Abstract
The Penelope family of retroelements was first described in species of the Drosophila virilis group. Intact elements encode a reverse transcriptase and an endonuclease of the UvrC type, which may play a role in Penelope integration. Penelope is a key element in the induction of D. virilis hybrid dysgenesis, which involves the mobilization of several unrelated families of transposable elements. We here report the successful introduction of Penelope into the germ line of Drosophila melanogaster by P element-mediated transformation with three different constructs. Penelope is actively transcribed in the D. melanogaster genome only in lines transformed with a construct containing a full-length Penelope clone. The transcript is identical to that detected in D. virilis dysgenic hybrids. Most newly transposed Penelope elements have a very complex organization. Significant proliferation of Penelope copy number occurred in some lines during the 24-month period after transformation. The absence of copy number increase with two other constructs suggests that the 5′ and/or 3′ UTRs of Penelope are required for successful transposition in D. melanogaster. No insect retroelement has previously been reported to be actively transcribed and to increase in copy number after interspecific transformation.
The activation of certain families of transposable elements (TEs) in some species of Drosophila produces a syndrome of aberrant traits collectively known as hybrid dysgenesis (1). In Drosophila melanogaster, three independent hybrid dysgenesis systems are associated with the activation of three different TE families: P, I, and hobo (2–4). Additional examples have subsequently been reported in Drosophila and other Diptera. Among these is an unusually interesting hybrid dysgenesis system described in Drosophila virilis (5) in which the Penelope TE family appears to play a pivotal role (6). Several additional, unrelated TE families, including Ulysses, Paris, Helena, Telemac, and Tv1, are simultaneously mobilized in dysgenic crosses. This comobilization of multiple families is surprising given the independent mobilization of P, I, and hobo elements in D. melanogaster hybrid dysgenesis (7, 8).
Penelope elements have an extremely complex, highly variable organization in all species of the virilis group studied (6). Phylogenetic analysis showed that the Penelope reverse transcriptase is not closely related to that of any characterized major retroelement group and possibly represents a novel branch of retroelements (9). Sequence profile analysis predicts that the C-terminal domain of the Penelope polyprotein is an active endonuclease that could be responsible for Penelope integration. It is related to intron-encoded endonucleases and the bacterial repair endonuclease UvrC, but no retroelement encoding the predicted endonuclease has been described (9).
Phylogenetic analysis indicates that two subfamilies of Penelope elements are present in species of the virilis group. One subfamily includes elements that differ in general organization, but are almost identical in sequence when homologous regions are compared. The other, more ancient subfamily consists of highly diverged defective copies (9). Several lines of evidence indicate that successive invasions of Penelope into species of the virilis group have previously occurred, possibly leading to gross genome reshuffling and speciation (10).
Penelope-like elements have recently been discovered in several fish, nematodes, Xenopus, and other organisms (9, 11, 12), suggesting that these elements may be present in many highly diverged species of animals. Such a patchy distribution is consistent with the occurrence of horizontal transfer between distantly related lineages. Furthermore, there is strong evidence for the recent horizontal transfer and subsequent invasion of Penelope elements in D. virilis (10). Although class II TEs, such as the P element and mariner families, are known to be relatively prone to horizontal transfer (13), there is also some evidence for this phenomenon involving class I elements, including retrotransposons (14), long interspersed nuclear elements (LINEs) (15), and short interspersed nuclear elements (SINEs) (16). A requisite for the transfer of Penelope-like elements under natural conditions would be their ability to integrate and transpose in a broad range of host organisms.
Previously, the only class I element to be introduced successfully into the D. melanogaster genome was the R2 non-LTR retrotransposon from Bombyx mori. This element integrated into 28S rRNA genes (17). In this case, however, ribonucleoprotein complexes containing in vitro-synthesized R2 RNA and R2 protein were used for injection into D. melanogaster embryos. Furthermore, the inserted R2 sequences were apparently not transcribed in D. melanogaster (17). This finding is not surprising because retrotransposons strongly depend on host transcriptional and processing functions for execution of their replication and transposition cycles (18).
We report here the introduction of full-sized Penelope retroelements into the D. melanogaster genome through conventional P-mediated germ-line transformation. We also show that Penelope is actively transcribed and undergoes massive copy number increase in some transformed lines. After transposition, new Penelope copies were found to be variable in structure and are distributed nonrandomly, predominantly in regions of late-replicating intercalary heterochromatin.
Materials and Methods
Fly Stocks.
The D. melanogaster strain y w67c23(2) was used as recipient in the transformation experiments. Flies were reared on standard resin-sugar-yeast-agar medium containing proprionic acid and methylparaben as mold inhibitors.
Construction of Transformation Vectors.
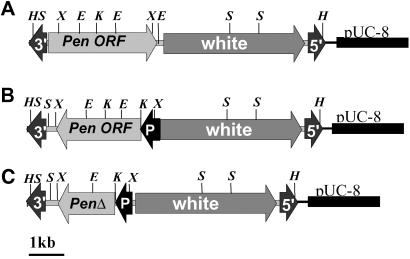
Three different Penelope-containing vectors were constructed (Fig. 1). The P-element transformation vector pCaSpeR-p6 carrying a full-length Penelope copy with two LTRs (construct A) was made by ligating the PstI–XbaI fragment from the p6 clone (6) into the PstI–XbaI sites of pCaSpeR (19). The pCaSpeR-hs-Pen ORF plasmid containing the full-length Penelope ORF (construct B) was made by ligating the Ecl136II–XbaI fragment from pUC-PenORF into the HpaI–XbaI sites of pCaSpeR-hs (19). The pCaSpeR-hs-ΔPen ORF plasmid containing the Penelope ORF with an internal 1,074-bp deletion of the 5′ region of the Penelope ORF (construct C) was made by KpnI digestion of the pCaSpeR-hs-Pen ORF and subsequent self-ligation.
Fig 1.
Schematic representation of the structure of Penelope copies integrated into the D. melanogaster genome. DNA constructs contain full-length Penelope clone p6 (A), full-length Penelope ORF under control of the D. melanogaster heat shock promoter (B), and Penelope ORF with a deletion of the 5′ region under the control of the D. melanogaster heat shock promoter (C). Arrows marked by 3′ and 5′ represent the positions of the P element inverted terminal repeats. The arrow marked with P shows the position of the Hsp70Bb, D. melanogaster heat shock promoter. Letters above the constructs mark the positions of the restriction sites X, H, E, S, and K representing XhoI, HindIII, EcoRI, SalI, and KpnI, respectively.
P Element-Mediated Transformation.
DNA for transformation of D. melanogaster was purified by CsCl equilibrium centrifugation and used for embryo injection as described (20). Transposase activity was provided by the helper plasmid Turbo Δ2–3 (21), and the recipient embryos were from the D. melanogaster y w67c23(2) strain (19). Adults emerging from the injected embryos were crossed with y w67c23(2) virgins of the opposite sex, and the eye color of their progeny was examined. Transformed lines that were homozygous for the transgene were established by full-sib mating. The presence of homozygous transgene copies was confirmed by Southern blotting and in situ hybridization. Each transformed line was routinely maintained en masse in five to six vials initiated with 20–30 flies per vial.
RNA Preparations and Analysis.
Total and poly(A) RNA was extracted from adult flies as described (6) and treated with RNase-free DNase I (Roche Molecular Biochemicals). The integrity of each RNA preparation was checked on ethidium bromide-stained 1% agarose/Mops-formaldehyde gels. Radiolabeled antisense transcripts were generated by using T7 RNA polymerase (Stratagene) and [32P]UTP. RNA probes were acrylamide gel-purified before hybridization. RNA was analyzed by Northern blot. Twenty micrograms was loaded in each lane. Hybridizations were performed overnight at 45°C in 50% formamide. For RT-PCR analysis, mRNA from the transformed flies was isolated by using an Oligotex Direct mRNA Purification Kit (Qiagen, Valencia, CA). Rapid amplification of 5′ cDNA end (RACE) was performed with a 5′RACE kit (GIBCO/BRL Life Technologies), and the PCR fragment was cloned into a pUC19 vector. The clones obtained were sequenced by using a SEQUENASE 2.0 kit (Stratagene).
DNA Manipulations and Southern Analysis.
DNA from D. melanogaster adults was prepared as described (22). Ten micrograms of each DNA sample was digested with the restriction endonucleases indicated in the particular experiment. After agarose gel electrophoresis, the gel was treated for 15 min in 0.25 M HCl and then incubated twice in denaturing buffer (3 M NaCl/0.4 M NaOH) for 30 min. After 30-min incubation in neutralization buffer, gels were capillary-blotted onto nylon membranes according to the manufacturer's protocol and fixed by UV cross-linking using the UV Stratalinker 2400 (Stratagene) protocol. Standard high-stringency hybridization and wash conditions were used for Southern blot analysis.
To detect Penelope sequences in the D. melanogaster transformed lines, we used the 2.8-kb XhoI–XhoI fragment of the p6 clone as probe. This represents a full-sized copy of the element (Fig. 1A). For detection of the white sequences, we used the PvuI 1.5-kb internal fragment of the white gene as probe.
A phage genomic library from the D. melanogaster Penelope-transformed line A1 was prepared by partial Sau3A digestion with subsequent ligation into the BamHI site of Lambda Dash phage arms (Stratagene), according to the manufacturer's instructions. A plasmid library from the D. melanogaster Penelope-transformed line, A1, was prepared by complete HindIII digestion with subsequent ligation into the HindIII site of pUC19. The libraries were probed with the white sequence-containing probe. After the label was removed, the same filters were rehybridized with the Penelope-containing probe to discriminate between the clones containing the original constructs (white and Penelope) and the clones resulting from insertions of Penelope into new genomic sites. A SEQUENASE 2.0 kit (Stratagene) was used to sequence Penelope-containing clones.
In Situ Hybridization to Polytene Chromosomes and Cytological Analysis.
Salivary glands were dissected from male and female third-instar larvae in 45% acetic acid and squashed according to the procedures in ref. 23. For in situ hybridization studies, larvae were grown at 18°C, and a live yeast solution was added to the culture 2 days before the larvae were analyzed. The DNA probes described above were biotinylated by nick translation using biotin 14-dATP (23). All chromosomal localizations were made by using cytological photographic maps of D. melanogaster (24).
Results and Discussion
Introduction of Penelope into the D. melanogaster Genome.
Three different Penelope-containing constructs were used in the transformation experiments (Fig. 1). Construct A contained the full-length Penelope clone p6 with two LTRs (6). Construct B contained a full-length Penelope ORF, but the 5′ and 3′UTRs were missing. Construct C contained a Penelope ORF having a deleted 5′ region. Both constructs B and C were under the control of the D. melanogaster heat shock promoter. The constructs were transferred into a P-element transformation vector and introduced into D. melanogaster y w67c23(2) embryos by microinjection (20). The eye color of transformed progeny varied from pale yellow to near WT and tended to darken with age, as is typical for P-element–miniwhite insertions. Ten independent transformants were recovered for construct A, six for construct B, and six for construct C. Each transformed fly was used to establish an individual line that was subsequently made homozygous for the construct.
Penelope Is Propagated in the Genome of D. melanogaster.
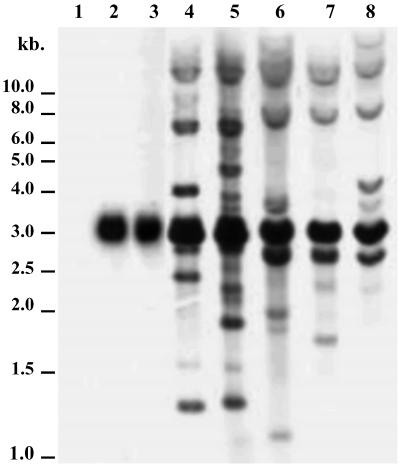
Southern blot hybridizations indicate that transposition of Penelope occurred in five transformed lines in which construct A, containing a full-sized Penelope element, was used in the transformation vector (Fig. 2, lanes 4–8). In addition to the major 2.8-kb band that was produced by XhoI digestion of the full-sized Penelope element contained in construct A, several minor bands of various sizes are present, indicating newly transposed elements. Furthermore, it appears that most transposed copies were rearranged because the intensity of the 2.8-kb band remains much the same in all lanes. In contrast, only the major 2.8-kb band is produced in two lines transformed with construct B, containing the full-length Penelope ORF under the control of the D. melanogaster heat shock promoter (Fig. 2, lanes 2 and 3). Proliferation of Penelope in the other four lines transformed with construct B was also not detected in preliminary Southern experiments. Transposition also did not occur in the six lines transformed with construct C, containing a deleted Penelope ORF under the control of the D. melanogaster heat shock promoter (data not shown). In situ hybridization experiments confirmed the absence of Penelope proliferation in all 12 of the lines transformed with the B and C constructs (data not shown). Construct B was sequenced to verify that its ORF remained intact. Therefore, it is concluded that the 5′ and/or 3′ UTRs of Penelope are required for successful Penelope transposition in the D. melanogaster genome.
Fig 2.
Southern blot hybridization of D. melanogaster lines. Lane 1, initial untransformed strain y w67c23(2); lanes 2 and 3, lines transformed with construct B (containing the Penelope ORF under control of the D. melanogaster heat shock promoter); lanes 4–8, the progeny of one of five individual females taken from lines transformed with construct A (lanes 4 and 5 from line A1; lanes 6–8 from line A2). In all lanes, DNA was digested by XhoI and hybridized with the Penelope XhoI–XhoI 2.8-kb internal fragment.
The number and genomic distribution of new Penelope insertion sites in five independently derived lines transformed with construct A were investigated by using in situ hybridization. One salivary gland of each examined larva was hybridized with the probe containing the structural part of the white gene (see Materials and Methods). The second gland of the same larva was hybridized with the internal 2.8-kb region of the Penelope element. This made it possible to distinguish between the insertion sites of the original construct and those of new transpositions. The chromosome bands containing the original construct exhibited hybridization with both probes whereas insertions resulting from subsequent transpositions hybridized with only the Penelope probe.
Table 1 summarizes the results of in situ hybridization experiments carried out with the five lines transformed with construct A. Each of the lines was examined several times for the presence of Penelope over a period of 2 years after transformation. The data presented in Table 1 represent the chromosomal locations in which Penelope insertions were detected during the whole period of investigation. It is evident that Penelope underwent multiple transpositions after transformation. Furthermore, there appear to be a few sites in which new transpositions were present in more than one line (e.g., 5C, 19E, and 21C), possibly representing “hot spots” for Penelope insertion in the D. melanogaster genome. Interestingly, ≈55% of all new Penelope insertions were located in late-replicating chromatin, probably in intercalary heterochromatic regions (ref. 25; I. F. Zhimulev, personal communication). The D. melanogaster genome is divided into 600 subsections. Of these, 230 (39%) belong to the late-replicating intercalary heterochromatin category (I. F. Zhimulev, personal communication). We cannot say at present whether the observed distribution of Penelope insertions reflects a preference for certain types of chromatin, or whether it can be merely attributed to varying DNA content of different subsections.
Table 1.
Chromosomal locations of Penelope insertions in five transformed sublines of D. melanogaster
| Line | Construct | Chromosome I | Chromosome II | Chromosome III | No. of sites (larvae) |
|---|---|---|---|---|---|
| A1 | 3B 18A | 1A 3C 4A 4B 5B 5C 6B 6C 6B/C 6E 7E/F 11C 19E 19E/F 20A 20B 20C/D | 21C 21C/D 22D 22F 23A 24E/F 25A 25F 26E 28D 28E 29B/C 30C 33B 34E/F 35C/D 36B 36C/D 40A 40B/C 40F 42A 44B 45A 46A 47B 48D 50A 51B 54A 54F 55A 56C 58A 58B 58A/B 60B 60E | 61A 61D 65D 69D 70D 70E 70F 78D 78D/E 79F 80B 82A 83A 84A 84B 84D 84E 85E/F 86C 87B 90D/E 92D 94B 95E 96C 96F 97A 97E 98C 99B 100B 100F | 87 (77) |
| A2 | 6E 17F 96F | 4C 5C 7C/D 8E 18B 18C 18F 19E | 21C 22A 22E 22F–23A 23D 24A 24D 26C/D 35B 38/39C 42A 51B 55F 57B 57F–58A 59E 60F | 61F 68B 76C 76C/D 79F 81F–82A 82E/F 85D 88C 96A | 35 (30) |
| A3 | 96F | 4A | 33A 54F–55A | 86B 99F | 5 (10) |
| A4 | 29–30A | 3D 4C 6C 7E/F 9B/C 11A/B 16D 18D 20B/C | 33B 35B 36C 37D/E 39E 41A/B 50D 51D/E 52A/B 52B 56F | 61E 66F 70D 71C 81F 82A 82E 83A 87C | 29 (27) |
| A5 | 59A | 6F 14B/C 14E 14F 19E/F | 21C/D 21D 22B 50C | 61E/F 84E 86B | 12 (9) |
Sites shown in bold belong to the late-replicating category.
Subsequently, the flanking regions of Penelope insertions were sequenced for all clones, and their positions were located in the D. melanogaster genome. Among 15 cloned and sequenced sites of Penelope insertion, we found only one in which a known gene was located. It is not clear whether Penelope inserts preferentially into these heterochromatic sites or whether the observed nonrandom distribution of sites results from selection against insertion into genes located in euchromatin. The location of intercalary heterochromatin in the D. virilis genome is not known. Therefore, we cannot say whether the distribution of newly transposed elements in P-like strains (i.e., those strains that contribute paternally to hybrid dysgenic crosses) and the progeny from dysgenic crosses is basically different to that seen here in transformed D. melanogaster lines. However, in D. virilis, Penelope elements were occasionally found to have inserted in specific genes such as yellow (26). In both D. virilis and D. melanogaster we failed to observe insertions of active Penelope copies in the heterochromatic chromocenter.
Table 2 provides a temporal breakdown of the in situ data for line A1, which contained two primary insertions in the X chromosome. It indicates that compared with rather modest increases during the first 18 months of the experiment, transposition rates increased significantly in this line between the 18th and 24th months after transformation. Furthermore, only seven insertion sites were found in common at the two times of analysis, and only one of these, 21C, was found in >50% of larvae examined in October 2001. Perhaps surprisingly, there appears to have been an increased rate of transposition in the A1 line over the last 6 months. Retardation in the rate of transposition might have been expected because of increased repression over time. Overall, these data indicate a very high level of insertion site polymorphism.
Table 2.
Chromosomal locations of Penelope insertions in D. melanogaster line A1
| Date | Original | New insertion sites | No. of sites (larvae) |
|---|---|---|---|
| 10/99 | 3B 18A | 0 (5) | |
| 4/01 | 3B 18A | Chromosome I: 5B 6B(1) 6E 11C 20B | 25 (34) |
| Chromosome II: 21C(7) 21C/D 22F(1) 23A(2) 28E 54A 54F 55A 56C | |||
| Chromosome III: 69D(2) 70D 79F 84B 86C 92D 95E 96C 98C 100B(1) 100F(1) | |||
| 10/01 | 3B 18A | Chromosome I: 1A 3C 4A 4B 5C 6B(2) 6B/C 6C 7E/F 19E 19E/F 20A 20C/D | 69 (43) |
| Chromosome II: 21C(24) 22D 22F(1) 23A(4) 24E/F 25A 25F 26E 28D 29B/C 30C 33B 34E/F 35C/D 36B 36C/D 40A 40B/C 40F 42A 44B 45A 46A 47B 48D 50A 51B 58A 58B 58A/B 60B 60E | |||
| Chromosome III: 61A 61D 65D 69D(5) 70E 78D 78D/E 80B 82F 83A 84A 84D 84E 85E/F 87B 90D/E 94A 94B 96F 97A 97E 99B 100B(1) 100F(1) |
Penelope sites localized in both series of analyses are shown in bold, along with the number of larvae examined (shown in parentheses).
Variable Structure of Penelope Copies in Transformed Lines of D. melanogaster.
As our Southern hybridization experiments suggested that Penelope copies were rearranged during transposition, it was of interest to study these structures further. To isolate Penelope copies amplified in the transformed D. melanogaster lines, genomic libraries were prepared as described (6). The libraries were screened repeatedly with Penelope-containing and white-containing probes. Thus, clones containing the original constructs and newly transposed Penelope copies were differentiated.
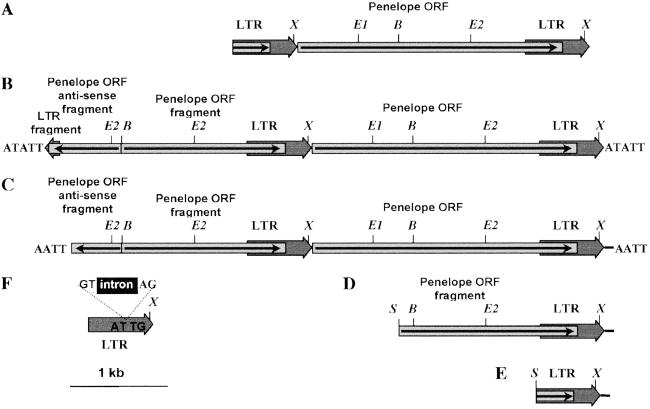
Fig. 3 shows the structure of Penelope elements isolated from D. melanogaster line A1 transformed with construct A. Elements D and E appear to be incomplete (because of Sau3A digestion during cloning). Although the 3′ ends of both of these elements are identical at the nucleotide level, they differ from the 3′ ends of Penelope copies isolated from D. virilis because they include an additional 20 bp that are present in the flanking DNA contained in clone p6. This structure is probably caused by a disturbance of normal termination of Penelope transcription in the foreign species. In elements C and D (Fig. 3) various parts of Penelope are present in antisense orientation and have a very complex structure. At the present time we cannot speculate on the molecular mechanism responsible for their origin, primarily because the mechanism of Penelope transposition is still unknown. Interestingly, elements with similar complex structures have been described in D. virilis and in other organisms in which Penelope-like elements were detected (6, 12). For example, element C (Fig. 3C) has an additional 34-bp repeat at the 3′ terminus present also at the very beginning of the Penelope ORF, exactly like that which was shown for some Penelope copies cloned from D. virilis and a few fish species (6, 12).
Fig 3.
Variable structure of Penelope copies isolated from the transformed D. melanogaster subline A1. (A) The reference original Penelope clone p6. (B–E) Penelope elements isolated from phage libraries. (F) An intronless Penelope element (solo LTR) isolated from a plasmid library. Elements D and E are Sau3A, are truncated at the 5′ end, and have an additional 20-bp sequence at the 3′ end that was derived from flanking sequences present in the original construct. Element C has an additional 34-bp repeat at the very beginning of the Penelope ORF. This 34-bp repeat was also detected at the 3′ end of some Penelope copies cloned from D. virilis. In clones B and C, the junction between the sense and antisense sequences is 6 nt upstream of the BamHI site, ruling out the possibility of a cloning artifact. Restriction enzyme abbreviations: B, BamHI; E, EcoRI; S, Sau3A; X, XhoI.
It has been demonstrated that many TE families are able to use different modes of integration depending on their host species (27). Element F (Fig. 3) consists essentially of a 5′ Penelope solo LTR with a spliced-out intron. The isolation of such a clone enables us to conclude that at least a fraction of Penelope copies propagated in D. melanogaster use an RNA intermediate for transposition. We failed to detect a poly(A) tail at the termini of any of the Penelope copies isolated from D. melanogaster. However, target site duplications of varying length were present at the ends of two Penelope elements that were sequenced. These data favor a model of transposition in which full-sized Penelope elements present in the original constructs can produce both active (full-sized) and inactive copies in the new host. We have isolated spliced Penelope RNAs from the genomes of D. virilis dysgenic hybrids, but failed to find any intronless copies (28). This finding suggests that such spliced RNA probably encodes the reverse transcriptase and endonuclease activities necessary for Penelope retrotransposition, but does not represent an RNA intermediate in D. virilis.
The evidence presented herein, coupled with previously obtained data (9), supports the hypothesis that Penelope and Penelope-like elements constitute a novel subclass of retroelements rather than belonging to either the LTR-retrotransposons or the non-LTR LINEs.
Penelope Is Actively Transcribed and Produces Normal-Sized Transcripts.
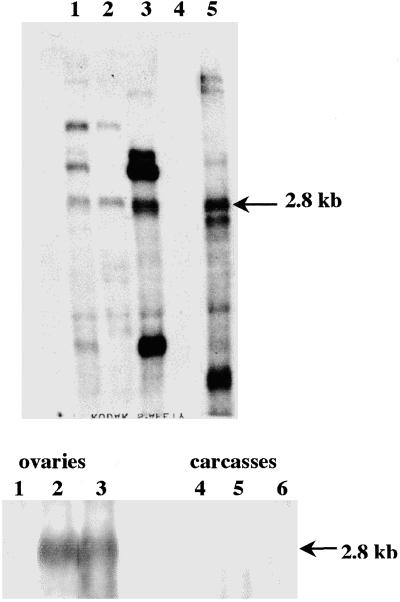
We had identified a polyadenylation site at the 3′ end of the D. virilis Penelope element and determined the structure of the 5′ end of Penelope-encoded transcripts present in the ovaries of D. virilis dysgenic females (25). All three cDNA sequences from D. virilis start at the same site of the reference D. virilis p6 clone and contain a 75-bp deletion, probably resulting from intron splicing (28). It was therefore of interest to find out whether Penelope is transcribed after introduction into D. melanogaster and whether or not it produces transcripts similar to those in D. virilis. RNA isolated from adult flies was used in Northern blot hybridization experiments. Fig. 4 Upper clearly shows that Penelope was actively expressed in all D. melanogaster lines transformed with construct A. However, in addition to the canonical 2.8-kb transcript, several other transcripts were present in all transformed lines tested, including some very abundant ones (Fig. 4 Upper, lanes 1–3). The presence of additional transcripts may be explained either by the occasional insertion of Penelope in the vicinity of strong promoters or by read-through transcription of Penelope and its flanking sequences in the host genome.
Fig 4.
(Upper) Northern analysis of poly(A) RNA. Lane 1, D. melanogaster line A1 at 5 months after transformation; lane 2, D. melanogaster line A1 at 11 months after transformation; lane 3, D. melanogaster line A2 at 11 months after transformation; lane 4, D. melanogaster control strain y w67c23(2); lane 5, D. virilis dysgenic hybrids. (Lower) Northern analysis of total RNA isolated from different tissues. Lanes 1–3, ovaries; lanes 4–6, carcasses; lanes 1 and 4, D. melanogaster control strain y w67c23(2); lanes 2 and 5, D. melanogaster line A1; lanes 3 and 6, D. virilis dysgenic hybrids. Arrows indicate the positions of the 2.8-kb transcript from the canonical Penelope element.
Tissue-Specific Expression and Transcript Structure.
Because we had shown that Penelope transcription in D. virilis dysgenic hybrids was restricted to ovaries (6) we investigated whether that is also the case for transformed D. melanogaster. Northern blot hybridization was carried out with total RNA isolated from ovaries and whole adult carcasses from D. melanogaster transformed lines in comparison with those of D. virilis dysgenic hybrids (Fig. 4 Lower). Penelope transcription was strongly induced only in ovaries of flies from the transformed line and dysgenic hybrids; no transcription occurred in carcasses of any lines tested or in the ovaries of the untransformed D. melanogaster line.
We performed rapid amplification of 5′ cDNA end analyses to compare the structure of the 5′ end of the Penelope transcripts in D. virilis dysgenic hybrids and in transformed lines of D. melanogaster. Using the p6 clone sequence as a reference (GenBank accession no. U49102), the results indicate that Penelope transcription starts at position 371 in D. melanogaster. It is therefore identical to that described in D. virilis dysgenic hybrids (28).
Prospects for the Development of Transformation Systems in Non-Drosophilid Insects.
During the last two decades, successful transformation of a number of species of non-Drosophilid insects of agricultural and medical importance has been achieved by using class II transposons. These include the Mos-1 (active mariner) element from D. mauritiana, the Hermes element from Musca domestica, the Minos element from Drosophila hydei, and the piggyBac element from Trichoplusia ni (27, 29). However, current technologies are still somewhat unwieldy and require further development (27). The frequency of successful germ-line transformation is often low and unpredictable. On one hand, the use of class I TEs, such as Penelope, could potentially overcome this problem because these elements transpose by means of an RNA intermediate. This mechanism could lead to relatively rapid accumulation of high copy numbers in the recipient genomes. On the other hand, the number of rearrangements found in transposed copies might be a limiting factor in the development of Penelope as a universal transformation vector. The narrow host range of the P element used as a vector for Penelope transformation in D. melanogaster should not severely limit the potential range for Penelope transformation because TEs other than P could be used in the construction of transformation vectors.
Conclusions
We have shown that after P element-mediated transformation Penelope is transcribed and able to propagate in the genome of D. melanogaster, a species that diverged from D. virilis at least 40 million years ago (30). Although transformation was successful with all three of the constructs tested, Penelope transcription and copy number increase were observed only in lines transformed with construct A, containing the full-length Penelope clone p6. This finding suggests that the 5′ and/or 3′ UTRs of Penelope are required for successful Penelope transposition in the heterologous host. In D. melanogaster, Penelope transcript structure is identical to that in D. virilis, as is the restriction of transcription to ovaries. The successful transformation of D. melanogaster opens up the possibility of answering the fascinating question of whether Penelope can comobilize other TE families in D. melanogaster, as it appears to do in D. virilis.
Acknowledgments
We thank Andrew Holyoake, Tom Eikbush, and Mike Simmons for critical reading of the manuscript and many helpful suggestions. We are also grateful to Igor Zhimulev from the Institute of Cytology and Genetics (Novosibirsk, Russia) for qualitative analysis of Penelope insertion sites in D. melanogaster. This work was supported by the Russian Foundation for Fundamental Research (Grants 00-04-48285 and 01-04-97009 to K.I.P. and M.B.E.), the Wellcome Trust (Grant 065221 to M.B.E. and D.J.F.), and the National Science Foundation (Grant DEB-9815754 to M.G.K.).
Abbreviations
TE, transposable element
References
- 1.Kidwell M. G., Kidwell, J. F. & Sved, J. A. (1977) Genetics 36, 813-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingham P. M., Kidwell, M. G. & Rubin, G. M. (1982) Cell 29, 995-1004. [DOI] [PubMed] [Google Scholar]
- 3.Bucheton A., Paro, R., Sang, H. M., Pelisson, A. & Finnegan, D. J. (1984) Cell 38, 153-163. [DOI] [PubMed] [Google Scholar]
- 4.Blackman R., Grimaila, R., Koehler, M. & Gelbart, W. (1987) Cell 49, 497-505. [DOI] [PubMed] [Google Scholar]
- 5.Lozovskaya E. R., Scheinker, V. S. & Evgen'ev, M. B. (1990) Genetics 126, 619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evgen'ev M. B., Zelentsova, H., Shostak, N., Kozitsina, M., Barsky, V., Lankenau, D.-H. & Corces, V. G. (1997) Proc. Natl. Acad. Sci. USA 94, 196-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidwell M. G. (1979) Genet. Res. 33, 105-117. [Google Scholar]
- 8.Stamatis N., Monastirioti, M., Yannopoulos, G. & Louis, C. (1989) Genetics 123, 379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyozin G. T., Makarova, K. S., Velikodvorskaja, V. V., Zelentsova, H. S., Khechumian, R. R., Kidwell, M. G., Koonin, E. V. & Evgen'ev, M. B. (2001) J. Mol. Evol. 52, 445-456. [DOI] [PubMed] [Google Scholar]
- 10.Evgen'ev M. B., Zelentsova, H., Mnjoian, L., Poluectova, H. & Kidwell, M. G. (2000) Chromosoma 109, 350-357. [DOI] [PubMed] [Google Scholar]
- 11.Volff J. N., Hornung, U. & Schartl, M. (2001) Mol. Genet. Genomics 265, 711-720. [DOI] [PubMed] [Google Scholar]
- 12.Dalle Nogare D. E., Clark, M. S., Elgar, G., Frame, I. G. & Poulter, R. T. (2002) Mol. Biol. Evol. 19, 247-255. [DOI] [PubMed] [Google Scholar]
- 13.Kidwell M. G. (1992) Genetica 86, 275-286. [DOI] [PubMed] [Google Scholar]
- 14.Jordan I. K., Matyunina, L. V. & McDonald, J. F. (1999) Proc. Natl. Acad. Sci. USA 96, 12621-12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kordis D. & Gubensek, F. (1998) Proc. Natl. Acad. Sci. USA 95, 10704-10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada M., Kido, Y., Himberg, M., Reist, J. D., Ying, C., Hasegawa, M. & Okada, N. (1997) Genetics 146, 355-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eickbush D. G., Luan, D. D. & Eickbush, T. H. (2000) Mol. Cell. Biol. 20, 213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeke J. D. & Corces, V. G. (1989) Annu. Rev. Microbiol. 43, 403-434. [DOI] [PubMed] [Google Scholar]
- 19.Pirrotta V. (1988) in Vectors: A Survey of Molecular Cloning Vectors and Their Uses, eds. Rodrigues, R. & Denhardt, D. (Butterworth, Boston), pp. 437–445.
- 20.Rubin G. M. & Spradling, A. C. (1982) Science 218, 348-353. [DOI] [PubMed] [Google Scholar]
- 21.Robertson H. M., Preston, C. R., Phillis, R. W., Johnson-Schlitz, D. M., Benz, W. K. & Engels, W. R. (1988) Genetics 118, 461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelentsova E. S., Vashakidze, R. P., Kraev, A. S. & Evgen'ev, M. B. (1986) Chromosoma 93, 469-476. [DOI] [PubMed] [Google Scholar]
- 23.Lim J. K. (1993) Drosophila Inf. Serv. 72, 73-77. [Google Scholar]
- 24.Lefevre G. (1976) in The Genetics and Biology of Drosophila, eds. Ashburner, M. & Novitski, E. (Academic, London), Vol. 1a, pp. 36–66. [Google Scholar]
- 25.Zhimulev I. F., Semeshin, V. F., Kulichkov, V. A. & Belyaeva, E. S. (1982) Chromosoma 87, 197-228. [Google Scholar]
- 26.Zelentsova H., Poluectova, H., Mnjoian, L., Lyozin, G., Veleikodvorskaja, V., Zhivotovsky, L., Kidwell, M. G. & Evgen'ev, M. B. (1999) Chromosoma 108, 443-456. [DOI] [PubMed] [Google Scholar]
- 27.Atkinson P. W., Pinkerton, A. C. & O'Brochta, D. A. (2001) Annu. Rev. Entomol. 46, 317-346. [DOI] [PubMed] [Google Scholar]
- 28.Pyatkov K. I., Shostak, N. G., Zelentsova, E. S. & Evgen'ev, M. B. (2001) Dokl. Russian Acad. Sci. 381, 268-270. [DOI] [PubMed] [Google Scholar]
- 29.Handler A. M. (2001) Insect Biochem. Mol. Biol. 31, 111-128. [DOI] [PubMed] [Google Scholar]
- 30.Russo C. A., Takezaki, N. & Nei, M. (1995) Mol. Biol. Evol. 12, 391-404. [DOI] [PubMed] [Google Scholar]