interactions, can be used to identify SRA-interacting proteins, without which the molecular mechanism of SRA action would remain unclear.
Two major methods are generally used to predict RNA structures: computer-based free energy calculations and the classic covariation analysis (11, 12). Free energy calculation, using sets of thermodynamic parameters and dynamic algorithms to find double-helical regions and various types of loops of a sequence, provide relatively reliable secondary structure predictions when applied to RNAs with the ability to fold by themselves to achieve a viable structure (13, 14). However, because constraints identified by comparative sequence analysis were generated by evolutionary selection, topological conservation reflects function and may also infer biological functions mediated by tertiary structures. A combination of both methods is mutually beneficial and has been proposed previously (15, 16).
This hybrid approach identified five substructures within the SRA core sequence to be important for SRA coactivation (Fig. 4B). It is unlikely that the lower transcription activity is attributable to the altered stability of their mutated transcripts. SRA variants with mutations adjacent to substructures involved in coactivation fully retained their ability to coactivate SRs (for example SDM3 and SDM4 nearby SDM9), whereas RT-PCR detected comparable levels of transcripts in transfected cells. However, the stability of RNA largely depends on interactions with other RNA and proteins. Because such interactions are mediated by the spatial folding of the RNA, without the SRA-interacting proteins available for experimentation, it is practically impossible to differentiate between RNA stability and function.
Additional evidence from low-resolution structure modeling has identified RNA motifs important for SRA coactivation. Structures shown to be sensitive to mutagenesis reside in regions of SRA that have previously been implied to play a role in coactivation by studies involving classical deletion analysis (Fig. 2). Moreover, a screen using an array of small antisense deoxy-oligonucleotides (ADOs) in transient transfection assays pinpointed four sequences in SRA that appear to be involved in its coactivation function (5, 17). The isolated sequences precisely map to three predicted functional substructures: the sequence of ADO1 is contained in STR10, ADO2 is in STR7, and ADO3 is in STR11 (Figs. 3 and 4B). ADO4 hybridizes to a portion at the very 3′ end of the SRA core sequence. This region of SRA failed to produce persistent motifs by using the mfold program. A role of this sequence in SRA function, however, is suggested by genomic deletion analysis (ΔPvuII in Fig. 2) and site-directed mutagenesis (not shown). Therefore, we conclude that the coactivation function of SRA is comprised of at least six distinct RNA motifs distributed over the entire core sequence of SRA.
The efficacy of SRA-specific ADOs with respect to inhibition of its coactivation of estrogen-receptor-mediated gene expression has been reported recently (17). A comparison of the inferred SRA substructures reflects the underlying principles of antisense–target interactions and supports our structure analysis. As demonstrated by ADO1/STR10, ADO2/STR7, and ADO3/STR11 (Fig. 3), initial oligonucleotide contacts require short linear regions adjacent to, or between structures (18–20).
It is widely accepted that RNA plays pleiotropic functions in the eukaryotic cell. Regardless of type or functional classification, the activity of RNA is determined by its interaction with other RNA and proteins. The sequences underlying the identified functional substructures of SRA are perfectly suitable for testing in a yeast-three hybrid assay (21) to screen for specific SRA-interacting proteins. The nature of RNA–protein interactions requires RNA double-helical regions to have distortions generated by internal loops or bulges (22–24). Proteins may also bind to helix termini by using adjacent single-stranded RNA regions (25). Because of their content of single-stranded regions, SRA substructures 1, 7, 10, and 11 are likely candidates to specifically bind to proteins. Alternatively, STR7 may be involved in tertiary structure formation, as indicated by its interplay with STR1. A possibly central role of this STR7 is also suggested by its topological conservation (Fig. 3A), and the requirement of an overall intact structure (Fig. 4A; SDM7a).
The identification of SRA-interacting proteins is fundamental for understanding the molecular mechanism of its coactivation of SRs. Because no evidence has been obtained yet for direct binding of SRA to the SRs, we have proposed a model in which SRA functions confer specificity on ribonucleoprotein complexes to modulate SR-mediated transcription (5). Recent results from other laboratories support this notion. SRA was shown to specifically bind to proteins containing different RNA-binding motifs and to selectively modulate SR transactivation. For instance, SRA associates with the DEAD-box proteins p72/p68 to act as an estrogen-receptor alpha-specific ribonucleoprotein coactivator complex by stimulating the amino-terminal activation function of the receptor while simultaneously integrating SRC/p160-mediated AF2 coactivator functions (26). SRA was also found to bind to the three RRM motifs of SHARP (7). The SHARP/SRA association modulates estrogen-receptor transactivation by rapidly attenuating the steroid response through sequestration of SRA, and simultaneously initiating repression by SMART. In both cases, SRA-containing ribonucleoprotein complexes accentuate transcription specificity by integrating coregulator activities on selective binding to SRs. Not all DEAD-box domain or RRM-containing proteins, however, bind SRA, indicating a level of specificity in the ribonucleoprotein complex formation. Specificity for RNA-protein interactions is likely provided by the spatial folding of the RNA, which is a function of its secondary and tertiary RNA structures. Unlike the well defined functional domains of proteins, the coactivation function of SRA is an integration of individual activities mediated through RNA structure motifs that are distributed over the entire core sequence of the molecule.
Recent cloning efforts by other laboratories produced SRA transcripts that support our previous observation of the existence of different SRA splice forms (for example AK024640), but also revealed 5′ extended SRA transcripts that include the putative starting ATG of the SRA ORF (AF318361 and AK054960). This reiterates the conceptual possibility of an SRA protein. Transcripts of 2–3 kb in size, however, have not yet been detected by Northern analysis (5), suggesting that the newly deposited mRNA must represent rare transcripts. In addition, expression analyses strongly support the idea of SRA being an autonomous genetic unit with an expression pattern significantly different from its neighboring genes (Fig. 1).
Although we do not totally exclude the existence of a translation product of SRA, we have presented additional strong evidence demonstrating that SRA exists and functions as an RNA transcript in coactivating SR transcriptional activity. The next important step to understand SRA biology is the identification of SRA-interacting proteins. The work described here, by hinting at potential docking sequences for SRA ligands, has provided a solid foundation on which the construction of a coherent model for SRA function can proceed.
Acknowledgments
We thank N. McKenna, S. Tsai, and M.-J. Tsai for critically reading the manuscript, and R. Sampath and D. Ecker (Ibis Therapeutics, Carlsbad, CA) for helpful discussions and use of Ibis' proprietary database. This work was supported by grants from the Texas Higher Education Advanced Technology Program (ATP 004949-0154-1999), from The CONCERN Foundation, SPORE P50CA58183 Breast Cancer, and the Edward Mallinckrodt Jr. Foundation (to R.B.L.), and by grants from the National Institutes of Health (to B.W.O.).
Abbreviations
SR, steroid receptor
SRA, SR RNA activator
PR, progesterone receptor
SDM, site-directed SRA mutant
STR, inferred SRA substructure
ADO, antisense deoxy-oligonucleotide
References
- 1.Mangelsdorf D. J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., et al. (1995) Cell 83, 835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai M. J. & O'Malley, B. W. (1994) Annu. Rev. Biochem. 63, 451-486. [DOI] [PubMed] [Google Scholar]
- 3.Glass C. K. & Rosenfeld, M. G. (2000) Genes Dev. 14, 121-141. [PubMed] [Google Scholar]
- 4.McKenna N. J. & O'Malley, B. W. (2002) Cell 108, 465-474. [DOI] [PubMed] [Google Scholar]
- 5.Lanz R. B., McKenna, N. J., Onate, S. A., Albrecht, U., Wong, J., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (1999) Cell 97, 17-27. [DOI] [PubMed] [Google Scholar]
- 6.Pruitt K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y., Downes, M., Xie, W., Kao, H. Y., Ordentlich, P., Tsai, C. C., Hon, M. & Evans, R. M. (2001) Genes Dev. 15, 1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross D. T., Scherf, U., Eisen, M. B., Perou, C. M., Rees, C., Spellman, P., Iyer, V., Jeffrey, S. S., Van de Rijn, M., Waltham, M., et al. (2000) Nat. Genet. 24, 227-235. [DOI] [PubMed] [Google Scholar]
- 9.Mathews D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911-940. [DOI] [PubMed] [Google Scholar]
- 10.Zuker M., Mathews, D. H. & Turner, D. H. (1999) RNA Biochemistry and Biotechnology in Proceedings of the NATO Advanced Research Workshop, eds. Barciszewski, J. & Clark, B. F. C. (Kluwer, Dordrecht, The Netherlands), pp. 11–43.
- 11.Gaspin C. & Westhof, E. (1995) J. Mol. Biol. 254, 163-174. [DOI] [PubMed] [Google Scholar]
- 12.Jaeger J. A., Turner, D. H. & Zuker, M. (1989) Proc. Natl. Acad. Sci. USA 86, 7706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M., Jaeger, J. A. & Turner, D. H. (1991) Nucleic Acids Res. 19, 2707-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konings D. A. & Gutell, R. R. (1995) RNA 1, 559-574. [PMC free article] [PubMed] [Google Scholar]
- 15.Konings D. A. & Hogeweg, P. (1989) J. Mol. Biol. 207, 597-614. [DOI] [PubMed] [Google Scholar]
- 16.Le S. Y. & Zuker, M. (1991) J. Biomol. Struct. Dyn. 8, 1027-1044. [DOI] [PubMed] [Google Scholar]
- 17.Cavarretta I. T., Mukopadhyay, R., Lonard, D. M., Cowsert, L. M., Bennett, C. F., O'Malley, B. W. & Smith, C. L. (2002) Mol. Endocrinol. 16, 253-270. [DOI] [PubMed] [Google Scholar]
- 18.Wu T. H., Liao, S. M., McClure, W. R. & Susskind, M. M. (1987) Genes Dev. 1, 204-212. [DOI] [PubMed] [Google Scholar]
- 19.Persson C., Wagner, E. G. & Nordstrom, K. (1988) EMBO J. 7, 3279-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer K. L. & McClure, W. R. (1997) RNA 3, 157-174. [PMC free article] [PubMed] [Google Scholar]
- 21.SenGupta D. J., Zhang, B., Kraemer, B., Pochart, P., Fields, S. & Wickens, M. (1996) Proc. Natl. Acad. Sci. USA 93, 8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye X., Kumar, R. A. & Patel, D. J. (1995) Chem. Biol. 2, 827-840. [DOI] [PubMed] [Google Scholar]
- 23.Puglisi J. D., Chen, L., Blanchard, S. & Frankel, A. D. (1995) Science 270, 1200-1203. [DOI] [PubMed] [Google Scholar]
- 24.Battiste J. L., Mao, H., Rao, N. S., Tan, R., Muhandiram, D. R., Kay, L. E., Frankel, A. D. & Williamson, J. R. (1996) Science 273, 1547-1551. [DOI] [PubMed] [Google Scholar]
- 25.Rould M. A., Perona, J. J. & Steitz, T. A. (1991) Nature 352, 213-218. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe M., Yanagisawa, J., Kitagawa, H., Takeyama, K., Ogawa, S., Arao, Y., Suzawa, M., Kobayashi, Y., Yano, T., Yoshikawa, H., et al. (2001) EMBO J. 20, 1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]