Abstract
The Dbf4/Cdc7 kinase acts at the level of individual origins to promote the initiation of DNA replication. We demonstrate through both immunoprecipitation and two-hybrid assays that a domain comprising the first 296 aa of Dbf4p interacts with Orc2p and Orc3p subunits of the origin recognition complex (ORC). Given that the activation of Rad53 kinase in response to the DNA replication checkpoint leads to the release of Dbf4p from an ORC-containing chromatin fraction, we also examined interaction between Dbf4p and Rad53p. This same domain of Dbf4p binds specifically to the forkhead homology-associated (FHA) domains of Rad53p. Cell cycle arrest in G2/M, provoked by the overexpression of the Dbf4 domain, is suppressed in a rad53 mutant. Moreover, its overexpression perturbs the regulation of late, but not early, origin firing in wild-type cells after treatment with hydroxyurea.
Initiation of DNA replication requires both the establishment of prereplicative complexes at origins and the activity of two kinases, Cdc28p and Cdc7p. In Saccharomyces cerevisiae, Cdc28p is activated by six distinct regulatory cyclin subunits, Clb1 to -6, the levels of which fluctuate in a cell cycle-dependent manner (reviewed in ref. 1). In contrast, Cdc7p has a single regulatory subunit, Dbf4p, which also exhibits cell cycle-programmed changes in level, peaking sharply at the G1-S transition (2–4). Individual S. cerevisiae origins initiate replication at characteristic times, with some firing early, most in mid-, and others in late-S phase (5, 6). Cdc7/Dbf4 activity is required throughout S phase for origin firing (7, 8), suggesting that this kinase complex acts at the level of individual origins, rather than providing a general signal that enables replication initiation at the G1-S transition.
Both genetic (9, 10) and biochemical (3, 11) evidence argue that mini-chromosome maintenance (MCM) proteins are physiological substrates of Dbf4/Cdc7. Mcm2-7 form a hexameric helicase that is loaded onto DNA at origins during G1, in preparation for origin firing (reviewed in ref. 12). Cdc45p, which associates with origins after Cdc28/Clb5 activation, is another potential target of the Cdc7/Dbf4 kinase (13–15). Consistent with its proposed role in activating the pre-RC, Dbf4p cofractionates with origin recognition complex (ORC) into an insoluble nuclear chromatin fraction from G1 phase cells, an association that is lost in an orc2-1 mutant (2). Furthermore, the punctate pattern of Dbf4p immunostaining in S-phase nuclei is ORC dependent (2), and one-hybrid data show that a Dbf4 fusion can activate from an origin consensus (16). These results support the hypothesis that Dbf4p targets Cdc7p to origins of replication through association with ORC.
The targeting of Cdc7p kinase to origins through Dbf4p may be a regulated event. First, Dbf4p seems to be phosphorylated, and the Dbf4/Cdc7 kinase down-regulated, after the activation of Rad53 kinase by the stalling of replication forks in hydroxyurea (HU; 3,11). Second, Rad53p activation by HU leads to the release of Dbf4p from an origin-enriched chromatin fraction (2). Here, we demonstrate that Dbf4p binds several ORC subunits and Rad53p through an N-terminal domain. In vivo studies provide evidence for a role for these interactions in regulating origin activation.
Methods
Yeast Strains.
GA-1020 (MATa, ade2-1, can1-100, trp1-1, his3-11, -15, ura3-1, leu2-3,-112, pep4:LEU2) was used for all immunoprecipitation (IP) studies involving plasmid-expressed proteins. For IP of proteins expressed from Myc-tagged copies of endogenous genes, GA-850 (MATa, ade2-1, can1-100, his3-11, -15, leu2-3, -112, trp1-1, ura3-1, GAL, psi+, dbf4:DBF4-Myc18/LEU2; formerly K6388 from K. Nasmyth, Institute of Molecular Pathology, Vienna), GA-1009 (yKu80-Myc13:kanMX4, MATa, ade2:hisG, his3-11, leu2, trp1, ura3-52, can1:hisG, DIA5-1), and GA-1099 (DNA2-Myc13:kanMX6, MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3, -112, pep4:LEU2, can1-100) were used, with GA-1020 as an untagged control. GA-1211 (MATa, his3, trp1, ura3-52, leu2:pro, LEU2-lexAop2, formerly EGY188 from R. Brent, Molecular Sciences Institute, Berkeley, CA) was used for two-hybrid analyses (17). GA-180 (MATa, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3, -112, can1-100), GA-850, GA-856 (MATa, tel1:HIS3, trp1, ade2, ura3, his3, can1-100), GA-1040 (Rad53-Myc13; ref. 18), GA-1491 (MATa, mec2-1 = rad53-11, ref. 19), GA-1701 (MATa, rad53:TRP1, sml1:KanMX6, Sgs1-Myc13:HIS, pep4:LEU2, ade2-1, trp1-1, his3-11, -15, ura3-1, leu2-3, -112), GA-1702 (as 1701 but mec1:TRP1 instead of rad53:TRP1), and GA-2012 (GA-180 with chk1:His3MX6) were hosts for the analysis of Dbf4p fragment overexpression. All strains except GA-1009, GA-1211, and GA-1040 are derivatives of W3031a.
Plasmid Construction.
pCM190-Dbf4-FL, encoding full-length Dbf4p with a C-terminal Myc18 tag, fused to a tetracycline (Tet) or doxycycline (Dox) repressible promoter was a gift of E. Schwob (Centre National de la Recherche Scientifique, Montpellier, France). pCM190-Dbf4-109, -178, -248, and -296 were constructed by PCR amplification and insertion of the regions encoding the indicated number of amino acids. pEG-Dbf4-FL, -296, and -Δ110–296 were constructed by cloning appropriate fragments into two-hybrid bait vector pEG-202 (17). pJG-Orc1 through -6 were made by cloning PCR amplified full-length ORC sequences into the two-hybrid prey vector pJG4-6 (ref. 20; gift of R. Brent, Molecular Sciences Institute, Berkeley, CA). RAD53 fragments including the coding regions for FHA1 (I4-S165) and FHA2 (M497-S821) were amplified by PCR from pJA92 (21). As template for the Rad53 kinase domain (G177-N599), pNB187-SPK1 (22) was used. PCR products were subcloned both into pJG4-6, resulting in pJG-FHA1, pJG-KIN, and pJG-FHA2, and into pGAL-lexA, which expresses lexA fusions under GAL1 control, replacing the constitutive ADH promoter in pEG-202. Point mutations in the FHA1 and FHA2 domains are as described (23). Correct fusion protein expression was analyzed by Western blot.
Two-Hybrid Analysis.
Liquid culture two-hybrid analysis was done as described (17). The lacZ-reporter pSH18-34, pEG-202-derived bait, and pJG4-6-derived prey plasmids were transformed into GA-1211. Exponentially growing, glucose-depleted yeast cells were then induced for 6 h on 2% galactose, and interactions between fusion proteins were detected by the quantitative β-galactosidase assay on permeabilized cells, by using four independent transformants per sample (24).
Co-IP, Western, and 2D Gel Analyses.
For the Dbf4/ORC interaction studies, GA-1020 cells, transformed with pCM190- and pJG4-6-derived expression vectors, were initially grown to 1 × 107 cells/ml, in 5 ml SD medium (25) without uracil and tryptophan and with 3 μg/ml Tet. Cells were centrifuged (1,000 × g, 5 min), and the pellet was resuspended in 10 ml S 2% raffinose/0.1% sucrose medium (25) without uracil and tryptophan, again with 3 μg/ml Tet. After 4 h growth at 30°C, cells were centrifuged as above, and the pellet was resuspended in 20 ml S 1% raffinose/2% galactose medium (25) lacking uracil, tryptophan, and Tet. Cells were centrifuged after a further 4 h incubation with shaking at 30°C. All subsequent steps were carried out at 4°C. Pellets were resuspended in 400 μl ice-cold lysis buffer (50 mM Hepes, pH 7.5/0.14 M NaCl/1 mM Na-EDTA/1% Triton X-100/0.1% sodium-deoxycholate, with protease inhibitors) and lysed with a bead beater by using 0.5 g glass beads per sample. Lysate was centrifuged (16,000 × g, 30 s), and the supernatant was called whole cell extract (WCE). WCEs (200 μl) were incubated on a rotating wheel (1 h, 4°C) with 40 μl anti-mouse IgG Dynabeads (Dynal, Great Neck, NY), saturated with both BSA and α-Myc monoclonal antibody (9E10). Beads were washed once in 400 μl lysis buffer and three times in wash buffer (0.1M Tris⋅Cl, pH 8.0/0.25 M NaCl/0.5% Nonidet P-40/0.5% sodium-deoxycholate/1 mM Na-EDTA), with a final resuspension in 40 μl SDS loading buffer. WCEs were prepared similarly for Rad53p-Myc Westerns. A polyclonal rabbit serum was raised against the N-terminal 173 residues of Orc2p. The 2D gel analysis of replication intermediates was performed as described (5) by using PCR-generated probes, on genomic DNA digested with PstI for ARS607 and BamHI for ARS603. Cell synchronization was as described (2).
Results
Dbf4p Interacts with Orc2p and Orc3p.
One-hybrid analysis suggests that Dbf4p recognizes, either directly or indirectly, the 11-bp ARS consensus sequence (ACS) found at all yeast origins of replication (16). To see whether this binding occurs through association with ORC subunits, we performed two-hybrid analysis using Orc1p to -6p as prey, and either full-length or the N-terminal 296 residues of Dbf4p as bait (Fig. 1A). Full-length Dbf4p demonstrated the strongest interactions with Orc2p and Orc3p (Fig. 1B), although weaker interactions with Orc5p and Orc6p could also be detected. These weaker interactions may be indirect, i.e., dependent on the other ORC subunits. However, because all ORC genes are essential, this possibility cannot be tested in living cells. Similar assays using the Dbf4p-296 fragment suggest that this region is responsible for association with Orc2p, and possibly with the other subunits, which give weaker but still significant interactions (Fig. 1B).
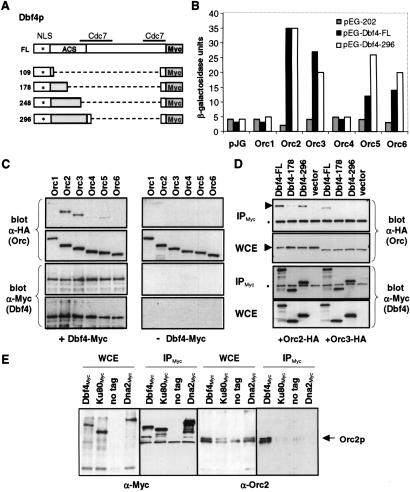
Fig 1.
Dbf4p interacts with multiple ORC subunits through its N-terminal 296-aa domain. (A) Shown are constructs of full-length DBF4 or the N-terminal 109, 178, 248, or 296 residues fused to the C-terminal 46 residues and a Myc tag. The two regions that bind Cdc7p (27) and that mediate origin consensus interaction (ACS target; ref. 16) are indicated. The asterisk denotes a putative nuclear localization signal (P55KKRSLE). (B) Two-hybrid assays are described in Methods, using bait constructs pEG-Dbf4-FL, pEG-Dbf4-296, and empty pEG-202. pJG-ORC1 through -ORC6 were used for prey, with empty pJG4-6 (pJG) as control. For all two-hybrid assays, SD among independent colonies was <10%. (C) The pJG plasmids express the ORC subunits fused to an N-terminal hemagglutinin tag. Yeast strain GA-1020 was cotransformed with one of the pJG-ORC clones and pCM190 either with Dbf4-FL (+Dbf4-Myc) or without (−Dbf4-Myc). Expression was induced from both promoters, and IP from WCEs with anti-Myc is described in Methods. Shown are immunoblots of IPs and of input WCEs detected with either monoclonal anti-HA (for ORC detection) or anti-Myc antibodies (for Dbf4p detection). In each case, 1/20 of the input WCE and 1/4 of the final bead resuspension was blotted. (D) IP assays were carried as described in C, with GA-1020 cells cotransformed with pJG-ORC2 or -ORC3 and pCM190-Dbf4-FL, -178, -296, or empty pCM190. Arrowheads indicate Orc2p or Orc3p bands; dots indicate the IgG heavy chain (55 kDa). (E) IP was carried out by using WCEs from strains carrying Myc-tagged genomic copies of DBF4 (GA-850), HDF1 (Ku80, GA-1009), and DNA2 (GA-1099), and a nontagged strain (GA-1020), as in C. Blots were reacted with anti-Myc IgG or polyclonal rabbit anti-Orc2p serum
To confirm this putative interaction between Dbf4p and the ORC subunits, co-IP assays were carried out in which Myc-tagged Dbf4p was overexpressed in combination with each of the six ORC subunits. Full-length DBF4 was cloned into the Tet-repressible expression vector pCM190 (ref. 26; Fig. 1A), and its expression was induced in the presence of the hemagglutinin-tagged, full-length Orc1 to -6 subunits (see above). When anti-Myc-coupled magnetic beads were added to WCEs from cells expressing Dbf4-Myc, Orc2p and Orc3p were reproducibly coimmunoprecipited (+Dbf4-Myc, Fig. 1C). Despite roughly equivalent expression levels for all ORC subunits and efficient IP of Dbf4-Myc (see WCE panels, Fig. 1C), Orc1, -4, -5, and -6 did not efficiently co-IP with Dbf4p. No ORC subunits were pulled down by using cells transformed with the pCM190 vector lacking Dbf4p (see −Dbf4-Myc, Fig. 1C).
To test whether the region of Dbf4p that binds Orc2p and Orc3p coincides with the residues that target the ACS, we compared co-IP efficiencies between full-length Dbf4p and constructs expressing either the N-terminal 178 aa or 296 aa residues of Dbf4p, fused in frame to the Myc tag (Fig. 1A). Confirming the two hybrid results, Orc2p was efficiently pulled down by using the 296 fragment (Fig. 1D, see arrow), whereas the 178 fragment was ineffective. This finding suggests that Dbf4p can bind Orc2p through its ACS-interaction domain. Neither the 178- nor the 296-residue Dbf4p fragment was sufficient to co-IP the Orc3p subunit. This finding may indicate that other parts of Dbf4p contribute to interaction with Orc3p, or, alternatively, that Orc3p binds Dbf4p through Orc2p, which would not be sufficiently abundant to mediate interaction with Orc3p in these assays.
We next tested for an interaction between Dbf4p and ORC when both are present at their endogenous levels of expression. WCEs were prepared from an exponentially growing culture of GA-850, in which the endogenous copy of Dbf4p is C-terminally Myc-tagged. As controls, strains were used in which the endogenous HDF1 gene, which encodes yKu80, or DNA2 was similarly fused to a 13-mer Myc epitope. These nuclear proteins are present in roughly equivalent amounts as Dbf4p (see WCE blotted with α-Myc, Fig. 1E). IP was carried out with α-Myc-coated magnetic beads. Immunodetection revealed that endogenous Orc2p could be precipitated with Dbf4p-Myc, but with neither yKu80-Myc nor Dna2-Myc (Fig. 1E), suggesting that the Dbf4/ORC interaction is specific and physiological.
Interactions with ORC and Rad53p Are Mediated by the Same Domain of Dbf4p.
The checkpoint kinase Rad53p acts as a signal transducer under conditions of DNA damage or replication fork stalling. Rad53p activation both prevents cells from entering mitosis (reviewed in ref. 28) and helps stabilize stalled replication forks (29, 30). In addition, Rad53p regulates the timing of origin firing during normal S phase progression, with some late origins firing earlier in rad53 mutants (31). Importantly, Dbf4p is phosphorylated in a RAD53-dependent manner (3), and, when modified, it is displaced from the insoluble ORC-containing nuclear pellet in chromatin fractionation assays (2). Full-length Dbf4p and Rad53p have also been shown to interact in two-hybrid assays (32).
In light of our findings for ORC, we characterized the domains of Rad53p and Dbf4p responsible for the reported interaction. In addition to its central kinase domain, Rad53p has two forkhead homology-associated domains (FHA1 and FHA2) that mediate protein–protein interactions by recognizing phosphothreonine- and phosphotyrosine-containing motifs (33, 34). FHA2 is primarily implicated in the interactions of Rad53p with Rad9p (23, 35). FHA1 mutations do not interfere with the Rad9p-mediated response to DNA damage, but render cells hypersensitive to HU (23). The relevant RAD53 fragments were cloned into pJG4-6, resulting in pJG-FHA1, -FHA2, and -KIN (Fig. 2A) for use in two-hybrid assays with various Dbf4p baits (Fig. 2B). A strong interaction between the Rad53p FHA1 domain and full-length Dbf4p was observed. A significant interaction was also observed between Dbf4p and Rad53p FHA2, although its β-galactosidase values were an order of magnitude lower than those obtained with FHA1. In both cases, the Dbf4 296-aa fragment was sufficient to mediate the interaction. Importantly, point mutations in the FHA1 and FHA2 domains that eliminate interaction with the Rad9p phosphoepitope (33) specifically eliminate interaction with full-length Dbf4p and reduce binding to the 296-aa fragment. In control assays, the N-terminal 248 aa of Dbf4p also retains avid interactions with the Rad53p FHA1 domain, whereas no significant binding could be detected by using the Dbf4p C-terminal 430 residues (data not shown). The Dbf4p-Rad53p interaction is thus specific and may require a phosphoepitope on the Dbf4p ligand. Further two-hybrid analyses using a Dbf4 deletion construct show that this amino terminal region is not only sufficient to bind both ORC and Rad53-FHA1, but that amino acid 110–296 are absolutely required for both interactions (Fig. 2C).
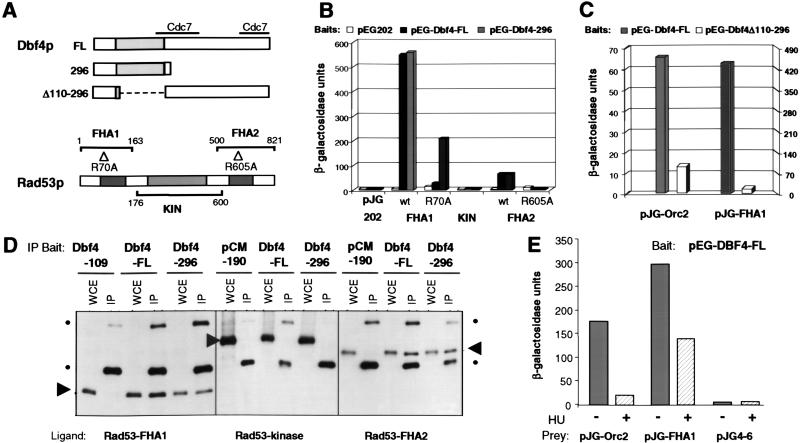
Fig 2.
Dbf4p interacts with the FHA1 and FHA2, but not the kinase domain of Rad53p. (A) Scheme of Dbf4 deletions and Rad53p fragments used in plasmid constructs for two hybrid and co-IP experiments. Fusions include amino acid 1–296 of Dbf4p, full-length Dbf4 (FL), and FL lacking amino acid 110 to 296. The Rad53p FHA1, kinase (KIN), and FHA2 domains are shown, including the R70A and R605A point mutations in FHA1 and FHA2 (33). (B) Two-hybrid assays were carried out as in Fig. 1B. Bait vectors and prey vectors are labeled as in A. Interactions with pJG-Dbf4-248 showed similar results as Dbf4-296, whereas pJG-Dbf4-274-704 bound none of the Rad53p domains (data not shown). (C) Two-hybrid assays were carried out as above, by using pEG-DBF4-FL and pEG-DBF4-Δ110–296 as baits. The scales at the left and right pertain to values obtained by using pJG-Orc2 and pJG-FHA1 as prey, respectively. (D) IP experiments were carried out as in Fig. 1C. WCEs were prepared from GA-1020 cells cotransformed with the indicated plasmids, as well as either pGAL-LexA-FHA1, -FHA2, or -KIN, as indicated. Immunoblots of the IP were probed with α-LexA antibody (CLONTECH). Arrows from top to bottom indicate Rad53 kinase, FHA2, and FHA1 LexA fusion proteins, whereas spots indicate the heavy and light IgG chains. (E) Two-hybrid analysis was performed as above, by using pEG-DBF4-FL as bait in each instance, along with the vector pJG4-6 expressing either ORC2 or FHA1. After 2 h of a 6-h galactose-induction, cultures were split, and 50 mM HU was added to one half of each.
To further confirm the two-hybrid results, we performed co-IP assays with cells that coexpress lexA-tagged FHA1, FHA2, and kinase domains of Rad53p, and either Dbf4-Myc-FL, or Dbf4-Myc-296. FHA1 and FHA2 successfully co-IP with both full-length Dbf4p and Dbf4-Myc-296, whereas the Rad53p KIN domain shows no interactions (Fig. 2D). In the case of the strongest interaction, i.e., Rad53-FHA1, we show that a smaller Dbf4p N-terminal fragment of 109 residues is unable to bind the Rad53p domain.
Because Dbf4p is displaced from chromatin under conditions of replication fork arrest, we next examined whether Orc2p association is altered when cells are treated with HU to activate Rad53p. Parallel two-hybrid analyses, in which half of each culture was treated with HU, resulted in a dramatic reduction in the Dbf4/Orc2 interaction as a consequence of HU exposure (Fig. 2E). The Dbf4/Rad53-FHA1 interaction persists in HU, but is reduced, suggesting that phosphorylation of Dbf4p may also weaken association with Rad53p. Alternatively, this reduction may reflect an increase of endogenous phosphorylated ligands that compete for the FHA1 domain.
Overexpression of the Dbf4 N Terminus Impedes Cell Cycle Progression.
Given that Dbf4p binds ORC through its N-terminal domain, it is likely that this interaction helps deliver the Cdc7 kinase, which Dbf4p binds through two conserved domains. Because the binding sites within Dbf4p for ORC/Rad53p and Cdc7p are distinct, it was worthwhile to test whether overexpression of the Dbf4p N-terminal domain would affect cell growth or checkpoint response. WT yeast were transformed with the Tet-repressible plasmids pCM190-Dbf4-109, -178, -248, and -296, or the unmodified vector (V), and grown to saturation in the presence of the Tet analogue Dox. Whereas all transformants grew well on Dox-containing plates, the induction of the 296- and 248-aa fragments of Dbf4p on plates lacking drug dramatically inhibited cell growth (Fig. 3A). No arrest was observed for cultures expressing the peptides of 178 or 109 aa, or by the vector alone (Fig. 3A). Liquid growth assays yielded similar results: the 296- and 248-expressing cultures grow very poorly relative to that observed for cultures transformed with pCM190 alone or expressing smaller Dbf4 domains (Fig. 3B). Growth inhibition was also observed when the Dbf4-248 fragment was overexpressed in synchronized cell populations released from either an α-factor or nocodazole block, with the strongest effect on cells recovering from nocodazole-induced mitotic arrest (Fig. 3B; quantitation shows a 6-fold increase in doubling time). In contrast, overexpression of full-length Dbf4p has no negative effect on cell growth or viability (ref. 13, and data not shown). In further analyses, we used the Dbf4-248 fragment, because it is more stable, binds both ORC and Rad53p, and has the same growth defects as Dbf4-296 (data not shown).
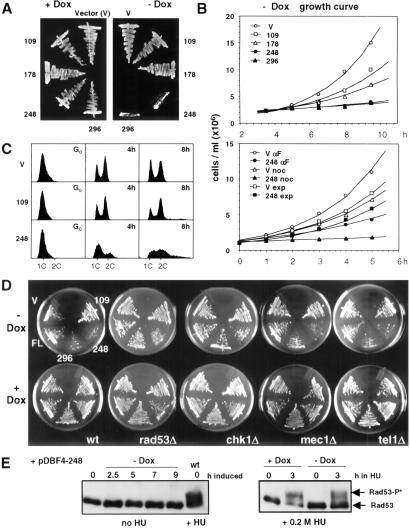
Fig 3.
Overexpression of the ARS-interacting domain of Dbf4p in WT cells slows down the cell cycle. (A) Induction of Dbf4p fragments from pCM190-derived plasmids as described in Fig. 1. Expression of Dbf4-248 and -296 in GA-850 cells, but not of -109 and -178, arrests cell growth on plates without uracil and Dox. (B) Growth curves of GA-1020 cells expressing the Dbf4p fragments. (Upper) Cells expressing the indicated Dbf4 fragment were grown to saturation in Dox before being diluted to 2 × 106 cells/ml (t0) in synthetic medium without Dox. (Lower) GA-1020 cells carrying the empty vector (V), or pCM190-Dbf4-248, were grown in media with Dox, before a 2-h induction of cell cycle arrest by α-factor or nocodazole on media -Dox (2). Cells were then released into media without Dox and without inhibitors, and growth rates were determined by using a Casy cell counter. (C) FACS analysis of GA-850 cells transformed either with vector alone, or expressing either Dbf4-109 or Dbf4-248 fragments at t0 (G0 cells) or after 4 or 8 h outgrowth in the absence of drug, from a stationary culture. (D) pCM-190 (V) and Dbf4 fragment expression vectors described in Fig. 1 were used to transform rad53Δ sml1Δ (GA-1701), mec1Δ sml1Δ (GA-1702), tel1Δ (GA-856), and chk1Δ (GA-2012) strains. These were streaked out along with GA-850 transformants (WT), on plates without uracil and Dox , as in A. (E) Rad53p phosphorylation (P*) response in cells expressing Dbf4-248 fragment (−Dox) with or without 0.2 M HU is monitored by SDS/PAGE. GA-1040 (Rad53-13Myc tagged) was transformed with pCM190-Dbf4-248. Cells were cultured in SD medium without uracil and with 3 μg/ml Dox, and released into media lacking the drug for indicated times. Where indicated, 0.2 M HU was added.
Fluorescence-activated cell sorter (FACS; Becton Dickinson) analysis of cells expressing the Dbf4-248 fragment (Fig. 3C) shows an accumulation of cells between 1C and 2C DNA content at 4 and 8 h, consistent with a defect in passage through S phase. In addition, cells with 2C DNA content accumulate as dumbbell-shaped pairs of cells containing partially segregated chromosomes (data not shown), suggestive of defects in mitotic progression. In contrast, cells expressing the Dbf4-109 fragment have relatively normal 1C and 2C peaks.
A possible explanation for both the delayed S phase progression and arrest with 2C DNA content, would be activation of the DNA damage checkpoint (36). We therefore induced Dbf4-248 expression in a number of strains carrying deletions in checkpoint genes to see whether these mutations eliminate the cell cycle arrest (Fig. 3D). Strikingly, in a rad53Δ sml1Δ background, growth inhibition by Dbf4-248 is bypassed. Similar results were obtained when Dbf4-248 was overexpressed in a rad53-11 strain (data not shown) and in strains deleted for the second effector kinase of the cell cycle checkpoint, chk1 (Fig. 3D). In contrast, cells lacking the ATM homologues Mec1p or Tel1p grow poorly when Dbf4-248 is overexpressed, like WT cells. The fact that MEC1 and TEL1 control Rad53p activation in response to DNA damage and replication blocks (36) suggests that the Dbf4-248 domain is not creating DNA damage. Rather, the rad53Δ result argues that the Dbf4-248-provoked block acts through Rad53p.
To see whether the Dbf4-248 overexpression slows growth by activating the Rad53p kinase, we tested the kinase's phosphorylation status by using SDS/PAGE. Even after 9 h of Dbf4-248 induction, the migration of Rad53p was unaltered (Fig. 3E). To see whether the replication checkpoint was intact in these cells, we added HU to cultures growing with or without Dox. Activation in the presence of HU occurs, but somewhat less efficiently, when Dbf4-248 is expressed (Fig. 3E). Thus, Dbf4-248 may partially impair Rad53p activation.
Further investigation led us to test the effects of Dbf4-248 in cells recovering from HU-induced checkpoint activation. We induced the fragment in α-factor-arrested cells, which were then released into 0.2 M HU. After 60 min, cells were released from the HU block, and DNA synthesis was examined both by FACS analysis and by 2D gel separation of replication intermediates (Fig. 4 A and B). By FACS, we see little or no effect of Dbf4-248 overexpression on early S phase progression, although there is a noticeable delay in passage from a G2 to G1 population (Fig. 4A). Consistently, we see that the early firing origin ARS607 initiates efficiently and with WT kinetics in cells expressing Dbf4-248. On the other hand, the appearance of the bubble arc at the late firing origin (ARS603) is significantly perturbed (Fig. 4B). Rather than a synchronous activation 40 min after HU release (e.g., cells transformed with empty pCM190), ARS603 is not fully inhibited on HU, and then fires in an unsynchronized fashion for up to 80 min after release (open arrowheads, Fig. 4B). Weak bubble arcs are evenly distributed over time, suggesting that the timing of initiation is altered in two ways. First, the down-regulation of late firing origins seems to be partially impaired, consistent with the fact that Rad53p activation is slightly reduced on Dbf4-248 overexpression (Fig. 3E). Second, the coordinated activation of late origins after recovery from HU is impaired. To explain this result, we propose that Dbf4-248 fragment interferes with the rebinding of Cdc7/Dbf4 to late origins, an event thought necessary to activate late initiation events on recovery from HU arrest (Fig. 5).
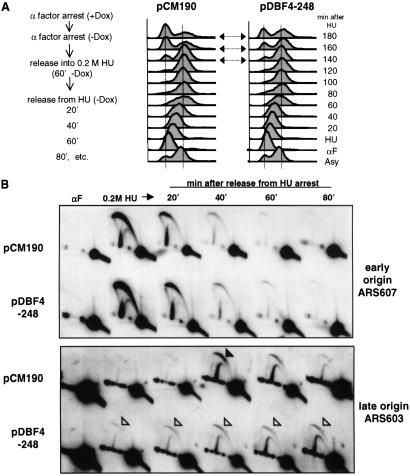
Fig 4.
Dbf4p N-terminal fragment overexpression results in deregulation of late-origin firing after HU arrest. (A) GA-1040 cells transformed with either pCM190 or pCM-Dbf4-248 were cultured in SD medium without uracil and with 3 μg/ml Dox and were arrested with α-factor for 1.2 h. During α-factor arrest, Dbf4 fragments were induced for 45 min by removing Dox. Cells were harvested and resuspended in SD medium without uracil and without Dox, but containing 0.2 M HU. After 1 h, cells were centrifuged and resuspended in SD medium without uracil and without Dox, and progression through S phase was monitored by FACS analysis at the indicated times. An asynchronous culture of the same cells (Asy) and the samples arrested in pheromone (αF) or HU were also analyzed. (B) The samples taken up to 80 min after release into HU were examined by 2D gel analysis as described in Methods. Probes specific for the early ARS607 and late ARS603 were created by PCR. Open arrowheads indicate weak bubble arcs resulting from the initiation of replication, whereas the filled arrowhead indicates efficient initiation events.
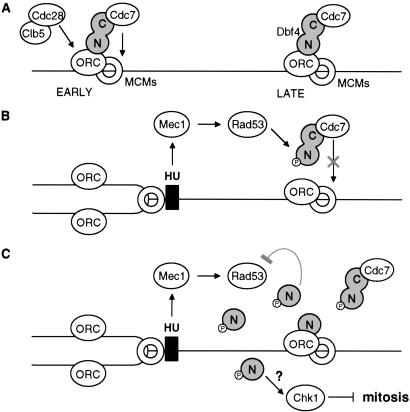
Fig 5.
Model for the role of the Dbf4-Rad53p and Dbf4-ORC interactions in the control of late-firing origins. (A) Under normal conditions, Dbf4p targets Cdc7 to origins of replication through an interaction with ORC, and the recruitment or activation of Cdc28/Clb5 promotes origin firing. (B) Exposure to HU results in replication fork stalling and activation of Rad53p. Rad53p phosphorylates Dbf4p, preventing its association with late replication origins, and attenuating kinase activity (3). During recovery, Dbf4 is dephosphorylated. We propose that it then rebinds ORC and phosphorylates Mcm2p, leading to initiation at late origins. We do not know whether the Dbf4N-ORC interaction mutually excludes binding to Rad53p. (C) If HU arrest occurs in the presence of overexpressed Dbf4-N terminus, its binding to Rad53p and ORC may interfere with the proper activation of the checkpoint and recovery after HU removal. Consistent with the idea that phosphorylated Dbf4p may signal to downstream checkpoint targets, we suggest here that it activates Chk1p kinase to impair mitotic progression.
Discussion
The Cdc7p kinase fulfills its replicative function at the level of individual origins, and its activity is required throughout S-phase for programmed, sequential initiation events to occur (7, 8). Previous work indicated that its regulatory subunit, Dbf4p, might target Cdc7p to origins in an ORC- and cell cycle-dependent manner (2). The present study shows that Dbf4p physically associates with ORC subunits through its N-terminal domain (16). This finding does not exclude the possibility that Dbf4p might also bind the pre-RC through interaction with its target, the MCM complex. Nonetheless, these data explain the published observation that a significant amount of Dbf4p can be bound to an ORC-containing chromatin fraction in G1 phase in the absence of MCM loading (i.e., in a cdc6 mutant; ref. 2). The fact that Dbf4p is highly labile, and is found in diminishing levels in stationary phase cells, confirms its key role as a limiting factor in the establishment of initiation competent origins of replication.
In contrast, work in Xenopus egg extracts has demonstrated that the association of Cdc7p with licensed origins does not require the continued presence of ORC, once the MCM complex is loaded (37, 38). This difference could simply reflect divergent pathways for Dbf4/Cdc7 localization (39), or the absence of Dbf4-ORC binding in embryonic extracts may reflect the shortened G1 phase in embryos, a stage in which this interaction may be favored. The affinity of the budding yeast Dbf4/Cdc7 complex for ORC may enhance the targeting of the kinase to origins, ensuring a highly localized action on MCM proteins. This pathway may be particularly important for the regulation of late firing origins during the recovery from checkpoint activation (Fig. 5).
It is well-established that the positioning of Dbf4/Cdc7 at individual origins affords “local control” over the timing of firing, because initiation of replication fails to occur at late origins in cdc7 mutants shifted to nonpermissive temperature mid-way through S-phase (7, 8). In cases of replication fork stalling or DNA damage, a block to late origin firing is likely to help cell survival by slowing S phase until dNTP levels are increased, or until repairs have been effected. The Rad53p checkpoint kinase is required for recovery from a replication block (40), and seems to stabilize stalled replication forks (30). Previous data have shown that Dbf4p is phosphorylated and displaced from chromatin in a Rad53-dependent manner after HU treatment (2, 3). Moreover, Rad53p-mediated phosphorylation reduces the kinase activity of Dbf4/Cdc7 (3). Finally, Rad53p was reported to bind Dbf4p in a two-hybrid assay (32) and helps inhibit late-firing origins under conditions of DNA damage or replication fork stalling (29, 31, 41).
Strong evidence for the physiological significance of the Rad53/Dbf4p interaction, whether it is direct or indirect, is provided by our in vivo studies. Overexpression of the N-terminal domain of Dbf4p, which is both sufficient and necessary to interact with the Rad53p FHA1 domain in pull-down and two-hybrid assays, results in an arrest of cell growth, defects in passage through S phase, and the accumulation of cells with pronounced mitotic defects (Fig. 3 and data not shown). Importantly, cells overexpressing Dbf4-248, show precocious firing of the late origin ARS603 during and after HU treatment (Fig. 4), consistent with the notion that interaction between this Dbf4 fragment and Rad53p can inhibit the latter's ability to regulate endogenous Dbf4/Cdc7 association with late-firing origins (Fig. 5C).
The Dbf4 N-terminal domain is necessary and sufficient to interact with Orc2p, and this interaction seems to be sensitive to conditions that activate the DNA replication checkpoint. We propose, therefore, that overexpressed Dbf4-248 may also perturb the proper loading of Dbf4/Cdc7 at origins, both in G1 and as cells recover from HU, a treatment that releases Cdc7/Dbf4 (2). This result is supported by the finding that cells blocked in nocodazole before fragment overexpression show a more dramatic inhibition of growth than those released from α-factor, or cells from an exponentially growing culture (Fig. 3B). Cells recovering from nocodazole arrest would have the largest opportunity for Dbf4-248 interference, because these cells do not synthesize endogenous Dbf4p until late G1 phase. In contrast, pheromone-arrested cells already contain a population of “licensed” pre-RCs that may bind Dbf4/Cdc7 stably, rendering the N-terminal fragment inefficient at displacing bound complex.
One explanation for the mitotic arrest observed in cells overexpressing Dbf4-248 could be the induction of damage or S-phase defects that activate a checkpoint response. However, Rad53p phosphorylation is not observed after Dbf4-248 overexpression (Fig. 3E), and growth remains poor in either a mec1Δ or tel1Δ background, which are sensors of DNA damage (Fig. 3D). This finding implies that the Dbf4-248 fragment acts downstream of the ATM homologues in a step that is linked to Rad53p. Dbf4-248 fragment overexpression in cells lacking Rad53p restores normal growth rates. One possibility is that the Rad53p-dependent phosphorylation of the Dbf4p N terminus causes the latter to act as a signal for metaphase arrest, working with other transducers such as Chk1p (Fig. 5C). Intriguingly, sequence comparisons have revealed that the Dbf4p homologue in Schizosaccharomyces pombe, Dfp1, has a conserved motif corresponding to amino acid 135–179 in Dbf4p (42, 43). Mutation of this S. pombe motif results in a checkpoint defect after HU treatment (44). There is also significant homology between this region and BRCT motifs found in numerous repair and replication proteins (44).
As previously reported by others, we found that overexpression of full-length Dbf4p was not inhibitory to cell cycle progression (Fig. 3D). This finding can be accounted for by the fact that full-length Dbf4p is rapidly degraded when it is not complexed with Cdc7p, whereas the Dbf4-248 fragment is relatively stable (ref. 27, and data not shown).
In conclusion, we present data supporting the existence of a direct interaction between the N-terminal domain of Dbf4p and the Orc2p and Orc3p subunits, as well as with Rad53p. The results of in vivo overexpression studies suggest that these interactions are involved in both the down-regulation of initiation and the resumption of replication initiation under conditions of replication fork stalling and DNA damage.
Acknowledgments
We thank M. Georgiou, M. Ramer, and D. Schnurr (University of Waterloo, Canada) for technical assistance. We thank R. Brent (Molecular Sciences Institute, Berkeley, CA) for two-hybrid strains and plasmids, K. Nasmyth (Institute of Molecular Pathology, Vienna) for strain K6388 (GA-850), and E. Schwob (Centre National de la Recherche Scientifique, Montpellier, France) for the Dbf4-Myc fusion. B.P.D. was a fellow of the National Cancer Institute of Canada, was supported by funds of the Terry Fox Run, and acknowledges support from the Canadian Natural Sciences and Engineering Research Council. P.P. thanks Association pour la Recherche sur le Cancer, the European Molecular Biology Organization, and Roche Foundation fellowships. K.S. thanks the International Agency for Research on Cancer for support. The Gasser laboratory is funded by the Swiss National Science Foundation and the Swiss Cancer League.
Abbreviations
ORC, origin recognition complex
HU, hydroxyurea
IP, immunoprecipitation
Tet, tetracycline
Dox, doxycycline
WCE, whole cell extract
ACS, ARS consensus sequence
FHA, forkhead homology-associated
FACS, fluorescence-activated cell sorter
MCM, mini-chromosome maintenance
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Diffley J. F. X. (1998) Curr. Biol. 8, R771-R773. [DOI] [PubMed] [Google Scholar]
- 2.Pasero P., Duncker, B. P., Schwob, E. & Gasser, S. M. (1999) Genes Dev. 13, 2159-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinreich M. & Stillman, B. (1999) EMBO J. 18, 5334-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira M. F., Santocanale, C., Drury, L. S. & Diffley, J. F. (2000) Mol. Cell. Biol. 20, 242-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman K. L., Diller, J. D., Ferguson, B. M., Nyland, S. V., Brewer, B. J. & Fangman, W. L. (1996) Genes Dev. 10, 1595-1607. [DOI] [PubMed] [Google Scholar]
- 6.Raghuraman M. K., Winzeler, E. A, Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J. & Fangman, W. L. (2001) Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- 7.Donaldson A. D., Fangman, W. L. & Brewer, B. J. (1998) Genes Dev. 12, 491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousset K. & Diffley, J. F. X. (1998) Genes Dev. 12, 480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson A. L., Pahl, P. M. B., Harrison, K., Rosamond, J. & Sclafani, R. A. (1993) Mol. Cell. Biol. 13, 2899-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei M., Kawasaki, Y., Young, M. R., Kihara, M., Sugino, A. & Tye, B. K. (1997) Genes Dev. 11, 3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kihara M., Nakai, W., Asano, S., Suzuki, A., Kitada, K., Kawasaki, Y., Johnston, L. H. & Sugino, A. (2000) J. Biol. Chem. 275, 35051-35062. [DOI] [PubMed] [Google Scholar]
- 12.Tye B. K. (1999) Annu. Rev. Biochem. 68, 649-686. [DOI] [PubMed] [Google Scholar]
- 13.Nougarede R., Della Seta, F., Zarzov, P. & Schwob, E. (2000) Mol. Cell. Biol. 20, 3795-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owens J. C., Detweiler, C. S. & Li, J. J. (1997) Proc. Natl. Acad. Sci. USA 94, 12521-12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou L. & Stillman, B. (2000) Mol. Cell. Biol. 20, 3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowell S. J., Romanowski, R. & Diffley, J. F. X. (1994) Science 265, 1243-1246. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel F. M., Brent, R., Kinston, R., Moore, D., Seidman, J. J., Smith, J. & Struhl, K., (1994) Current Protocols in Molecular Biology (Wiley, New York).
- 18.Frei C. & Gasser, S. M. (2000) Genes Dev. 14, 81-96. [PMC free article] [PubMed] [Google Scholar]
- 19.Weinert T. A., Kiser, G. L. & Hartwell, L. H. (1994) Genes Dev. 15, 652-665. [DOI] [PubMed] [Google Scholar]
- 20.Gyuris J., Golemis, E., Chertkov, H. & Brent, R. (1993) Cell 75, 791-803. [DOI] [PubMed] [Google Scholar]
- 21.Allen J. B., Zhou, Z., Siede, W., Friedberg, E. C. & Elledge, S. J. (1994) Genes Dev. 8, 2401-2415. [DOI] [PubMed] [Google Scholar]
- 22.Zheng P., Fay, D. S., Burton, J., Xiao, H., Pinkham, J. L. & Stern, D. F. (1993) Mol. Cell. Biol. 13, 5829-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z., Hsiao, J., Fay, D. S. & Stern, D. F. (1998) Science 281, 272-274. [DOI] [PubMed] [Google Scholar]
- 24.Adams A., Gottschling, D. E., Kaiser, C. A. & Stearns, T., (1997) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 25.Rose M. D., Winston, F. & Hieter, P., (1990) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 26.Gari E., Piedrafita, L., Aldea, M. & Herrero, E. (1997) Yeast 13, 837-848. [DOI] [PubMed] [Google Scholar]
- 27.Hardy C. F. J. & Pautz, A. (1996) Mol. Cell. Biol. 16, 6775-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowndes N. F. & Murguia, J. R. (2000) Curr. Opin. Genet. Dev. 10, 17-25. [DOI] [PubMed] [Google Scholar]
- 29.Tercero J. A & Diffley, J. F. (2001) Nature 412, 553-557. [DOI] [PubMed] [Google Scholar]
- 30.Lopes M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C. S. & Foiani, M. (2001) Nature 412, 557-561. [DOI] [PubMed] [Google Scholar]
- 31.Shirahige K., Hori, Y., Shiraishi, K., Yamashita, M., Takahashi, K., Obuse, C., Tsurimoto, T. & Yoshikawa, H. (1998) Nature 395, 618-621. [DOI] [PubMed] [Google Scholar]
- 32.Dohrmann P. R., Oshiro, G., Tecklenburg, M. & Sclafani, R. A. (1999) Genetics 151, 965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durocher D., Henckel, J., Fersht, A. R. & Jackson, S. P. (1999) Mol. Cell 4, 387-394. [DOI] [PubMed] [Google Scholar]
- 34.Liao H., Byenon, I. J. L. & Tsai, M. D. (1999) J. Mol. Biol. 294, 1041-1049. [DOI] [PubMed] [Google Scholar]
- 35.Liao H., Yuan, C., Su, M. I., Yongkiettrakul, S., Qin, D., Li, H., Byenon, I. J. L., Pei, D. & Tsai, M. D. (2000) J. Mol. Biol. 304, 941-951. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez Y., Desany, B. A., Jones, W. J., Liu, Q., Wang, B. & Elledge, S. J. (1996) Science 271, 357-360. [DOI] [PubMed] [Google Scholar]
- 37.Jares P. & Blow, J. J. (2000) Genes Dev. 14, 1528-1540. [PMC free article] [PubMed] [Google Scholar]
- 38.Walter J. C. (2000) J. Biol. Chem. 275, 39773-39778. [DOI] [PubMed] [Google Scholar]
- 39.Jares P., Donaldson, A. & Blow, J. J. (2000) EMBO Reports 1, 319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desany B. A., Alcasabas, A. A., Bachant, J. B. & Elledge, S. J. (1998) Genes Dev. 12, 2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santocanale C. & Diffley, J. F. X. (1998) Nature 395, 615-618. [DOI] [PubMed] [Google Scholar]
- 42.Landis G. & Tower, J. (1999) Development (Cambridge, U.K.) 126, 4281-4293. [DOI] [PubMed] [Google Scholar]
- 43.Ogino K., Takeda, T., Matsui, E., Iiyama, H., Taniyama, C., Arai, K. & Masai, H. (2001) J. Biol. Chem. 276, 31376-31387. [DOI] [PubMed] [Google Scholar]
- 44.Takeda T., Ogino, K., Matsui, E., Cho, M. K., Kumagai, H., Miyake, T., Arai, K. & Masai, H. (1999) Mol. Cell. Biol. 19, 5535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]