Abstract
It has been asserted that recent mtDNA phylogenies support the plausibility of sympatric speciation, long considered a controversial mechanism of the origin of species. If such inferences are reliable, mtDNA phylogenies should be congruent with phylogenies based on other data. In previous work, a mtDNA phylogeny suggested that diversification of the Hawaiian cricket genus Laupala was initiated by single invasions into each of several Hawaiian islands, followed by multiple sympatric divergences within each island. In contrast, a systematic hypothesis based on morphology argues that speciation in Laupala has occurred primarily in allopatry, with two independent species radiations diversifying across the archipelago. In this study, I analyze nuclear DNA (nDNA) sequences from Laupala to compare with sequences from the mtDNA. The nDNA phylogeny corroborates the hypothesis of allopatric divergence and multiple invasions, and when compared with mtDNA patterns, suggests that interspecific hybridization is a persistent feature of the history of Laupala. The discrepancy between mtDNA and nDNA phylogenies reveals that speciation histories based on mtDNA alone can be extensively misleading.
Isolated biotas, such as those in island archipelagos or lake habitats, often harbor many complexes of closely related, endemic species coexisting in sympatry. Mayr (1), the well-known proponent of “allopatric” speciation, argued that such biotas are likely the consequence of multiple invasions. Under a multiple-invasions model, closely related species originate in allopatry and come to occupy a common area secondarily by parallel, independent invasions. A single-invasion model, controversial because it requires primary divergence and speciation in sympatry, offers an alternative to the multiple-invasions scenario. Under this alternative, a single invasion by an exclusive common ancestor and in situ divergence leads to the coexistence of two sister species. As Mayr (1) noted, limited levels of morphological or behavioral variation often exist among closely related “island” species and their putative source populations, thwarting efforts to estimate the phylogenetic relationships necessary to test these alternatives. Recently, mtDNA genes have been found to vary considerably among closely related species, making phylogenetic estimates of recent species radiations possible.
The native rainforests of the Hawaiian archipelago harbor numerous cases of closely related species coexisting in sympatry (2–6). The endemic genus Laupala Otte (2) is part of a large radiation of >150 flightless species of swordtail crickets (subfamily Trigonidiinae), one of the many native Hawaiian species flocks. The 37 morphologically cryptic species of Laupala are each single-island endemics restricted to the rainforested slopes of the high islands of the Hawaiian archipelago, often living in sympatric communities with one to three acoustically distinct congeners (2, 3, 7). Laupala offers an ideal opportunity to test the likelihood of a multiple- versus single-invasion model.
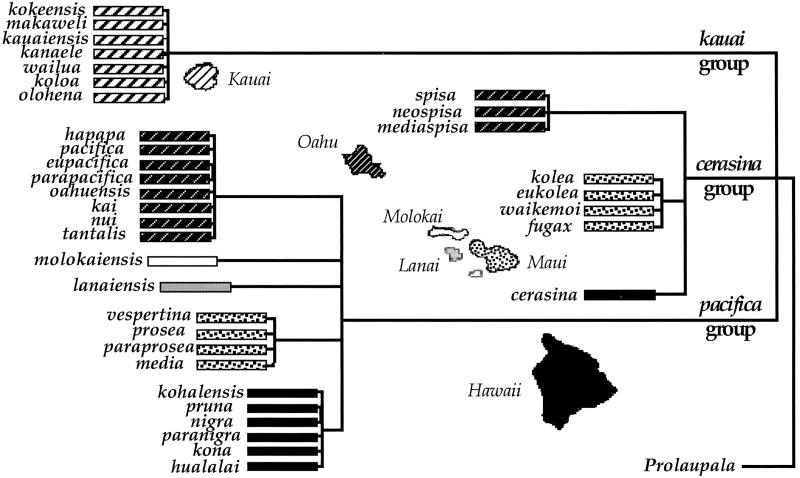
Otte (2, 8) hypothesized that there are three species groups of Laupala (the kauai, pacifica, and cerasina groups) and that sympatric communities from Oahu, Maui, and Hawaii each have representatives from two of these groups (pacifica and cerasina). Otte (2, 8) suggested that each species group shares exclusive common ancestry on the basis of slight metric differences of the male genitalia. Therefore, sympatric Laupala communities must have been established by multiple invasions (Fig. 1). In contrast, phylogenetic relationships inferred from the mtDNA sequences of the three hypothesized species groups (3) contradict Otte's (2) hypothesized relationships. Instead, the mtDNA phylogeny suggests a single, older-to-younger biogeographical species radiation across the Hawaiian archipelago coincident with the chronological age of island formation (3). Furthermore, in many instances the mtDNA sequences suggest that sympatric species pairs representing Otte's different species groups frequently were each other's closest relatives (3, 9), revealing patterns consistent with sympatric speciation. The patterns evident in the mtDNA phylogeny support a single-invasion model of speciation.
Fig 1.
Otte's (2) phylogenetic hypothesis of the endemic Hawaiian cricket genus Laupala. The 35 species described by Otte are shown, arranged into the kauai, cerasina, and pacifica groups. The islands of Oahu, Maui, and Hawaii harbor sympatric communities with representatives from both cerasina and pacifica species groups. Under this phylogenetic hypothesis, sympatric communities arose through multiple invasions. Branch lengths are for graphical purposes and do not reflect degree of relationship.
Here, I present a nuclear DNA (nDNA) phylogeny of the genus Laupala that corroborates Otte's (2) hypothesized relationships and the multiple-invasions model. This nDNA phylogeny dramatically conflicts with the mtDNA phylogeny. A comparison of nDNA and mtDNA patterns suggests repeated episodes of hybridization and interspecific mtDNA gene flow in the recent history of Laupala and cautions against interpretations based on mtDNA phylogenies in young species radiations.
Materials and Methods
Collection of Samples.
Mature males of 23 species of Laupala from Kauai, Oahu, Molokai, Maui, and Hawaii (Fig. 1), representing Otte's (2) three species groups, were sampled and identified by published methods (2, 3). Another endemic Hawaiian swordtail cricket, Prolaupala kukui, was used as an outgroup. Most individuals had been previously sampled for mtDNA (3, 9, 10).
DNA Extraction, Amplification, and Sequencing.
DNA samples were isolated by standard methods (3). Nuclear primers for the PCR (11, 12) were designed from a randomly chosen Laupala cDNA clone. PCR yielded a noncoding product of ≈1,200 bp under the following conditions: 94°C for 30 s, 56°C for 30 s, and 72°C for 90 s. PCR fragments were agarose gel-purified by using Geneclean (Bio 101). The two external and four internal PCR primers were used to generate sequence in both directions. Sequences were collected by automation on Applied Biosystems 373 or 377 at Harvard University (Boston) or the Brigham Young University (Salt Lake City) sequencing facility.
To augment previous studies (3, 9, 10), additional mtDNA samples were sequenced as above, including the 3′ end of the 12S rRNA, the tRNAval, and 5′ end of the 16S rRNA genes, in one direction only. Published primers and PCR conditions were used (3).
Sequence Analysis.
Base calls were edited with SEQUENCHER 3.1.1 (Gene Codes, Ann Arbor, MI). The high degree of similarity and rarity of gaps in both data sets allowed alignments to be achieved easily by eye. Two exceptions were a variable number of semiperfect CATA repeats in the nuclear sequence (sites 806–835) and a 6-bp region of TA repeats in the mtDNA sequence (sites 434–439). These sites were excluded from all analyses.
Phylogeny Estimation.
Phylogenetic analyses were conducted separately for nDNA or mtDNA data sets by using maximum parsimony (MP) and maximum likelihood (ML) in PAUP* 4.08b (13). All unique sequences were included. In MP analyses, gaps were scored as new states and assumed to be 1 bp in length. All variable characters were weighted equally. Searches for MP trees included 1,000 random addition, heuristic replicates with tree bisection and reconnection branch-swapping. One thousand bootstrap replicates were conducted with the heuristic search option, with maxtrees set to 100 and random addition sequence (n = 1) in effect.
ML analysis of the nDNA data were conducted by assuming the HKY85 model with no among-site rate variation. Model and parameter values were estimated by using MODELTEST 3.04 (ref. 14; base frequencies: A = 0.3085, C = 0.1737, G = 0.2066, and T = 0.3112; transition/transversion ratio = 0.78). ML analysis of mtDNA assumed the HKY85+G+I model of substitution. Model parameters were estimated by using modeltest as above (estimated proportion of invariable sites = 0.5678; gamma shape parameter α = 0.7612; transition/transversion ratio = 4.02; estimated base frequencies: A = 0.3664, C = 0.0449, G = 0.1653, and T = 0.4235). In both nDNA and mtDNA ML analyses, gaps were treated as missing data, and the molecular clock was not enforced.
Tree structure based on nDNA was compared with Otte's hypothesis and to trees derived from mtDNA. Tree comparisons were statistically evaluated with a Shimodairo–Hasegawa likelihood ratio test (15) or Templeton's Wilcoxon signed-rank test (16), as implemented in PAUP*. Minimal patterns of colonization and ancestral geographical localities were inferred by using the nDNA and mtDNA phylogenies and the parsimony algorithm for discrete characters with macclade (17). Only ingroup geographical states were considered in the reconstruction.
Results
The data consist, in total, of 1,085 nuclear nucleotide characters derived from 22 species (including 34 populations and 116 individuals) of Laupala. Among the 39 distinct sequences, there were 143 variable sites (31 were parsimony informative). Coding gap characters as missing data, the range of uncorrected pairwise sequence divergences was 0–1.6% within the ingroup (maximum of 6.3% to the outgroup).
For comparison, 531 mtDNA nucleotide characters were sampled from 23 species of Laupala (including 37 populations and 97 individuals) and one outgroup species, P. kukui. Forty mtDNA sequences are new to this study; an additional 16 partial sequences including the 3′ end of the 12S rRNA and all of the tRNAval supplement those previously published (9). The remaining 46 mtDNA sequences have been published (3, 10). Among 57 distinct mtDNA sequences, there were 77 variable sites (58 were parsimony informative). Coding gap characters as missing, there was an uncorrected pairwise sequence divergence range of 0–5.9% within the ingroup (9.1% maximum to the outgroup).
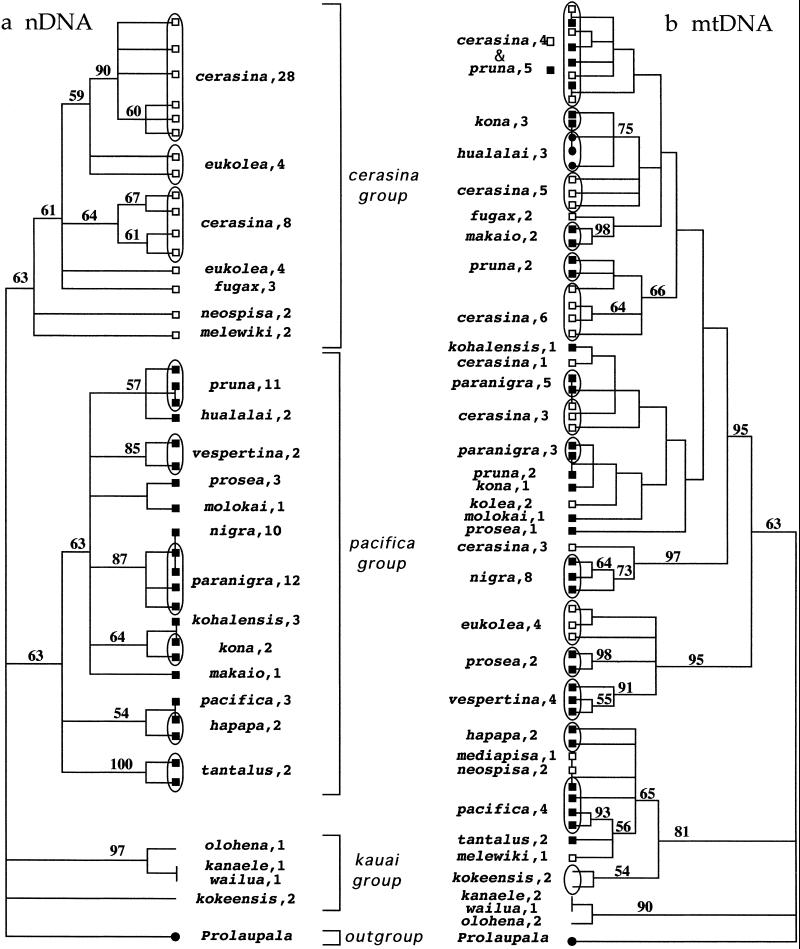
MP analysis of the nDNA resulted in 40 trees. A strict consensus of these trees is shown in Fig. 2. Consistency and retention indices of parsimony-informative characters were high (consistency index = 0.868; retention index = 0.962). Most characters showed complete consistency and 84–87% (depending on the topology) changed only once. Bootstrap support was modest because of a low overall level of variation. Two ML trees were recovered, both of which occurred in the set of MP trees. Although not revealed in the ML analysis, six additional MP trees had the same likelihood score; a strict consensus of the eight ML trees is shown in Fig. 3.
Fig 2.
Contrasting phylogenies for (a) 1,085 bp of nDNA (based on anonymous, noncoding sequence) and (b) 531 bp of mtDNA (based on partial 12s, 16s, and tRNAval rDNA) for species of the genus Laupala, estimated by MP (PAUP* 4.0). The nDNA phylogeny is the strict consensus of 40 most parsimonious trees; the mtDNA phylogeny is the strict consensus of 35 most parsimonious trees. Bootstrap values >50% are shown above branches from 1,000 replicates (maxtrees = 100). At the terminal position on each tree, cerasina group species are noted by open boxes, pacifica group species by closed boxes, and kauai species by the absence of a symbol. Multiple sequences sampled from the same species are circumscribed; species name, followed by sample size, is shown opposite. P. kukui served as an outgroup in both analyses.
Fig 3.
(a) Strict consensus of ML phylogenies for nDNA and (b) the MP phylogeny with the highest likelihood for mtDNA of species in the genus Laupala. Geographical affinities of species are shaded as in Fig. 1, and populations from which DNA sequences were sampled are shown to the right of species names, followed by sample size. To reconstruct ancestral colonization patterns, geographical assignments of interior branches were made based on a minimum colonization scenario, coding geographical location as a character in MACCLADE 3.0 (17). An older-to-younger pattern of colonization is evident in both nDNA and mtDNA phylogenies. Collection locality abbreviations are as in ref. 3. Boxes surrounding groups of mtDNA haplotypes indicate sympatric localities (defined as collections within a 0.5-km radius).
MP and ML mtDNA trees were very similar, differing only in the placement of sequences from Laupala makaio and Laupala fugax; their position does not affect conclusions drawn below. Trees are in basic agreement with previous results (3, 9). A strict consensus of the 35 MP trees (consistency index = 0.577; retention index = 0.882) is shown in Fig. 2. Although not in the set of MP trees, the ML tree was not significantly better at explaining the data than were any of the 35 MP trees by the Shimodairo and Hasegawa (15) likelihood ratio test (P ≥ 0.30; 1,000 RELL bootstrap replicates). Gap characters were treated as missing data in ML analyses. With changes caused by gap characters included, the ML and MP trees also did not differ significantly (Templeton's test, P ≥ 0.30, with equal character weighting). Including gap characters in tree-length calculations found the ML tree to be five steps longer (tree length = 158) than any of the MP trees (tree length = 153). Accordingly, the tree in Fig. 3 and used in subsequent comparisons is the MP tree with the highest likelihood under the parameter set specified for mtDNA above.
Using the nDNA sequence data, I compared the nDNA topology to the hypothesis of Otte (2). L. makaio, Laupala melewiki, and Laupala kolea were as they were absent from one or the other topologies. Meaningful statistical comparisons require that the trees being compared are equally resolved. To resolve the polytomies in Otte's tree, parsimony heuristic searches were conducted with the nDNA data set under Otte's topological constraints. The resulting four equally parsimonious trees, representing Otte's hypothesis, were compared with the 40 MP nDNA trees (Fig. 2). None of these trees differed significantly (tree length of 149 vs. 151 for MP vs. Otte topologies, respectively; Templeton's test, P = 0.3173, with equal character weighting). A resolved phylogeny consistent with Otte's hypothesis also was achieved through a heuristic ML search, conducted with the nDNA data and parameter values estimated for the nDNA above. No significant difference was found between Otte's tree and the eight ML trees (Fig. 3; Shimodairo–Hasegawa test, P ≥ 0.213, 1,000 RELL bootstrap replicates). These results reflect the fact that the nDNA phylogeny is largely congruent with Otte's hypothesis.
In contrast, trees based on nDNA and mtDNA showed statistically significant disagreement. MP nDNA trees were compared with trees consistent with the mtDNA MP trees but including only those nodes with bootstrap support ≥70% (Fig. 2). Further resolution of the mtDNA tree for statistical tests was achieved by conducting parsimony heuristic searches using the nDNA data, with these mtDNA constraints in effect. Templeton's test showed that the number of character changes required of the nDNA data set was significantly greater when under the constraint of the well-supported nodes in the mtDNA topology (tree length of 153 vs. 177 for MP topologies vs. constrained topologies, respectively; P = 0.0017, under equal character weighting). Similarly, the nDNA ML topologies were a significantly better explanation of the nDNA data than was the mtDNA topology (Shimodairo–Hasegawa test, P < 0.006; 1,000 RELL bootstrap replicates). These results reflect the fact that the nDNA and mtDNA phylogenies are largely incongruent.
Despite disagreement in topologies, both nDNA and mtDNA trees suggest compelling biogeographical histories, based on minimum colonization inferences (Fig. 3). The mtDNA result is consistent with previous reports (3), with inferred biogeographical patterns suggesting a history of older-to-younger island species radiation in Laupala. A similar older-to-younger island colonization pattern is also evident in the nDNA trees. The basal portion of the nDNA tree contains four lineages, two of which correspond to Kauai (the oldest island) species. The remaining two groups correspond to Otte's cerasina and pacifica groups. Within each group, the basal position is occupied by Oahu taxa, with distal clades containing taxa from the younger islands of Molokai, Maui, and Hawaii. Although lack of resolution prevents a strong biogeographical inference, the similarity between the pacifica and cerasina groups is compellingly consistent with an older-to-younger island progression to the Laupala species radiation.
Discussion
Phylogenies of closely related species groups provide powerful tools for testing alternative hypotheses of species origins. However, limited levels of morphological or behavioral variation often exist among closely related species, making phylogenetic relationships difficult to estimate. Recently, mtDNA variation has yielded unprecedented levels of character variation, suggesting sister group relationships among sympatric species in several island systems [e.g., sticklebacks of the Pacific Northwest (18), Bonin Island land snails (19), Caribbean anoles (20), African cichlids (21)]. These mtDNA phylogenies have played a major role in convincing biologists of the plausibility of sympatric speciation (21–23).
The present study of Hawaiian Laupala relationships also focuses on a closely related species group endemic to an island system. Previous estimates of interspecific relationships from mtDNA (3) implied a “single invasion” origin for each island's sympatric diversity because (i) close mtDNA relationships from different species within the same island suggested intraisland species radiations, and (ii) sympatric species often shared closely related or identical mtDNA haplotypes, suggesting primary divergence in sympatry. Morphological variation, although minimal because of the generally cryptic morphological condition of these taxa, led Otte (2) to propose that sympatric communities arose through multiple invasions by separate lineages, and accordingly that closely related species are allopatrically distributed, often on different islands.
These alternatives were tested by estimating species relationships from a marker other than mtDNA, as mtDNA has been known to obscure species boundaries in other taxa (ref. 24, pp. 252–305). In contrast to mtDNA, the Laupala nDNA phylogeny shows broad support for Otte's (2) species groups. MP and ML nDNA trees show a highly consistent pattern of character change in support of a split between Otte's pacifica and cerasina species groups (Fig. 2). An analysis of amplified fragment length polymorphisms among four species comprising a subset of the taxa included in this study (one cerasina and three pacifica group species) corroborate these findings (25).
The inferred biogeographical history (Fig. 3) implied by the nuclear phylogeny suggests that the cerasina and pacifica groups have independently radiated in parallel across the Hawaiian archipelago. The evidence suggests that after independent colonizations of Oahu, Maui, and Hawaii, representatives from each species group established sympatric communities together. This multiple-invasions scenario also means that interisland speciation has played an important role in the diversification and community origins of Laupala.
An older-to-younger island colonization history is consistent with the nDNA phylogeny (Fig. 3). Although the basal member of the genus remains uncertain, two members of the kauai species group, endemic to the oldest island of Kauai, occur in an unresolved position with the cerasina and pacifica lineages at the base of the tree. Moreover, the cerasina and pacifica species groups each show a pattern of phylogenetic grades: the basal position in each of the cerasina and pacifica groups is held by taxa from the older island of Oahu, whereas more distal taxa from the younger islands of Molokai, Maui, and the youngest island of Hawaii share a close relationship with each other. ML results suggest that several interisland colonizations occurred between Maui and Hawaii in both species groups, although polarity of these events remains unresolved at present. The interisland divergence histories in Laupala, native Hawaiian Drosophila, and many other taxa (5) may indicate common mechanistic processes at the heart of speciation phenomena in the Hawaiian Islands.
The nDNA phylogeny clearly supports Otte's (2) two distinct species groups of Laupala and consistently shows that sister species are distributed allopatrically. In addition, the exclusive relationships of nDNA sequences observed within most species supports Otte's hypothesized species boundaries. However, no evidence for Otte's species groups appear, nor are species boundaries generally well supported, in the mtDNA phylogeny. Instead, sympatric pairs of pacifica and cerasina group species (Fig. 3) frequently share closely related, and sometimes identical, mtDNA haplotypes. The most likely explanation for the conflict between mtDNA and nDNA phylogenies is interspecific hybridization and mtDNA gene flow between sympatric species pairs. There are many isolated cases of mtDNA “capture” (24). However, the repeated occurrences in Laupala, with complete introgression at as many as six localities over three islands (Fig. 3), represents the most extensive case yet documented. Despite this mtDNA introgression, song differences remain distinct in sympatry (25, 26), and at least part of the nuclear genome indicates separate genetic histories between pacifica and cerasina groups. Repeated cases of mtDNA capture across the genus, coupled with a deep polymorphism in one population of Laupala prosea (Fig. 3), suggest that interspecific gene flow has been, and continues to be, an important process in this group.
Two processes could explain the geographically local mtDNA capture found in Laupala. First, rare hybridization events, followed by selection on mtDNA haplotypes in the alternate species' background, could cause the displacement of a species' mtDNA haplotype by that of its sympatric congener. Under a rare hybridization scenario, the mtDNA molecule of each species is hypothesized to evolve by genetic drift for some time independently. Upon contact and hybridization, each species' mtDNA is exposed to the genetic backgrounds of the alternate species. One species' mtDNA variant may have higher fitness than the other, and a selective sweep across the interspecific boundary could result from this interaction.
Second, frequent interspecific hybridization and strong selection against hybrids could provide a continual conduit for mtDNA across the species boundary. Genetic drift could then bring selectively neutral mtDNA variants to fixation in the genetic background of the other species. This process requires strong reinforcing selection to resurrect the parental genotypes in the zone of hybridization and could be assisted by biases in the direction of interspecific mating. Definitive data to test between these alternatives do not presently exist. However, a lack of intermediate song variants in nature (25, 26) makes the hypothesis of frequent hybridization less tenable. The likelihood of these processes or other viable alternatives cannot be addressed until further information on the extent of genic introgression between hybridizing taxa is available.
This study is significant in showing how extensively misleading mtDNA patterns of variation can be about species relationships in closely related species groups. The nDNA phylogeny, along with preliminary evidence from amplified fragment length polymorphism data (25), corroborates Otte's (2) hypothesis of multiple invasions, providing additional details that elucidate the patterns and processes of speciation in Hawaiian Lauapla. In addition, comparisons of nDNA and mtDNA suggest that interspecific hybridization has been a persistent feature in the history of this group.
Acknowledgments
I thank B. Duwe, T. Sangster, N. Singh, and S. Lesnick for technical assistance and P. Danley, M. Hare, T. Mendelson, S. Lesnick, J. Coyne, and two anonymous reviewers for comments on earlier versions of the manuscript. This work was supported by National Science Foundation Grant DEB-9729325 while I was at Harvard University.
Abbreviations
nDNA, nuclear DNA
MP, maximum parsimony
ML, maximum likelihood
References
- 1.Mayr E., (1963) Animal Species and Evolution (Belknap, Cambridge, MA).
- 2.Otte D., (1994) The Crickets of Hawaii: Origin, Systematics, and Evolution (The Orthopterists' Society, Academy of Natural Sciences of Philadelphia, Philadelphia).
- 3.Shaw K. L. (1996) Evolution (Lawrence, Kans.) 50, 256-266. [DOI] [PubMed] [Google Scholar]
- 4.Funk V. A. & Wagner, W. L. (1995) in Hawaiian Biogeography: Evolution on a Hot Spot Archipelago, eds. Wagner, W. L. & Funk, V. A. (Smithsonian Institution Press, Washington, DC), pp. 379–451.
- 5.Roderick G. K. & Gillespie, R. G. (1998) Mol. Ecol. 7, 519-531. [DOI] [PubMed] [Google Scholar]
- 6.Craddock E. M. (2000) in Evolutionary Biology, eds. Hecht, M. K., Wallace, B. & Prance, G. H. (Plenum, New York), Vol. 31, pp. 1–53. [Google Scholar]
- 7.Shaw K. L. (1995) in Hawaiian Biogeography: Evolution on a Hot Spot Archipelago, eds. Wagner, W. L. & Funk, V. A. (Smithsonian Institution Press, Washington, DC), pp. 39–56.
- 8.Otte D. (1989) in Speciation and Its Consequences, eds. Otte, D. & Endler, J. A. (Sinauer, Sunderland, MA), pp. 482–526.
- 9.Shaw K. L. (1999) Mol. Phylogenet. Evol. 11, 332-341. [DOI] [PubMed] [Google Scholar]
- 10.Shaw K. L. (2000) Zool. J. Linnean Soc. 129, 73-91. [Google Scholar]
- 11.Saiki R. K., Scharf, S., Faloona, F., Mullis, K. B., Horn, G. T. & Erlich, H. A. (1985) Science 230, 1350-1354. [DOI] [PubMed] [Google Scholar]
- 12.Mullis K., Faloona, F., Scharf, S., Saiki, R., Horn, G. & Erlich, H. (1986) Cold Spring Harbor Symp. Quant. Biol. 51, 263-273. [DOI] [PubMed] [Google Scholar]
- 13.Swofford D. L., (1998) PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 14.Posada D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 15.Shimodairo H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 1114-1116. [Google Scholar]
- 16.Templeton A. R. (1983) in Statistical Analysis of DNA Sequence Data, ed. Weir, B. S. (Dekker, New York), pp. 151–179.
- 17.Maddison W. P. & Maddison, D. R., (1992) macclade: Analysis of Phylogeny and Character Evolution (Sinauer, Sunderland, MA). [DOI] [PubMed]
- 18.Taylor E. B. & McPhail, J. B. (1999) Biol. J. Linnean Soc. 66, 271-291. [Google Scholar]
- 19.Chiba S. (1999) Evolution (Lawrence, Kans.) 53, 460-471. [DOI] [PubMed] [Google Scholar]
- 20.Losos J. B., Jackman, T. R., Larson, A., de Queroz, K. & Rodriguez-Schettino, L. (1998) Science 279, 2115-2118. [DOI] [PubMed] [Google Scholar]
- 21.Schliewen U. K., Tautz, D. & Paabo, S. P. (1994) Nature 368, 629-632. [DOI] [PubMed] [Google Scholar]
- 22.Kondrashov A. S. & Kondrashov, F. A. (1998) Nature 400, 351-354. [DOI] [PubMed] [Google Scholar]
- 23.Shaw P. W., Turner, G. F., Idid, M. R., Robinson, R. L. & Carvalho, G. R. (2000) Proc. R. Soc. London Ser. B 267, 2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avise J. C., (1994) Molecular Markers, Natural History, and Evolution (Chapman & Hall, New York).
- 25.Parsons Y. M. & Shaw, K. L. (2001) Mol. Ecol. 10, 1765-1772. [DOI] [PubMed] [Google Scholar]
- 26.Mendelson, T. C. & Shaw, K. L. (2002) Genetica, in press. [PubMed]