Abstract
Genome rearrangements, especially amplifications and deletions, have regularly been observed as responses to sustained application of the same strong selective pressure in microbial populations growing in continuous culture. We studied eight strains of budding yeast (Saccharomyces cerevisiae) isolated after 100–500 generations of growth in glucose-limited chemostats. Changes in DNA copy number were assessed at single-gene resolution by using DNA microarray-based comparative genomic hybridization. Six of these evolved strains were aneuploid as the result of gross chromosomal rearrangements. Most of the aneuploid regions were the result of translocations, including three instances of a shared breakpoint on chromosome 14 immediately adjacent to CIT1, which encodes the citrate synthase that performs a key regulated step in the tricarboxylic acid cycle. Three strains had amplifications in a region of chromosome 4 that includes the high-affinity hexose transporters; one of these also had the aforementioned chromosome 14 break. Three strains had extensive overlapping deletions of the right arm of chromosome 15. Further analysis showed that each of these genome rearrangements was bounded by transposon-related sequences at the breakpoints. The observation of repeated, independent, but nevertheless very similar, chromosomal rearrangements in response to persistent selection of growing cells parallels the genome rearrangements that characteristically accompany tumor progression.
An important general question is how organisms alter their genomes in response to selective pressure from their environment. Paquin and Adams (1) originally described how budding yeast evolve in a simple, continuous, resource-limited environment, such as a chemostat. Under such conditions, microbial evolution is held to occur by sequential accumulation of mutations conferring higher fitness (2). Isogenic haploid and diploid clones were propagated asexually for 100–500 generations in glucose-limited continuous culture. Paquin and Adams (1) found that diploids evolve more rapidly than haploids; other research found that usually the “evolved” clones assimilate glucose more rapidly than the ancestral strains from which they were derived (3, 4). More recently, Ferea et al. (5) used DNA microarrays to study genomewide gene expression differences associated with the evolution of one of the Paquin and Adams cultures as well as two others similarly derived by Rosenzweig (5). Comparisons between evolved clones and the common diploid ancestor showed changes in transcription patterns consistent with a shift from fermentative to oxidative metabolism.
In this study, we used microarray-based comparative genomic hybridization (array CGH) to screen eight similarly derived evolved clones for changes in DNA copy number. We were particularly interested in amplifications, deletions, and genome rearrangements because they have long been thought to play important roles in various evolutionary contexts, including “adaptive” mutation (6) and transcriptional regulation in prokaryotes (7), speciation (8, 9), and cancer progression (10). Classical genetics and electrophoretic techniques such as pulsed-field gel electrophoresis (PFGE), karyotyping, and restriction fragment length polymorphism analysis have revealed changes in DNA copy number in both experimentally evolved yeast (4, 11–13) and bacteria (14–16). Recently, Reihle et al. (17) studied rearrangements in bacteria at single gene resolution and found certain regions repeatedly amplified and deleted among strains that evolved in serial dilution culture at elevated temperature.
Previous work on one of the Paquin and Adams strains using standard techniques strongly implicated gene amplification as a mechanism by which the capacity to assimilate a limiting substrate is increased (4). In this study, using array CGH, we found additional characteristic genome rearrangements in six of eight strains. These rearrangements include previously reported gene-local amplifications, changes in chromosome copy number, and intrachromosomal and interchromosomal translocations. Several of the regions are affected in multiple independent strains, including three events with the same breakpoint. Most of the rearrangements seem to be caused by ectopic recombination involving transposon-related sequences.
The genomic changes observed in response to strong selection in yeast are similar not only to those found in experimental evolution studies in microorganisms, but also notably similar to those found regularly in tumors. Amplification of oncogenes, loss of heterozygosity, and the creation of novel breakpoints leading to oncogenic fusions have all been particularly well documented in cancers (18). Indeed, genome instability leading to widespread aneuploidy is a nearly ubiquitous feature of all cancers (19). Our results suggest that experimental evolution studies in yeast may usefully model the process by which mammalian genomes evolve during tumorigenesis and tumor progression.
Materials and Methods
Yeast Strains and Media.
Strains used in this study are listed in Table 1. Yeast media and sporulation are as described (21). Detailed recipes and techniques are available at http://genome-www.stanford.edu/rearrangements/.
Table 1.
Strains used
| Strain | Source | Isolated at generation |
|---|---|---|
| CP1AB | 20 | N/A (ancestral strain) |
| E1 | 1 | 460 |
| E2 | 5 | 250 |
| E3 | 5 | 250 |
| E4 | 1 | 301 |
| E5 | 1 | 264 |
| E6 | 1 | >325 |
| E7 | 1 | 105 |
| E8 | 1 | 243 |
| E1, E2, E3, and E4 spores | This study |
PFGE.
Yeast chromosome plugs were prepared by standard methods (22). One percent agarose gels were run in a PFGE apparatus at 6 V/cm for 15 h at a switch time of 70 s and 11 h at a switch time of 120 s. Gels were then stained with ethidium bromide and photographed, and relevant bands were excised. The sizes of novel bands were estimated by fitting their electrophoretic migration distance to the regression equation derived from plotting the known size of yeast chromosome marker bands (New England Biolabs) versus migration distance. Chromosome 12 was excluded from the regression because of known anomalous electrophoretic behavior.
Array CGH.
Genomic DNA was prepared according to the protocol of Hoffman and Winston (23). DNA (0.5–2 μg) was labeled with fluorescent dUTP by using Klenow and hybridized to a microarray. Detailed protocols are available at http://genome-www.stanford.edu/rearrangements/. Microarrays were produced, postprocessed, scanned, and analyzed by using genepix software as described (24). Some microarrays were purchased from Corning.
Pulsed-field gels were run as described above to isolate novel chromosome bands for array CGH analysis. Excised bands and plugs from the ancestral strain CP1AB were equilibrated in restriction enzyme buffer and digested overnight with HaeIII. DNA was purified from the agarose by using the Qiagen (Chatsworth, CA) QIAquick gel extraction kit, amplified, labeled, and hybridized as described (25).
Data were filtered for regression correlations >0.6 and signal intensity in at least one channel more than 350 units and 2.5-fold above background. Spots corresponding to failed PCRs, unmapped clones, and physical array artifacts were also eliminated from the analysis. Replicates were averaged after filtering. Raw data are deposited in the Stanford Microarray Database (http://genome-www.stanford.edu/microarray).
PCR.
Thermal asymmetric interlaced PCR was performed as described (26). Primer sequences and detailed PCR conditions can be found at http://genome-www.stanford.edu/rearrangements/.
Results
To study genetic events that might underlie the heritable changes in gene expression found by Ferea et al. (5) in response to 100–500 generations of growth under glucose limitation in chemostats, we analyzed the phenotypes and DNA content of eight clones derived from the evolved cultures, designated strains E1–E8. All but one (E6) of these eight strains had acquired the ability to grow in the chemostat at a relatively higher density than their common ancestor (CP1AB). Analysis of E1–E3 revealed an apparent increase in metabolic efficiency as manifested in higher yield (g biomass/g glucose) and lower residual concentrations of glucose and ethanol at steady state.
Phenotypes of Ancestral and Evolved Strains.
To look for additional acquired phenotypes, we sporulated these strains and tested spores for growth on various carbon sources. Only E1–E4 were able to sporulate. E1 and E4 exhibited 2:0 segregation of viability/inviability, with two spores from each tetrad failing to yield a colony. Strains E2 and E3 both exhibited 2:2 segregation of a new phenotype, the ability to grow at 37°C on acetate as the sole carbon source (http://genome-www.stanford.edu/rearrangements/). Crosses of spores from E2 and E3 indicate that the mutation(s) are likely to be in the same gene, as no recombinants were observed in 36 complete tetrads; spore viability overall was 97.5%.
DNA Content of Ancestral and Evolved Strains.
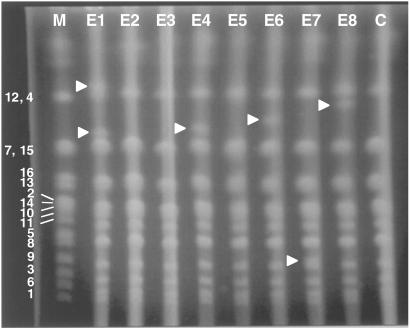
PFGE was used to determine the electrophoretic karyotype of each strain. Fig. 1 shows a display of intact chromosomes isolated from the evolved strains (lanes E1–E8) and their ancestor, CP1AB (lane C). Five of the eight strains (E1, E4, E6, E7, and E8) contain bands not found in the ancestor (arrowheads). Our results are similar qualitatively to those reported by Adams et al. (13) who investigated genomic changes in replicate yeast populations that had evolved in phosphate-limited continuous culture.
Fig 1.
Pulsed-field gel electrophorogram of the intact chromosomes from each yeast strain. M, yeast chromosome markers. Marker bands are labeled by chromosome. E1–E8, evolved strains; C, CP1AB.
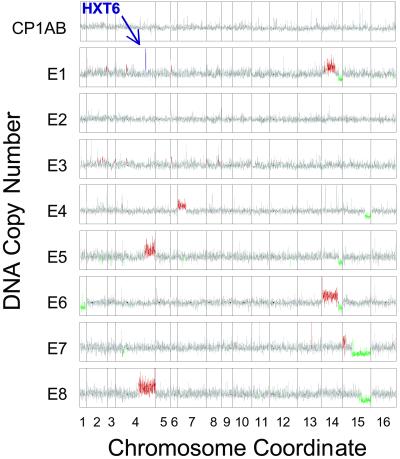
Array CGH measures DNA copy number by making use of DNA microarrays consisting of essentially every ORF in an organism's genome (27–29). In our application of this technique, genomic DNA isolated from the ancestor and labeled with one fluorescent dye was mixed with DNA from one of the evolved strains labeled with a different dye; this mixture was cohybridized on a yeast DNA microarray. The resulting intensity ratios were converted to copy numbers by multiplying the ratios by 2, because the ancestor is diploid. The results (each comparison is an average of two experiments) are shown in Fig. 2 for each gene separately. They are depicted in genome order such that regions with different average copy numbers are colored red (three or more copies, calculated from a running average over nine genes), green (one copy or less), and gray (the expected two copies).
Fig 2.
Genomewide copy number in the evolved strains. Genes are plotted by coordinate along each chromosome. Copy numbers were obtained by multiplying the average of two evolved strain/CP1AB copy number ratios by 2, because CP1AB is a WT diploid. The confirmed amplification of HXT6 in E1 is highlighted in purple. A measurement was colored red or green if the running average of nine genes centered at the query gene was beyond 99.9% of the data in the control experiment. Small black circles along each plot represent the centromeres. Copy number scale ranges from 0 to 6.
The sensitivity of this technique is confirmed by comparison of one of our results to those obtained by conventional Southern analysis. Brown et al. (4) reported, in strain E1, amplification of a chimeric gene encoding a high-affinity hexose transporter that consisted of the coding region of HXT6 and the promoter of HXT7. The HXT6 amplification can be clearly observed by array CGH (even though, being a single gene, it failed to raise the running average over the threshold) and is colored purple in Fig. 2. We observed other possible localized changes in gene copy number, whose significance remains to be assessed.
It is obvious from Fig. 2 that gross chromosomal aberrations have occurred, resulting in large segments of lesser or greater copy number, which we refer to for simplicity as deletions and amplifications (see below). Of the eight similarly evolved strains, only E2 and E3 had no major changes of this kind. Among the aberrations we found nested deletions on chromosome 15 in three strains (E4, E7, and E8) and a shared chromosomal breakpoint on chromosome 14 (E1, E5, and E6). The chromosome 14 aberrations all share a deletion of the right arm. E1 and E6 also have amplifications of most of the left arm of chromosome 14. Two strains (E5 and E8) contain amplifications of the right arm of chromosome 4, in which HXT6, amplified locally in E1, lies. Three events were unique to a strain; amplification of part of chromosome 7 occurred only in E4 and deletion of an entire copy of chromosome 1 occurred only in E6. E7 alone amplified the extreme left end of chromosome 15.
Chromosome Size Estimates.
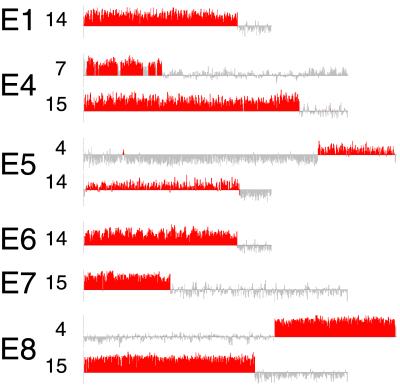
A striking feature of these results is that each of the strains with extensive chromosomal aberrations contains a large segment having an increased copy number and another with a decreased copy number. The most straightforward and parsimonious explanation for this is the generation of a new chromosome in each strain by a single simple translocation. On this assumption, we estimated the expected size of each new chromosome by adding the length of the duplicated segment to the length of the remaining fragment of the partner in the translocation event (see the example in Fig. 3). In every case, sizes estimated in this way were entirely consistent with the sizes obtained from the PFGE analysis, as shown in Table 2. Additionally, our estimates predicted that E5 also carries a novel chromosome, and that the PFGE band for this chromosome overlaps with the chromosomes 7 and 15 doublet.
Fig 3.
Inferred karyotype of E4. Only chromosomes 7 and 15 are shown; all other chromosomes are diploid. In E4 the dashed section of chromosome 7 has become triploid by translocating to chromosome 15, causing a deletion of one copy of the dashed section of 15. The number of centromeres remains 32. The full-length chromosome 15 and the translocation would segregate 2:2 in tetrads.
Table 2.
Chromosome size estimates
| Strain | Segments with altered copy number | Predicted size, kb | PFG band, kb |
|---|---|---|---|
| E1 (lower band) | Amp. of YNL telomere-YNL066W | 503 | |
| Del. of YNR002C-YNR telomere | + 631 | ||
| 1,134 | 1,280 | ||
| E4 | Amp. of YGL telomere-YGL098W | 318 | |
| Del. of YOR290C-YOR telomere | + 854 | ||
| 1,172 | 1,290 | ||
| E5 | Amp. of YDR327W-YDR telomere | 407 | |
| Del. of YNR002C-YNR telomere | + 631 | ||
| 1,038 | No band | ||
| E6 | Amp. of YNL telomere-YNL018C | 602 | |
| Del. of YNR002C-YNR telomere | + 631 | ||
| 1,233 | 1,340 | ||
| E7 | Amp. of YOL telomere-YOL107W | 113 | |
| Del. of YOR012W-YOR telomere | + 354 | ||
| 467 | 430 | ||
| E8 | Amp. of YDR211W-YDR telomere | 647 | |
| Del. of YOR193W-YOR telomere | + 701 | ||
| 1,348 | 1,460 |
Amp., amplification; Del., deletion.
Array CGH on Bands Enriched from Pulsed-Field Gels.
To confirm that the novel bands represented translocations, we extracted the DNA from the new PFGE bands to allow us to characterize them more exactly. The novel chromosome bands marked in Fig. 1 and the predicted E5 band were excised and the extracted DNA was labeled, mixed with differentially labeled DNA eluted from an ancestral DNA plug, and hybridized to a microarray. The log2 fluorescence ratios of novel band DNA to ancestral DNA are plotted, again in chromosome order, in Fig. 4. Only chromosomes with discontinuous averages are plotted, with enriched regions colored red and unenriched regions in gray. In some cases, nearby chromosomal bands contaminated the excised band and were also enriched throughout their entire length (see http://genome-www.stanford.edu/rearrangements/ for the complete data).
Fig 4.
Log2 transformed ratios from DNA microarrays of PFGE band DNA compared with a constant reference of CP1AB plug DNA. Log2 ratios for each gene are plotted in chromosome order; the vertical scales are different for each comparison (see http://genome-www.stanford.edu/rearrangements/ for details). Only chromosomes with enriched regions are shown. The section of each chromosome enriched in the excised band is colored red, with the rest of the chromosome colored gray. The boundary defining these classifications was determined by finding the minimum between the two peaks of the bimodal histogram of a running average of three genes along each chromosome.
The results of the PFGE band enrichments allowed us to confirm that the new chromosomal bands consisted of the chromosomal regions expected from hypothesized translocation events. The analysis showed that E4, E5, and E8 all contain interchromosomal translocations. In E4, the amplified piece of the left arm of chromosome 7 is translocated to the piece of chromosome 15 remaining from the deletion of part of the right arm (see Fig. 3). Similarly, the band from E5 contains DNA from both chromosome 4 and chromosome 14. The novel chromosome in E8 is part chromosome 4 and part chromosome 15.
E1 and E6 both have a band containing only DNA from the left arm of chromosome 14. This result agrees with the observation that these bands are of the approximate size expected for an isochromosome that fused two left arms of chromosome 14. The E7 band is the fragment of chromosome 15 remaining after the deletion of most of the right arm, probably fused to the amplified bit of the left arm. The uncertainty arises solely from the difficulty in distinguishing doubly enriched from simply enriched regions using this technique.
A second novel band was identified in E1. When applied to the array, it was found to contain only DNA from chromosomes 12 and 4, chromosomes that migrate to the same general location in the ancestral strain. Therefore, the band must contain a slightly expanded version of one of these two chromosomes. We suggest that this band could be the result of either a small, unconfirmed amplification internal to chromosome 4 (possibly near the centromere; see Fig. 2), which contains several genes involved in glucose metabolism. Alternatively, this chromosome could result from expansion in the highly reiterated ribosomal DNA cluster on chromosome 12.
DNA Copy Number Contribution to Phenotype.
As previously mentioned, tetrads from E1 and E4 segregate lethality 2:2. Referring to the results of the yeast genome deletion project (30, 31), the arm of chromosome 14 deleted in E1 contains 11 essential genes and the deleted region of chromosome 15 in E4 contains 14 required genes. The segregating lethality in these strains could result from the 2:2 segregation of the translocation chromosomes, with the two live spores carrying normal chromosomes and the two dead spores carrying the translocation and thus deletions of essential genes (see Fig. 3). We carried out array CGH on 12 spores from E1 and eight spores from E4, with results entirely consistent with this idea: all live spores contained intact copies of the chromosomes affected in the rearrangements (http://genome-www.stanford.edu/rearrangements/).
Breakpoint Identification.
DNA from the excised bands was enriched ≈10-fold over sequences not contained in the band. This degree of enrichment is adequate to identify most of the breakpoints by a sudden change in average log2 ratio from near 3 to near −0.5. Table 3 shows the log2 ratios from Fig. 4 of three genes to either side of each supposed breakpoint. Intrachromosomal breakpoints not identified in Fig. 4 are estimated with lower confidence from Fig. 2. Most strikingly, the chromosome 14 breakpoint shared by E1, E5, and E6 seems to be between FUN34 and CIT1, which encodes the citrate synthase from the mitochondrial tricarboxylic acid cycle. Many of the proposed breakpoints, including the one on chromosome 14, contain transposon sequences: entire Ty elements or just the terminal repeats. Both E4 breakpoints also contain identical tRNAs. Only three of the 12 breakpoints do not contain a transposon immediately adjacent. Two of these are not as well defined because they were intrachromosomal by the above analysis.
Table 3.
Proposed breakpoints
| Strain | Chromosome | Gene-3 | Gene-2 | Gene-1 | Breakpoint | Gene + 1 | Gene + 2 | Gene + 3 |
|---|---|---|---|---|---|---|---|---|
| E1 | 14 (Fig. 4) | RLP7 1.393 | DOM34 1.971 | CIT1 3,309 | SUF10, YNRCdelta7, YNRCtau3, tN(GUU)N2 | FUN34 −0.64 | RPC34 −0.962 | YNR004W −0.22 |
| E1 | 14 (Fig. 2) | YNL063W 0.121 | YDJ1 0.4545 | AQR1 0.2995 | None (YNLWTy1-2, tD(GUC)N nine genes away) | SUN4 0.533 | RPL9B 0.0045 | FKH2 0.821 |
| E4 | 7 (Fig. 4) | SEH1 | LSG1 | USE1 | YGLWdelta4, YGLCdelta5, | SRM1 | TOS8 | VPS45 |
| 1.84 | 2.33 | 2.73 | YGLCsigma1, tH(GUG)G2, YGLCtau3 | 0.57 | 0.92 | 0.27 | ||
| E4 | 15 (Fig. 4) | YOR292C | YOR291W | SNF2 | YORWsigma3, tA(UGC)O | YOR289W | MPD1 | YOR287C |
| −0.127 | −0.162 | 0.283 | 1.561 | 1.811 | 2.214 | |||
| E5 | 14 (Fig. 4) | HRB1 | PET8 | CIT1 | SUF10, YNRCdelta7, YNRCtau3, tN(GUU)N2 | FUN34 | YNR004W | YNR006W |
| 1.116 | 0.174 | 1.136 | −0.998 | −0.388 | −1.094 | |||
| E5 | 4 (Fig. 4) | YDR317W | MCM21 | YDR319C | None (YDRWTy1–4, tF(GAA)D 3 genes away) | YDR327W | SKP1 | PEX3 |
| −1.741 | −1.659 | −1.237 | 1.076 | 1.223 | 2.555 | |||
| E6 | 14 (Fig. 4) | RLP7 | DOM34 | CIT1 | SUF10, YNRCdelta7, YNRCtau3, tN(GUU)N2 | FUN34 | RPC34 | YNR004W |
| 1.922 | 1.863 | 3.319 | −0.574 | −0.777 | −0.629 | |||
| E6 | 14 (Fig. 2) | PB12 | PUB1 | YNL017C | tI(AAU)N2 | YNL018C | YNL019C | ARK1 |
| 0.0275 | −0.292 | 0.367 | 0.8425 | 0.595 | 0.776 | |||
| E7 | 15 (Fig. 4) | TIR4 | TIR2 | AUS1 | tT(AGU)O2, YOLWsigma2, YOLWdelta10 | YOR012W | YOR013W | RTS1 |
| 2.918 | 3.7 | 3.865 | −0.974 | 0.008 | −0.98 | |||
| E7 | 15 (Fig. 2) | NDJ1 | WSC3 | YOL106W | YOLCdelta3, tT(AGU)O1 | YOL107W | INO4 | YOL109W |
| 0.294 | 0.124 | −0.2185 | 0.4175 | 0.667 | 1.049 | |||
| E8 | 4 (Fig. 4) | MSS4 | YDR209C | YDR210W | YDRWTy2–2, YDRCTy1–2, tI(UAU)D | GCD6 | TCP1 | UPC2 |
| 0.132 | −0.666 | 0.07 | 3.38 | 3.069 | 2.926 | |||
| E8 | 15 (Fig. 4) | SPR1 3.781 | RIS1 2.904 | YOR192C 2.845 | YORCdelta18, YORWdelta19, YORWtau2, YORCTy2-1, YORWtau3, IMT1 | YOR193W −0.796 | TOA1 −0.932 | SLK19 −1.538 |
Log2 ratios are from Figs. 2 and 4. Breakpoint candidates were determined as described in Figs. 2 and 4 and then adjusted by at most one position to center around any local Ty element. Measurements are given for the three measured genes on each side of the breakpoint. Genes contiguous in rearranged chromosomes are in bold.
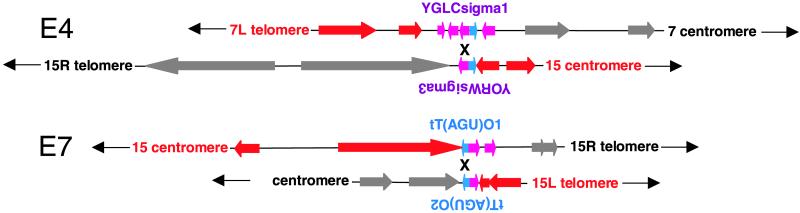
To confirm our strong suspicion that repetitive elements reside at most of the breakpoints, we designed primers across the translocation breakpoints of E4 and E7. Both of the translocations could have been formed by recombination or strand invasion involving the Ty or tRNA sequences at the breakpoint. In both cases, a band amplified only from the evolved strain. Sequencing the PCR products revealed fusion products containing sequence from both chromosomes and intervening sequences, placing the breakpoints at terminal repeat (sigma) sequences in E4 and tRNAs in E7 (Fig. 5; sequences can be found at http://genome-www.stanford.edu/rearrangements/).
Fig 5.
E4 and E7 rearrangements. The breakpoints of the rearrangements described in Table 3 are schematized by using map information from the Saccharomyces Genome Database. Red and gray coloring is as in Fig. 4. Light purple arrows are Ty terminal repeats. Blue arrows are tRNA genes. The depicted crossover regions were determined by sequencing as described in the text.
We also used an asymmetric PCR technique, thermal asymmetric interlaced PCR, to amplify from CIT1 toward the breakpoint in E1, E5, and E6. In each case, we obtained evolved strain-specific chimeric PCR products with one end composed of the promoter region of CIT1 and the other end sequence from a transposon or transposon remnant (http://genome-www.stanford.edu/rearrangements/). Because CP1AB is a derivative of S288C, the sequenced strain, we were able to use the published sequence to see that in the WT strain only a delta terminal repeat sequence resides between these two genes. In E5, the translocation appears tohave resulted not only in the fusion of two chromosome arms, but also in the reconstitution, presumably by recombination, of an intact transposon. In E1 and E6, more extensive sequencing will be required to clarify the novel joint fully.
Discussion
We identified genome rearrangements in six of eight strains evolved in continuous culture under glucose limitation. Although events of this type have been identified in experimentally evolved microorganisms (13, 17), to our knowledge, they have never been studied at single gene resolution in eukaryotes. Several of the rearrangements recur in multiple strains. We propose that these rearrangements underlie some of the observed increases in fitness in the evolved strains (1).
The repeated observation (in E1, E5, and E6) of chromosomal rearrangement at the same breakpoint suggests that the resulting genotype has adaptive value and/or this sequence is particularly susceptible to DNA damage. Both possibilities are reminiscent of events associated with tumor progression, such as the translocation that produces the famous Philadelphia chromosome, which creates the BCR/ABL oncogenic fusion protein (32). In yeast, transposons have been observed to activate expression of nearby genes (e.g., ref. 33). CIT1 is one of the key regulated points in the tricarboxylic acid cycle (34). It is possible that the observed rearrangement recruits such an activating transposon sequence to CIT1 resulting in CIT1 derepression in the presence of glucose. Activation of this key locus may indirectly promote derepression of other genes in the tricarboxylic acid cycle, as observed in E1 (5). It is also interesting to note that what was once thought to be the yeast “chromosome 17” was later shown to be because of the presence, in some strains, of a chromosome 14 fragment very similar to the one we have observed (35).
Previous studies have also implicated rearrangements of this sort as the cause of increases in fitness. Adams et al. (13) argued convincingly that nonselected companion events (e.g., “hitchhiking”) are unlikely explanations because measured rates of chromosomal rearrangement are orders of magnitude too low to account for their appearance by chance in so many adaptive lineages. This argument is particularly compelling in our study because we see particular regions of the genome repeatedly affected, rather than a more random distribution.
Also relevant is the finding by Hughes et al. (36) that ≈8% of 300 yeast deletion mutants examined had acquired a detectable aneuploidy. In six of the cases they examined, the amplified chromosome contained a close homolog of the deleted gene, implying that characteristic aneuploidies can act as dominant suppressors and under some circumstances lead to increased fitness. Several of these segmental amplifications are flanked by Ty sequences.
All of our rearrangements could plausibly be traced to ectopic rearrangement between transposons, transposon fragments, or tRNAs. Because there are only 331 Ty fragments total in the yeast genome sequence (37), such a high coincidence with the breakpoints is unlikely to have occurred by chance. Even the less well-defined breakpoints have nearby Ty elements or tRNAs, common targets of Ty transposition (37).
Ectopic recombination events involving Tys have been noted before in closely related yeast species (9), lab strains (38), and industrial strains (39), and their occurrence has been studied extensively by a number of groups (40–42). Cha and Kleckner (43) recently provided a potential mechanistic reason that transposon sequences are so strongly correlated with chromosome breakpoints. In their mec1 mutant strain, they found chromosomal breakage preferentially at transposon-related sequences during replication, which they suggested might be the result of slowing of replication at those sites. A similar explanation has been proposed for chromosomal breakage at “fragile sites” in mammalian chromosomes.
Our data suggest that transposons and transposon remnants may be the principal source of changes in chromosome structure in yeast that are growing under strong selection. This interpretation adds depth to the previous observation that some degree of transposon load is adaptive (44, 45). Changes of an uncharacterized nature in transposons have been noted in other experimentally evolved microorganisms (15, 16, 46) and could in part be explained by rearrangements at these sites. Because of their generally catastrophic effects on fertility, translocations would be particularly effective in cells growing autonomously under strong, relatively simple selection, and clonal growth, conditions encountered in nutrient-limited chemostat growth and also in cancer progression.
Finally, our results encourage the idea, still necessarily a speculation, that at least some of the ≈300 transposon-related sequences that are found in the sequenced strain of Saccharomyces cerevisiae are in positions that provide a selective advantage at the population level. Such an advantage at the population level for potentially reversible generation of diversity through recombination has been suggested before, for example, in bacteriophage evolution (47–49). By allowing relatively high-frequency (and potentially reversible) and adaptively useful chromosomal rearrangements, appropriately positioned transposon-related sequences could facilitate preferential survival of those lineages in which the transposon sequences remain. This concept is supported by the recent observation, in the sequences of several close relatives of S. cerevisiae (diverged by ≈100 million years) of the conserved presence of transposons or transposon remnants at these positions (M. Kamvysselis, personal communication).
Acknowledgments
We are grateful to Nancy Kleckner for her very helpful comments on the manuscript. M.J.D. is a Stanford Graduate Fellow and a Howard Hughes Medical Institute Predoctoral Fellow. P.O.B. is an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants GM46406 (to D.B.) and HG00983 (to P.O.B.), University of Florida Research Foundation grants (to H.B. and F.R.), and National Science Foundation Grant EPS-00-91995 (to F.R.).
Abbreviations
CGH, comparative genomic hybridization
array CGH, microarray-based CGH
PFGE, pulsed-field gel electrophoresis
References
- 1.Paquin C. & Adams, J. (1983) Nature 302, 495-500. [DOI] [PubMed] [Google Scholar]
- 2.Dykhuizen D. E. & Hartl, D. L. (1983) Microbiol. Rev. 47, 150-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams J., Paquin, C., Oeller, P. W. & Lee, L. W. (1985) Genetics 110, 173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C. J., Todd, K. M. & Rosenzweig, R. F. (1998) Mol. Biol. Evol. 15, 931-942. [DOI] [PubMed] [Google Scholar]
- 5.Ferea T. L., Botstein, D., Brown, P. O. & Rosenzweig, R. F. (1999) Proc. Natl. Acad. Sci. USA 96, 9721-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrickson H., Slechta, E. S., Bergthorsson, U., Andersson, D. I. & Roth, J. R. (2002) Proc. Natl. Acad. Sci. USA 99, 2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero D. & Palacios, R. (1997) Annu. Rev. Genet. 31, 91-111. [DOI] [PubMed] [Google Scholar]
- 8.White M., (1978) Modes of Speciation (Freeman, San Francisco).
- 9.Fischer G., James, S. A., Roberts, I. N., Oliver, S. G. & Louis, E. J. (2000) Nature 405, 451-454. [DOI] [PubMed] [Google Scholar]
- 10.Cahill D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (1999) Trends Cell Biol. 9, M57-M60. [PubMed] [Google Scholar]
- 11.Hansche P. E. (1975) Genetics 79, 661-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansche P. E., Beres, V. & Lange, P. (1978) Genetics 88, 673-687. [PMC free article] [PubMed] [Google Scholar]
- 13.Adams J., Puskas-Rozsa, S., Simlar, J. & Wilke, C. M. (1992) Curr. Genet. 22, 13-19. [DOI] [PubMed] [Google Scholar]
- 14.Bergthorsson U. & Ochman, H. (1999) J. Bacteriol. 181, 1360-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulos D., Schneider, D., Meier-Eiss, J., Arber, W., Lenski, R. E. & Blot, M. (1999) Proc. Natl. Acad. Sci. USA 96, 3807-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider D., Duperchy, E., Coursange, E., Lenski, R. E. & Blot, M. (2000) Genetics 156, 477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riehle M. M., Bennett, A. F. & Long, A. D. (2001) Proc. Natl. Acad. Sci. USA 98, 525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lengauer C., Yu, J., Riggins, G. J., Willson, J. K., Markowitz, S. D., Kinzler, K. W. & Vogelstein, B. (1998) Nature 392, 300-303. [DOI] [PubMed] [Google Scholar]
- 19.Sen S. (2000) Clin. Cancer Res. 6, 112-126. [PubMed] [Google Scholar]
- 20.Paquin C. & Adams, J. (1982) Curr. Genet. 6, 21-24. [DOI] [PubMed] [Google Scholar]
- 21. (1991) Methods Enzymol. 194, 3-131. [DOI] [PubMed] [Google Scholar]
- 22.Birren B., Green, E. D., Klapholz, S., Myers, R. M. & Roskams, J., (1997) Genome Analysis: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Hoffman C. S. & Winston, F. (1987) Gene 57, 267-272. [DOI] [PubMed] [Google Scholar]
- 24.DeRisi J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- 25.Iyer V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533-538. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y. G., Mitsukawa, N., Oosumi, T. & Whittier, R. F. (1995) Plant J. 8, 457-463. [DOI] [PubMed] [Google Scholar]
- 27.Pollack J. R., Perou, C. M., Alizadeh, A. A., Eisen, M. B., Pergamenschikov, A., Williams, C. F., Jeffrey, S. S., Botstein, D. & Brown, P. O. (1999) Nat. Genet. 23, 41-46. [DOI] [PubMed] [Google Scholar]
- 28.Lashkari D. A., DeRisi, J. L., McCusker, J. H., Namath, A. F., Gentile, C., Hwang, S. Y., Brown, P. O. & Davis, R. W. (1997) Proc. Natl. Acad. Sci. USA 94, 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinkel D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., Zhai, Y., et al. (1998) Nat. Genet. 20, 207-211. [DOI] [PubMed] [Google Scholar]
- 30.Winzeler E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 31.Giaever G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 32.Stam K., Heisterkamp, N., Grosveld, G., de Klein, A., Verma, R. S., Coleman, M., Dosik, H. & Groffen, J. (1985) N. Engl. J. Med. 313, 1429-1433. [DOI] [PubMed] [Google Scholar]
- 33.Chisholm G. E. & Cooper, T. G. (1992) J. Bacteriol. 174, 2548-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stryer L., (1995) Biochemistry (Freeman, New York).
- 35.Klapholz S. & Esposito, R. E. (1982) Mol. Cell. Biol. 2, 1399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes T. R., Roberts, C. J., Dai, H., Jones, A. R., Meyer, M. R., Slade, D., Burchard, J., Dow, S., Ward, T. R., Kidd, M. J., et al. (2000) Nat. Genet. 25, 333-337. [DOI] [PubMed] [Google Scholar]
- 37.Kim J. M., Vanguri, S., Boeke, J. D., Gabriel, A. & Voytas, D. F. (1998) Genome Res. 8, 464-478. [DOI] [PubMed] [Google Scholar]
- 38.Camasses A. (1996) Curr. Genet. 30, 218-223. [DOI] [PubMed] [Google Scholar]
- 39.Rachidi N., Barre, P. & Blondin, B. (1999) Mol. Gen. Genet. 261, 841-850. [DOI] [PubMed] [Google Scholar]
- 40.Liebman S., Shalit, P. & Picologlou, S. (1981) Cell 26, 401-409. [DOI] [PubMed] [Google Scholar]
- 41.Sugawara N. & Szostak, J. W. (1983) Proc. Natl. Acad. Sci. USA 80, 5675-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kupiec M. & Petes, T. D. (1988) Genetics 119, 549-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cha R. S. & Kleckner, N. (2002) Science 297, 602-606. [DOI] [PubMed] [Google Scholar]
- 44.Wilke C. M., Maimer, E. & Adams, J. (1992) Genetica 86, 155-173. [DOI] [PubMed] [Google Scholar]
- 45.Wilke C. M. & Adams, J. (1992) Genetics 131, 31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams J. & Oeller, P. W. (1986) Proc. Natl. Acad. Sci. USA 83, 7124-7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Botstein D. & Herskowitz, I. (1974) Nature 251, 584-589. [DOI] [PubMed] [Google Scholar]
- 48.Botstein D. (1980) Ann. N.Y. Acad. Sci. 354, 484-491. [DOI] [PubMed] [Google Scholar]
- 49.Campbell A. & Botstein, D. (1983) in Lambda II, eds. Hendrix, R., Roberts, J., Stahl, F. & Weisberg, R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 365–380.