Abstract
The low density lipoprotein (LDL) receptor plays a pivotal role in cholesterol metabolism. Inherited mutations that disturb the activity of the receptor lead to elevations in plasma cholesterol levels and early-onset coronary atherosclerosis. Defects in either the LDL receptor or apolipoprotein B, the proteinaceous component of LDL particles that binds the LDL receptor, elevate circulating LDL-cholesterol levels in an autosomal-dominant fashion, with heterozygotes displaying values between homozygous and normal individuals. Rarely, similar clinical phenotypes occur with a recessive pattern of inheritance, and several genetic lesions in the autosomal recessive hypercholesterolemia (ARH) gene on chromosome 1 have been mapped in this class of patients. ARH has an N-terminal phosphotyrosine-binding (PTB) domain evolutionarily related to that found in Disabled-2 and numb, two endocytic proteins. PTB domains bind to the consensus sequence FXNPXY, corresponding to the internalization motif of the LDL receptor. We show here that in addition to the FXNPXY sequence, ARH binds directly to soluble clathrin trimers and to clathrin adaptors by a mode involving the independently folded appendage domain of the β subunit. At steady state, ARH colocalizes with endocytic proteins in HeLa cells, and the LDL receptor fluxes through peripheral ARH-positive sites before delivery to early endosomes. Because ARH also binds directly to phosphoinositides, which regulate clathrin bud assembly at the cell surface, our data suggest that in ARH patients, defective sorting adaptor function in hepatocytes leads to faulty LDL receptor traffic and hypercholesterolemia.
Transmembrane nutrient receptors, like the low density lipoprotein (LDL) receptor, deliver extracellular macromolecules to the cell interior by the process of endocytosis. Preferential recognition of internalization information, in the form of specific sequence motifs embedded within the cytosolic domain, packages the receptor into a transport intermediate, the clathrin-coated vesicle. The LDL receptor utilizes an atypical tyrosine-based internalization sequence, FDNPVY, to direct cellular uptake of LDL (1). Although this was the first tyrosine-based endocytic motif characterized (1, 2), in mammals, the majority of endocytosed proteins use internalization sequences based upon the consensus YXXØ, where Ø represents a residue with a bulky hydrophobic side chain (3). The YXXØ sequence engages the μ2 subunit of the clathrin-associated, heterotetrameric AP-2 adaptor complex (4), but several lines of evidence argue that the FDNPVY signal engages an alternate protein surface at the clathrin bud site. First, overexpression of transferrin receptors (with a YTRF internalization sequence) does not effect the rate of LDL receptor uptake (5). Second, in contrast to the canonical YXXØ motif, the Tyr residue in the FDNPVY sequence can be altered to Phe with no measurable effect on internalization rate (2). In fact, the minimal LDL receptor internalization sequence requires six residues (6) and cannot be accommodated by the YXXØ binding site on μ2 (4, 7). Third, lymphoblasts from patients with non-LDL receptor, non-apolipoprotein B (apoB) hypercholesterolemia (autosomal recessive hypercholesterolemia, ARH) do not internalize LDL but take up transferrin normally and have no mutations in the AP-2 μ2-subunit gene (8). Genetic lesions located at 1p36–1p35 occur in this class of patients (9–11), and the mutated gene, ARH, encodes a 308-aa protein undetectable in several ARH patients' fibroblasts (10).
Tracer studies show that ARH patients have seriously diminished hepatic uptake of circulating LDL (12). As ARH patients phenocopy familial hypercholesterolemia, it seems likely that ARH might participate in the endocytic activity of the LDL receptor in hepatocytes (9), which remove the bulk of plasma LDL. ARH is characterized by a phosphotyrosine-binding (PTB) domain (9) (Fig. 1A). The generic PTB fold binds to FXNPXpY motifs (where pY is phosphotyrosine) (13) but some, like that of Disabled-1 and -2 (Dab1/2) (14, 15), have a much better selectivity for the nonphosphorylated FXNPXY sequence, the LDL receptor internalization sequence (1, 13). We have focused on ARH because the PTB domain is most closely related to the PTB domains of numb, Dab1 and -2 (14–16), and GULP (17). Both numb and Dab2 are endocytic proteins that appear to expand the sorting repertoire of clathrin-coated regions at the cell surface (15, 16, 18, 19). In this study, we show that ARH is similarly a component of the endocytic machinery that likely contributes to the LDL-uptake-deficient phenotype of ARH patients.
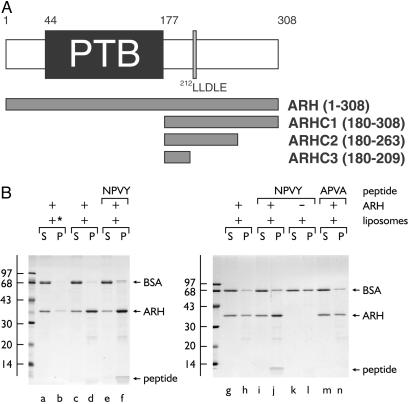
Fig 1.
ARH binds phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] and FXNPXY motifs synchronously. (A) Schematic illustration of the overall domain organization of ARH and the various GST-ARH constructs used. (B) Thrombin-cleaved ARH-(1–308) was added to 3 μM together with carrier BSA and 0.4 mg/ml control (asterisk, lanes a and b) or PtdIns(4,5)P2-containing liposomes alone (lanes c, d, g, and h) or liposomes plus either 15 μM NWRLKNINSIFDNPVYQKTT (lanes e, f, i–l) or NWRLKNINSIFDAPVAQKTT (lanes m and n) peptide as indicated and incubated on ice for 60 min. After centrifugation, aliquots of 1/25th of each supernatant (S) or one-fourth of each pellet (P) were analyzed by SDS/PAGE and stained with Coomassie blue. The bound peptide migrates at the dye front and the position of the molecular mass standards (in kDa) is indicated on the left.
Materials and Methods
DNA Manipulations.
All ARH clones were prepared by PCR using appropriate primers and full-length placental EST AL545069 (Invitrogen) as a template. Products were ligated into pGEX-4T-1 and mutations were introduced into the appropriate plasmids by using the QuikChange (Stratagene) system with the required mutagenic primer pairs. A glutathione S-transferase (GST)-β2 appendage (residues 714–951) fusion was prepared similarly by using the rat β2 cDNA as a template. Plasmids encoding GST fused to the epsin 1 DPW domain and the αC appendage have been described (20, 21), and the GST-γ1 appendage (mouse residues 703–822) in pGEX-5X-3 (22) was kindly provided by Stuart Kornfeld. All clones and mutations were verified by automated sequencing.
Protein Preparation.
GST and the various GST-fusion proteins were prepared exactly as described (16, 20, 23). Rat brain, liver, and placental cytosol was prepared from frozen tissue in 25 mM Hepes-KOH, pH 7.2/250 mM sucrose/1 mM EDTA and adjusted to assay buffer (25 mM Hepes-KOH, pH 7.2/125 mM potassium acetate/5 mM magnesium acetate/2 mM EDTA/2 mM EGTA/1 mM DTT) before use. Clathrin-coated vesicles from rat brain or liver were purified as described (24), using discontinuous sucrose gradients to separate the liver coated vesicles from contaminating vaults.
Binding Assays.
Pull-down-type assays, liposome-binding assays, and inhibition assays were performed in assay buffer exactly as published (16, 20, 21, 23) and detailed in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Antibodies.
Polyclonal serum directed against GST-ARH was raised in rabbits (Research Genetics), and affinity-purified antibodies were prepared by using thrombin-cleaved ARHC1 immobilized on CNBr-activated Sepharose 4B. Other antibodies used have been described (16, 20, 21, 23) and are detailed in the supporting information on the PNAS web site.
Cell Culture and Immunofluorescence Microscopy.
HeLa SS6 cells were cultured in DMEM supplemented with 10% FCS and 2 mM l-glutamine at 37°C in a 10% CO2/90% air atmosphere. For experiments with mAb IgG-C7, cells were cultured in 8% lipoprotein-deficient serum (LPDS) for 44 h to up-regulate the LDL receptor. ARH3 family fibroblasts GM00695A (father), GM00696 (affected sister), and GM00697 (affected brother) were obtained from the Human Genetic Cell Repository at the Coriell Institute. The cells were cultured in Eagle's MEM supplemented with 15% FCS, 2 mM l-glutamine, and 2× nonessential amino acids at 37°C in a 5% CO2/95% air atmosphere. Prefixation permeabilization and formaldehyde fixation (16, 23) were used before incubation with primary antibody mixtures. Complete details are provided in the supporting information on the PNAS web site.
Results
Over a core segment of about 160 residues, the ARH PTB domain is 31% identical (46% similar) and 22% identical (46% similar) to the endocytic proteins numb and Dab2, respectively. As many of the conserved residues are Lys or Arg, and both the numb (25) and Dab2 (16) PTB domains bind to acidic phosphoinositides, we first assessed whether ARH can bind to synthetic liposomes containing phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2]. In sedimentation assays, a portion of added ARH-(1–308) cofractionates with the liposomes (Fig. 1B, lanes d and h), whereas in the absence of PtdIns(4,5)P2 the recovery of ARH is diminished (lane b). In both Dab1 (14) and Dab2 (16), PtdIns(4,5)P2 binding occurs concurrently with engagement of the nonphosphorylated form of the FXNPXY sequence. The same is true for ARH. Addition of a peptide corresponding to the first 20 residues of the cytosolic domain of the LDL receptor, encompassing the FDNPVY sequence, results in recovery of the peptide, together with ARH, in the liposome pellet (lanes f and j). No peptide sediments with the liposomes in the absence of added ARH (lane l) or when the FDNPVY is altered to an endocytically inactive sequence FDAPVA(lane n). These experiments confirm that, like the endocytic proteins numb (18, 19) and Dab2 (16), ARH can engage the FXNPXY internalization sequence and inositol phospholipids.
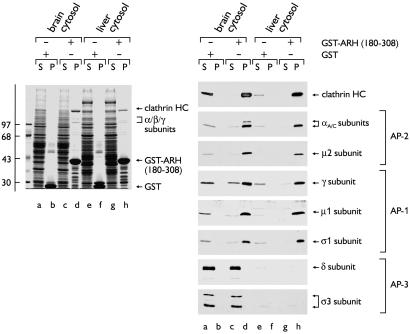
We noticed an additional endocytic-like feature within the ARH sequence; similar to AP180, epsin (21, 26), and Dab2, the C-terminal region (amino acids 183–308) is predicted to be relatively unstructured and harbors a putative type-I clathrin box (Fig. 1A). The sequence, 212LLDLE, adheres to the general clathrin-binding consensus L(L/V/I)(D/E/N)(L/F)(D/E) (27, 28). When fused to GST, the C-terminal segment (residues 180–308) of ARH (GST-ARHC1) binds specifically to soluble clathrin trimers from both brain (Fig. 2, lane d) and liver cytosol (lane h); the prominent ≈180-kDa band in the pellet fractions is the clathrin heavy chain (HC). In addition, a substoichiometric group of ≈100-kDa proteins also cosediment with the immobilized GST-ARH fusion but not with GST. Subunit-specific antibodies identify these polypeptides as the large α, β1/2, and γ subunits of the heterotetrameric AP-1 and AP-2 adaptor complexes. On immunoblots, the μ2 and both μ1 and σ1 subunits are also detected in the GST-ARH-bound fraction, confirming that the adaptors associate as intact heterotetrameric complexes. The ARH fusion does not associate with the AP-3 complex, however (Fig. 2). Full-length ARH fused to GST also binds clathrin, AP-1, and AP-2, excluding that these interactions occur only with a partial segment of the protein.
Fig 2.
ARH interacts with clathrin, AP-1, and AP-2. Approximately 150 μg of either GST (lanes a, b, e, and f) or GST-ARH-(180–308) (lanes c, d, g, and h) immobilized on GSH-Sepharose was incubated with either rat brain (lanes a–d) or liver (lanes e–h) cytosol. After centrifugation, aliquots corresponding to 1/60th of each supernatant (S) and one-fifth of each washed pellet (P) were resolved by SDS/PAGE and either stained with Coomassie blue (Left) or transferred to nitrocellulose (Right). Portions of the blots were probed with mAb TD.1 [clathrin heavy chain (HC)], mAb 100/2 (α subunits), anti-μ2 serum, affinity-purified AE/1 (γ subunit), RY/1 serum (μ1 subunit), affinity-purified DE/1 (σ1 subunit) or KQ/1 (δ subunit), or anti-σ3 serum. The position of the molecular mass standards (in kDa) is indicated on the left, and only the relevant portion of each blot is shown.
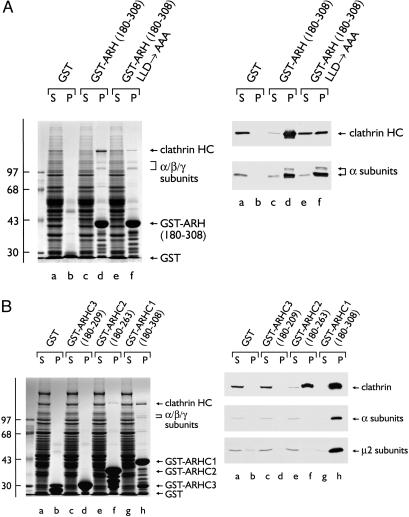
Variations of the type I sequence LLDLE, which bind to a surface groove in the β-propeller of the clathrin HC terminal domain (29), are found in many endocytic proteins (30, 31). To test the contribution of this ARH sequence to clathrin binding, the effect of Ala substitutions (LLD → AAA) in the context of the ARH-(180–308) fusion was determined. Disruption of the LLDLE sequence markedly attenuates, but does not abolish, clathrin binding (Fig. 3A). Yet the mutated fusion still displays unaltered adaptor-binding capacity, demonstrating that, as in several other so-called endocytic accessory proteins (amphiphysin, AP180, Dab2, epsin, and HIP1) (32), the clathrin- and adaptor-binding determinants are adjacent but separate (31). Additional evidence for discrete binding sites comes from truncation analysis of the C-terminal ARH segment (Fig. 3B). Removal of the last 45 aa of ARH (180–263, GST-ARHC2) abolishes adaptor interactions while preserving clathrin binding. The difference in the clathrin-binding capacity of the GST-ARHC2 compared with GST-ARHC1, and the residual clathrin binding seen with the LLD → AAA substitution, indicate that the region between residues 263 and 308 plays some role in maximal clathrin binding. The C-terminal DLL-like triplet 304DLF might contribute to clathrin binding (33). This analysis clearly shows that, like several endocytic proteins, ARH has separate clathrin- and adaptor-binding determinants. In other experiments (see Supporting Results and Fig. 9, which are published as supporting information on the PNAS web site), we established that ARH binds directly to the independently folded β2 appendage domain, most probably accounting for the ability of ARH to engage both AP-1 and AP-2. In sum, our experiments reveal that ARH shares a modular architecture and several functional attributes with endocytic proteins, including Dab2.
Fig 3.
(A) A type-I clathrin box. Approximately 150 μg of GST (lanes a and b), GST-ARH-(180–308) (lanes c and d), or GST-ARH-(180–308) LLD → AAA (lanes e and f) immobilized on GSH-Sepharose was incubated with rat brain cytosol. After centrifugation, aliquots corresponding to 1/60th of each supernatant (S) and one-fifth of each washed pellet (P) were resolved by SDS/PAGE and either stained with Coomassie blue (Left) or transferred to nitrocellulose (Right). Portions of the blots were probed with the anti-clathrin HC mAb TD.1 or anti-AP-2 α-subunit mAb 100/2. (B) Approximately 100 μg of GST (lanes a and b), GST-ARHC3-(180–209) (lanes c and d), GST-ARHC2-(180–263) (lanes e and f), or GST-ARHC1-(180–308) (lanes g and h) immobilized on GSH-Sepharose was incubated with rat liver cytosol. After centrifugation, aliquots corresponding to 1/60th of each supernatant (S) and one-fifth of each washed pellet (P) were resolved by SDS/PAGE and either stained with Coomassie blue (Left) or transferred to nitrocellulose (Right). Portions of the blots were probed with the anti-clathrin HC mAb TD.1, anti-AP-2 α-subunit mAb 100/2, or anti-AP-2 μ2-subunit antiserum.
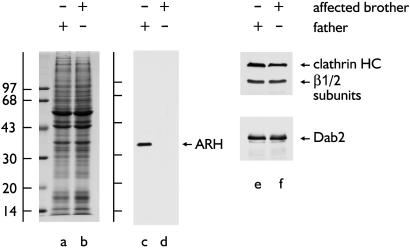
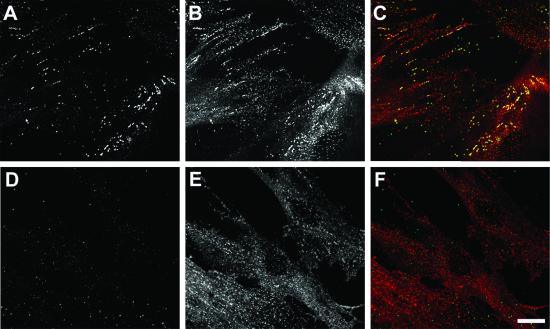
To verify the association of ARH with the endocytic machinery in vivo, we generated affinity-purified anti-ARH antibodies and first checked for immunoreactivity in cultured fibroblasts from the Lebanese ARH3 family (34) used in the initial characterization of ARH (9). On immunoblots, a single polypeptide of ≈35 kDa is present in the father (Fig. 4, lane c) but is undetectable in cell extracts from an affected (homozygous) brother (lane d) or sister (not shown). The overall protein profile (lanes a and b) and abundance of clathrin, AP-1/2, and Dab2 (lane e and f) in the two lysates is indistinguishable. These results agree well with only trace mRNA levels in the affected ARH3 siblings (9). In fibroblasts from the heterozygous ARH3 father, ARH is distributed in bright, scattered puncta, often arranged in linear arrays (Fig. 5A). These structures colocalize precisely with clathrin on the basal plasma membrane (Fig. 5 B and C). Interestingly, not all clathrin-containing sites at the cell surface are ARH positive; only a subset of clathrin label, which is generally brighter, contains ARH. This does not appear to be a gene-dosage effect, as similar results are seen in normal fibroblasts (not shown).
Fig 4.
ARH expression. Equal aliquots of total cell lysate from father (GM00965A) or affected brother (GM00697) were fractionated by SDS/PAGE and either stained with Coomassie blue (lanes a and b) or transferred to nitrocellulose (lanes c–f). Blots were probed with anti-ARH (c and d) or anti-clathrin HC mAb TD.1 and anti-β-subunit mAb 100.1 or anti-Dab2 antibodies (e and f).
Fig 5.
Intracellular localization. Cultured fibroblasts from either the asymptomatic heterozygous father (A–C) or ARH-affected (homozygous) brother (D–F) were permeabilized, fixed, and probed with affinity-purified anti-ARH antibodies (green) and anti-clathrin HC mAb X22 (red), followed by Alexa 488-labeled anti-rabbit IgG and Cy3-labeled anti-mouse IgG secondary antibodies. A single optical section at the base of the cells is shown. (Bar = 20 μm.)
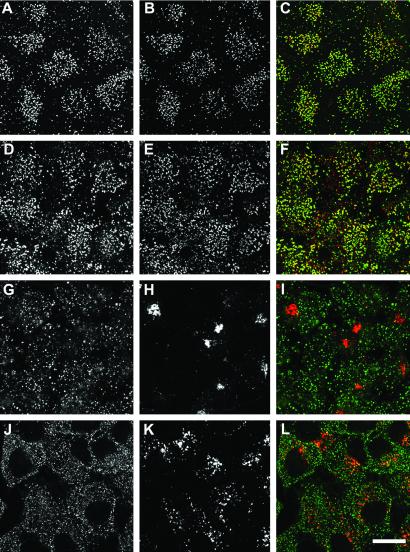
The clathrin distribution in the homozygous ARH3 siblings is similar to that in the father, but the ARH labeling is essentially background (Fig. 5 D and F). Close subcellular colocalization of ARH with clathrin can also be clearly seen on the basal surface membrane of HeLa cells that were mechanically sheared during staining (Fig. 6 A–C). In the HeLa cells, the correlation between ARH and clathrin is more complete and is very similar to the intracellular colocalization of ARH with AP-2 at the ventral cell surface of nonruptured cells (Fig. 6 D–F). We notice that the close association of ARH with the endocytic components is always greatest at the basal cell surface. Despite its being able to bind to AP-1 subunits in vitro, we find no evidence of ARH at the Golgi apparatus at steady state (Fig. 6 G–I). The distribution of ARH is also clearly distinct from that of EEA1, an early endosome marker (Fig. 6 J–L). Thus, the intracellular distribution of ARH agrees well with the in vitro binding partner analysis.
Fig 6.
Subcellular colocalization. Fixed and permeabilized HeLa cells were incubated with anti-ARH antibodies (A, D, G, and J) (green) and anti-clathrin HC mAb X22 (B), anti-AP2 α-subunit mAb AP.6 (E), an anti-AP-1 γ-subunit mAb (H), or an anti-EEA1 mAb (K) (red). Single optical sections near the base of the adherent cells are shown with merged images in C, F, I, and L. (Bar = 20 μm.)
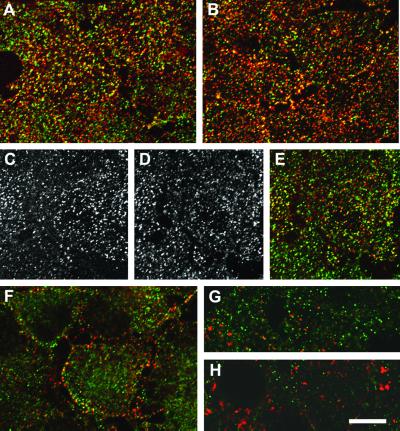
These localization studies reveal that ARH is suitably positioned to participate in LDL receptor uptake. Indeed, the LDL receptor fluxes through an ARH-positive compartment on the way from the plasma membrane to the cell interior. To label the surface LDL receptor population, we first cultured HeLa cells in LPDS to up-regulate receptor expression and then incubated the cells with an anti-receptor mAb (IgG-C7) on ice. On warming the cells to 37°C for 30 sec, a high degree of colocalization of the LDL receptor and ARH is observed (Fig. 7A). At 1 min, while still largely colocalized, the receptor can be seen to diverge from the ARH-positive elements (Fig. 7 B–E) and, at 2 min, further separation of the two proteins is evident (Fig. 7G). After 10 min there is virtually no overlap between the two proteins (Fig. 7 H and I); the LDL receptor signal has moved into larger, more perinuclear endosomal structures reminiscent of EEA1-positive early endosomes.
Fig 7.
LDL receptor internalization. HeLa cells precultured in LPDS were incubated with anti-LDL receptor mAb IgG-C7 (red) on ice for 60 min before warming to 37°C for 30 sec (A), 1 min (B–E), 2 min (F), or 10 min (G and H). After fixation, the cells were incubated with anti-ARH antibodies (green). A single optical section at the base of the cells is shown except in H, which is a medial section. (Bar = 20 μm.)
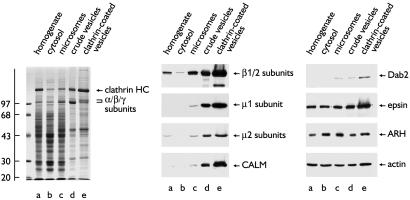
The kinetics of the intracellular juxtaposition of the LDL receptor and ARH are consistent with this occurring within clathrin-coated buds and/or vesicles. To determine whether ARH is enriched within clathrin-coated vesicles purified from liver, where ARH function seems most critical for cholesterol homeostasis, we assessed the distribution of various proteins in fractions from a coated vesicle preparation. As expected, the major polypeptide in the purified clathrin-coated vesicles is the clathrin HC (Fig. 8). AP-1 and AP-2, as shown by the β1/β2 and respective μ1 and μ2 subunits are also enriched in the coated vesicles, as is the nonneuronal AP180 orthologue, CALM, and Dab2. Surprisingly, although present, ARH is not concentrated in the coated vesicle fraction; the distribution mirrors that of actin. ARH shows a similar distribution in brain clathrin-coated vesicle preparations (not shown) suggesting that, like epsin, which also fails to be substantially enriched in coated vesicles (23, 35), ARH can be classified as an endocytic accessory protein.
Fig 8.
ARH in liver clathrin-coated vesicles. Aliquots of 20 μg of liver homogenate (lanes a), cytosol (lanes b), microsomes (lanes c), and 10 μg of crude vesicles (lanes d) or purified clathrin-coated vesicles (lanes e) were fractionated by SDS/PAGE and either stained (Left) or transferred to nitrocellulose (Center and Right). Portions of the blots were probed with mAb 100.1 (anti-β1/2-subunits), RY/1 serum (μ1 subunit), anti-AP-2 μ2 serum, mAb anti-CALM, affinity-purified anti-Dab2, anti-epsin, anti-ARH antibodies, or mAb C2 (anti-actin).
Discussion
The age of onset and overall clinical features of ARH are extremely similar to familial hypercholesterolemia, despite no defect in either LDL receptor or apoB genes (8–10, 12, 34, 36). One explanation for this could be that a key component of the endocytic machinery, dedicated primarily to the hepatic uptake of LDL, is compromised by inherited mutation. In this model, ARH phenocopies the class 4, internalization-defective, mutant form of familial hypercholesterolemia. We show here that ARH possesses an intrinsic capacity to interact with the internalization motif of the LDL receptor and harbors endogenous determinants that facilitate direct protein–protein interactions with the core endocytic components clathrin and AP-2. These biochemical results are in complete accord with a synchronous study (37). We also provide important information on the intracellular location of ARH and show that the LDL receptor encounters ARH-containing structures very early along its trajectory from the cell surface. Curiously, ARH patient fibroblasts, unlike those from familial hypercholesterolemia patients, do not show major defects in LDL uptake and degradation (9, 10, 12, 36, 38). This observation must mean that ARH is not singularly responsible for LDL uptake in all tissue types. We find that, although lacking detectable ARH, patient fibroblasts do express Dab2 at high levels. Dab2 and ARH exhibit striking structural and functional similarities. Overexpression of the PTB domain of Dab2 potently, but selectively, blocks LDL uptake (16), and the protein localizes to AP-2-containing clathrin coats at steady state (15, 16). We have proposed that Dab2 acts as a specific intermediate adaptor for LDL uptake (16) but Dab2-null mice have only a mild phenotype (39). Therefore, ARH and Dab2 could be functionally redundant; the normal LDL receptor activity in ARH patient fibroblast might be due to Dab2, or possibly numb; small inhibitory RNAs might be used to investigate this question.
If ARH is an endocytic sorting adaptor, why is the protein not enriched in either liver or brain clathrin-coated vesicles, like AP-2? We have considered two possibilities. First, ARH may not be a minor component, as our data do not establish the mass of ARH within the isolated coated vesicle fraction and we cannot infer the stoichiometry of ARH to either AP-2 or clathrin. Alternatively, ARH might actively participate in sorting, but not progress along with the cargo into the budded vesicle. There are several examples of this behavior, including eps15 and epsin. ARH would act then as a molecular usher, gathering FXNPXY-bearing cargo, like the LDL receptor, into the clathrin lattice, thereby enhancing the efficiency of packaging into the carrier vesicle. This idea is in accord with several ultrastructural studies showing LDL particles clustered around the perimeter of flat clathrin arrays and the rim of coated pits on the cell surface (40–42). In this way, ARH might transfer cargo to an intermediate adaptor, like Dab2, which is enriched in liver coated vesicles. The FXNPXY sequence has also been shown to bind weakly to a surface of the AP-2 μ2 subunit distinct from the YXXØ interaction surface (7). As the μ2 subunit requires phosphorylation to induce cargo-binding competence (43, 44), ARH could also pass the LDL receptor to AP-2 activated focally at the bud site. A similar mechanism has been proposed for GGA (Golgi-localized, γ-ear-containing, ADP-ribosylation factor-binding protein)-assisted sorting of mannose 6-phosphate receptors into AP-1-containing vesicles assembling at the trans-Golgi network (45). GGAs, although binding clathrin (46) and clearly colocalized with AP-1 in budding profiles on the trans-Golgi (45), are not present in large amounts in clathrin-coated vesicle preparations (47).
The distinct modes of adaptor engagement used by ARH and Dab2 could be a manifestation of differing roles in cargo selection. A predilection for engaging the β appendage could bias ARH for early exit from the assembling lattice. The β appendage and hinge, an unstructured linker that connects the globular appendage to the adaptor core, play an important role in recruiting clathrin the membrane bud site (48–50). A type I clathrin box, 632LLNLD, in the hinge works in conjunction with the β appendage to promote optimal clathrin binding and lattice assembly (50). Importantly, by engaging the β appendage/hinge, the clathrin HC displaces other appendage–protein interactions (50). This displacement might promote release of ARH from the clathrin lattice, whereas Dab2, which binds the α appendage, remains at the bud site and becomes incorporated into the budded coated vesicle.
Hepatocytes express much higher levels of LDL receptor than other cells, yet the steady-state protein level of ARH in liver is similar to that in other tissues (see Supporting Results and Fig. 10, which are published as supporting information on the PNAS web site). Compared with fibroblasts, Dab2 levels are also low in liver. While this fact could potentially explain the tissue-specific defects seen in ARH, it is not clear why, given the volume of LDL traffic in hepatocytes, adaptor protein levels are not commensurately elevated. Interestingly, ARH has been recently suggested to be a digenic disorder, with lesions in both ARH alleles on chromosome 1 and a second, as-yet-uncharacterized gene at 13q22-q32 required for disease (11). The second gene product, proposed to be functionally redundant with ARH (11), might be a liver-specific component. Further experiments will be required to define the precise role of PTB domain proteins in hepatic LDL receptor uptake, but our results clearly implicate ARH in endocytic cargo selection events.
Supplementary Material
Acknowledgments
We thank our colleagues for generously providing important reagents, Gerard Apodaca and members of the Traub laboratory for discussion, and Helen Hobbs for alerting us to the ARH3 fibroblasts in the Coriell repository. This work was supported in part by National Institutes of Health Grant R01 DK53249 (to L.M.T.).
Abbreviations
apoB, apolipoprotein B
ARH, autosomal recessive hypercholesterolemia
Dab, Disabled
GST, glutathione S-transferase
HC, heavy chain
LDL, low density lipoprotein
LPDS, lipoprotein-deficient serum
PTB, phosphotyrosine binding
PtdIns(4,5)P2, phosphatidylinositol 4,5-bisphosphate
References
- 1.Chen W. J., Goldstein, J. L. & Brown, M. S. (1990) J. Biol. Chem. 265, 3116-3123. [PubMed] [Google Scholar]
- 2.Davis C. G., Lehrman, M. A., Russell, D. W., Anderson, R. G., Brown, M. S. & Goldstein, J. L. (1986) Cell 45, 15-24. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino J. S. & Dell'Angelica, E. C. (1999) J. Cell Biol. 145, 923-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen D. J. & Evans, P. R. (1998) Science 282, 1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren R. A., Green, F. A., Stenberg, P. E. & Enns, C. A. (1998) J. Biol. Chem. 273, 17056-17063. [DOI] [PubMed] [Google Scholar]
- 6.Collawn J. F., Kuhn, L. A., Liu, L. F., Tainer, J. A. & Trowbridge, I. S. (1991) EMBO J. 10, 3247-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boll W., Rapoport, I., Brunner, C., Modis, Y., Prehn, S. & Kirchhausen, T. (2002) Traffic 3, 590-600. [DOI] [PubMed] [Google Scholar]
- 8.Norman D., Sun, X. M., Bourbon, M., Knight, B. L., Naoumova, R. P. & Soutar, A. K. (1999) J. Clin. Invest. 104, 619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia C. K., Wilund, K., Arca, M., Zuliani, G., Fellin, R., Maioli, M., Calandra, S., Bertolini, S., Cossu, F., Grishin, N., et al. (2001) Science 292, 1394-1398. [DOI] [PubMed] [Google Scholar]
- 10.Arca M., Zuliani, G., Wilund, K., Campagna, F., Fellin, R., Bertolini, S., Calandra, S., Ricci, G., Glorioso, N., Maioli, M., et al. (2002) Lancet 359, 841-847. [DOI] [PubMed] [Google Scholar]
- 11.Al-Kateb H., Bahring, S., Hoffmann, K., Strauch, K., Busjahn, A., Nurnberg, G., Jouma, M., Bautz, E. K., Dresel, H. A. & Luft, F. C. (2002) Circ. Res. 90, 951-958. [DOI] [PubMed] [Google Scholar]
- 12.Zuliani G., Arca, M., Signore, A., Bader, G., Fazio, S., Chianelli, M., Bellosta, S., Campagna, F., Montali, A., Maioli, M., et al. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 802-809. [DOI] [PubMed] [Google Scholar]
- 13.Yan K. S., Kuti, M. & Zhou, M. M. (2002) FEBS Lett. 513, 67-70. [DOI] [PubMed] [Google Scholar]
- 14.Howell B. W., Lanier, L. M., Frank, R., Gertler, F. B. & Cooper, J. A. (1999) Mol. Cell. Biol. 19, 5179-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris S. M. & Cooper, J. A. (2001) Traffic 2, 111-123. [DOI] [PubMed] [Google Scholar]
- 16.Mishra S. K., Keyel, P. A., Hawryluk, M. J., Agostinelli, N. R., Watkins, S. C. & Traub, L. M. (2002) EMBO J. 21, 4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su H. P., Nakada-Tsukui, K., Tosello-Trampont, A. C., Li, Y., Bu, G., Henson, P. M. & Ravichandran, K. S. (2002) J. Biol. Chem. 277, 11772-11779. [DOI] [PubMed] [Google Scholar]
- 18.Santolini E., Puri, C., Salcini, A. E., Gagliani, M. C., Pelicci, P. G., Tacchetti, C. & Di Fiore, P. P. (2000) J. Cell Biol. 151, 1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berdnik D., Torok, T., Gonzalez-Gaitan, M. & Knoblich, J. (2002) Dev. Cell 3, 221-231. [DOI] [PubMed] [Google Scholar]
- 20.Traub L. M., Downs, M. A., Westrich, J. L. & Fremont, D. H. (1999) Proc. Natl. Acad. Sci. USA 96, 8907-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brett T. J., Traub, L. M. & Fremont, D. H. (2002) Structure (Cambridge, MA) 10, 797-809. [DOI] [PubMed] [Google Scholar]
- 22.Doray B. & Kornfeld, S. (2001) Mol. Biol. Cell 12, 1925-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra S. K., Agostinelli, N. R., Brett, T. J., Mizukami, I., Ross, T. S. & Traub, L. M. (2001) J. Biol. Chem. 276, 46230-46236. [DOI] [PubMed] [Google Scholar]
- 24.Campbell C., Squicciarini, J., Shia, M., Pilch, P. F. & Fine, R. E. (1984) Biochemistry 23, 4420-4426. [DOI] [PubMed] [Google Scholar]
- 25.Dho S. E., French, M. B., Woods, S. A. & McGlade, C. J. (1999) J. Biol. Chem. 274, 33097-33104. [DOI] [PubMed] [Google Scholar]
- 26.Kalthoff C., Alves, J., Urbanke, C., Knorr, R. & Ungewickell, E. J. (2002) J. Biol. Chem. 277, 8209-8216. [DOI] [PubMed] [Google Scholar]
- 27.Krupnick J. G., Goodman, O. B., Jr., Keen, J. H. & Benovic, J. L. (1997) J. Biol. Chem. 272, 15011-15016. [DOI] [PubMed] [Google Scholar]
- 28.Dell'Angelica E. C., Klumperman, J., Stoorvogel, W. & Bonifacino, J. S. (1998) Science 280, 431-434. [DOI] [PubMed] [Google Scholar]
- 29.ter Haar E., Harrison, S. C. & Kirchhausen, T. (2000) Proc. Natl. Acad. Sci. USA 97, 1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dell'Angelica E. C. (2001) Trends Cell. Biol. 11, 315-318. [DOI] [PubMed] [Google Scholar]
- 31.Aridor M. & Traub, L. M. (2002) Traffic 3, 537-546. [DOI] [PubMed] [Google Scholar]
- 32.Slepnev V. I. & De Camilli, P. (2000) Nat. Rev. Neurosci. 1, 161-172. [DOI] [PubMed] [Google Scholar]
- 33.Morgan J. R., Prasad, K., Hao, W., Augustine, G. J. & Lafer, E. M. (2000) J. Neurosci. 20, 8667-8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khachadurian A. K. & Uthman, S. M. (1973) Nutr. Metab. 15, 132-140. [DOI] [PubMed] [Google Scholar]
- 35.Chen H., Fre, S., Slepnev, V. I., Capua, M. R., Takei, K., Butler, M. H., Di Fiore, P. P. & De Camilli, P. (1998) Nature 394, 793-797. [DOI] [PubMed] [Google Scholar]
- 36.Zuliani G., Vigna, G. B., Corsini, A., Maioli, M., Romagnoni, F. & Fellin, R. (1995) Eur. J. Clin. Invest. 25, 322-331. [DOI] [PubMed] [Google Scholar]
- 37.He, G., Gupta, S., Michaely, P., Hobbs, H. H. & Cohen, J. C. (2002) J. Biol. Chem. 277, in press. [DOI] [PubMed]
- 38.Goldstein J. L. & Brown, M. S. (2001) Science 292, 1310-1312. [DOI] [PubMed] [Google Scholar]
- 39.Morris S. M., Tallquist, M. D., Rock, C. O. & Cooper, J. A. (2002) EMBO J. 21, 1555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heuser J. & Anderson, R. G. (1989) J. Cell Biol. 108, 389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanan D. A., Van der Westhuyzen, D. R., Gevers, W. & Coetzee, G. A. (1987) Histochemistry 86, 517-523. [DOI] [PubMed] [Google Scholar]
- 42.Sanan D. A. & Anderson, R. G. (1991) J. Histochem. Cytochem. 39, 1017-1024. [DOI] [PubMed] [Google Scholar]
- 43.Collins B. M., McCoy, A. J., Kent, H. M., Evans, P. R. & Owen, D. J. (2002) Cell 109, 523-535. [DOI] [PubMed] [Google Scholar]
- 44.Olusanya O., Andrews, P. D., Swedlow, J. R. & Smythe, E. (2001) Curr. Biol. 11, 896-900. [DOI] [PubMed] [Google Scholar]
- 45.Doray B., Ghosh, P., Griffith, J., Geuze, H. J. & Kornfeld, S. (2002) Science 297, 1700-1703. [DOI] [PubMed] [Google Scholar]
- 46.Puertollano R., Randazzo, P. A., Presley, J. F., Hartnell, L. M. & Bonifacino, J. S. (2001) Cell 105, 93-102. [DOI] [PubMed] [Google Scholar]
- 47.Hirst J., Lui, W., Bright, N., Totty, N., Seaman, M. & Robinson, M. (2000) J. Cell Biol. 149, 67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahle S. & Ungewickell, E. (1989) J. Biol. Chem. 264, 20089-20093. [PubMed] [Google Scholar]
- 49.Galluser A. & Kirchhausen, T. (1993) EMBO J. 12, 5237-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owen D. J., Vallis, Y., Pearse, B. M., McMahon, H. T. & Evans, P. R. (2000) EMBO J. 19, 4216-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.