Abstract
Peripheral lymphocyte deletion is required for reduction of lymphocyte numbers after expansion in response to antigen. Peripheral deletion is mediated in part by the activation of apoptosis by engagement of the death receptor, Fas (CD95), by its ligand, Fas ligand (FasL; CD95L), among other mechanisms. Here we used T cell receptor (TCR) transgenic animals to examine the role of inducible expression of nonlymphoid FasL in response to peptide antigen. Antigenic challenge of TCR transgenic mice resulted in increased expression of FasL in a number of nonlymphoid tissues including the epithelium of the small intestine. Similar results were obtained in an adoptive transfer system in which TCR transgenic T cells were transferred into recipient animals. The functional relevance of nonlymphoid FasL in peripheral deletion is supported by the observation that FasL-deficient gld animals showed a significantly reduced rate of clearance of transferred antigen-specific lymphocytes, although the lymphocytes themselves were wild type for FasL. These observations were supported further by studies in a transgenic mouse model where lacZ was expressed under the control of the proximal promoter of the FasL gene. Using these transgenic mice, we observed induced activity of the FasL promoter in intestinal epithelial cells throughout the crypts and villi, where we also observed infiltration of activated T cells. These data demonstrate that nonlymphoid FasL is expressed in response to peripheral T cell activation and participates in the regulation of T cells that infiltrate peripheral tissues.
Elimination of activated lymphocytes after an immunogenic stimulus is necessary for the maintenance of a functional immune system. Peripheral lymphocyte deletion is believed to occur predominantly through autocrine and paracrine interactions between lymphocytes. Activation-induced cell death occurs in T cells after repeated stimulation of the T cell receptor (TCR) complex (for review see ref. 1). Resting lymphocytes are resistant to killing via receptor-induced apoptotic mechanisms but develop increased sensitivity to apoptosis due to the coordinated up-regulation of the death-signaling surface receptor, Fas (CD95) (2) and its corresponding ligand, Fas ligand (FasL; CD95L) (3–6) along with regulation of the Fas-inhibitory protein, c-Flip (7–9).
Inadequate peripheral deletion results in the accumulation of activated lymphocytes and subsequent development of autoimmune pathologies. Dysfunctional Fas signaling is the underlying defect in autoimmune lymphoproliferative syndrome (ALPS types Ia and Ib) (10, 11) in humans and the corresponding mouse strains lpr and gld, respectively (12, 13). Although a primary means of peripheral lymphocyte deletion depends on lymphocyte FasL, evidence suggests that there is a contribution from inducible FasL expression in nonlymphoid tissues after antigen-driven lymphocyte expansion (14).
Peripheral lymphocytes undergo rapid expansion after encountering antigen, after which the majority of the expanded population undergoes clonal contraction to restore homeostasis (1). The latter phenomenon is referred to as “peripheral deletion” and is seen as a decrease in the numbers of antigen-specific T cells in peripheral lymphoid organs (1, 15). Although some of this decrease may be due to redistribution of the cells into peripheral nonlymphoid tissues, assessment throughout the animal has indicated that true deletion occurs (15, 16), probably via apoptosis of the activated T cells (1). Transgenic mice possessing a dominant lymphocyte population with a TCR for a known peptide have been used to activate a specific subset of peripheral T cells that subsequently expand and then decrease in number after clearance of antigen (17). These features make TCR transgenic mice a useful system in which to study peripheral deletion either directly in the transgenic animals or by transfer of transgenic lymphocytes into nontransgenic recipients.
Materials and Methods
Cell Lines and Animals.
C57BL/6, BALB/cByJ, BALB/c-TgN(DO11.10)10Loh (DO11.10), B6Smn.C3-Tnfsf6gld (B6-gld) BALB/cByJSmn-Prkdcscid (severe combined immunodeficient, SCID), B6.SJL-PtprcaPep3b/BoyJ (B6.SJL), and B10.D2-HclH2dH2-T18c/nSnJ (B10.D2) were purchased from The Jackson Laboratory. OT-2 [C57BL/6-Tg(TcraTcrb)425Cbn] have been described (18). hFlp-lacZ transgenic mice (C57BL/6/C3H) were generated at La Jolla Institute for Allergy and Immunology by introduction of the lacZ coding sequence under the control of the proximal promoter (1.2 kb) of the human FasL gene. Double transgenic (F1) mice were generated by hFlp-lacZ × OT-2 cross. Animals were housed in pathogen-free conditions at La Jolla Institute for Allergy and Immunology. DO11.10 TCR transgenic mice were assessed by analysis of peripheral blood using the clonotypic antibody KJ126-FITC (PharMingen) and anti-CD4 (phycoerythrin conjugate, PharMingen). OT-2 transgenic mice were assessed by three-color analysis with anti-CD4-CyChrome, anti-Vα2-phycoerythrin, and anti-Vβ8-FITC. Lymphocytes were isolated from spleens and lymph nodes, and primary intestinal epithelial cells (IECs) were isolated as outlined (19).
Adoptive Transfer and Antigenic Stimulation.
CD4+ lymphocyte subsets were enriched by complement lysis of unwanted cells, and 5-carboxy-fluorescein diacetate succinimidyl ester (CFSE)-labeling was performed as described (20). Lymphocytes (5 × 106) were introduced into the tail vein of recipient animals. Superantigen Staphylococcus enterotoxin B (SEB, 100 μg per mouse, Sigma) was injected i.p., OT-2 and hFlp-lacZ.OT-2 TCR transgenic mice were given ovalbumin (OVA) 323–339 peptide (100 μg i.p.) in PBS, and T cell-transfer recipients were given peptide antigen (300 μg s.c.) in complete Freund's adjuvant (CFA) as described (21).
mRNA Expression Analysis.
RNA was isolated from primary cells by Qiagen (Valencia, CA) RNeasy column or Trizol (GIBCO/BRL) extraction according to manufacturer instructions. First-strand cDNA was generated by using Superscript reverse transcriptase (GIBCO/BRL). Standard PCR was performed with Taq DNA polymerase (GIBCO/BRL) and assayed by agarose gel electrophoresis. Amplification of full-length FasL cDNA was performed with 5′ UTR (gAg Aag gAA ACC CTT TCC Tg) and 3′ UTR (Tgg AAg TgA gTg TAA Agg T) primers according to the conditions of Kayagaki et al. (22). Real-time PCR was performed with AmpliTaq Gold polymerase in a Perkin–Elmer Biosystems 5700 thermocycler by using the SyBr green detection protocol outlined by the manufacturer. Specific primers for murine FasL (forward primer, 5′-TgA ATT ACC CAT gTC CCC Ag-3′; reverse primer, 5′-AAA CTg ACC CTg gAg gAg CC-3′) and β-actin (forward primer, 5′-TTC gTT gCC ggT CCA CA-3′; reverse primer, 5′-ACC AgC gCA gCg ATA TCg-3′) were used. Quantitative measurements were performed in triplicate and standardized to the internal control of β-actin mRNA for each sample. Functionality of IEC FasL was analyzed by killing of L1210- and L1210-Fas target cells as described (19).
Histology.
Tissues from hFlp-lacZ transgenic and control littermates were stained for β-galactosidase activity with 5-bromo-3-indolyl β-D-galactopyranoside (Bluo-Gal) substrate (Sigma) followed by paraffin embedding. Briefly, tissues were fixed (2% formaldehyde/O.1 M Pipes, pH 6.9/2 mM MgCl2/1.25 mM EGTA) for 15 min at room temperature and stained in KFeCN solution [5 mM K3Fe(CN)6/5 mM K4Fe(CN)6⋅3H2O/1 mM MgCl2/0.01% sodium-deoxycholate/0.02% Nonidet P-40, pH 7.5) with Bluo-Gal (1 mg/ml) at 37°C for 4–12 h. Tissues were rinsed thoroughly in KFeCN solution before being embedded, sectioned, and counterstained with nuclear fast red for viewing by light microscopy with a Nikon microscope and digital capture device. Fluorescent images were captured with a Bio-Rad MRC1024ES confocal laser-scanning microscope.
Results
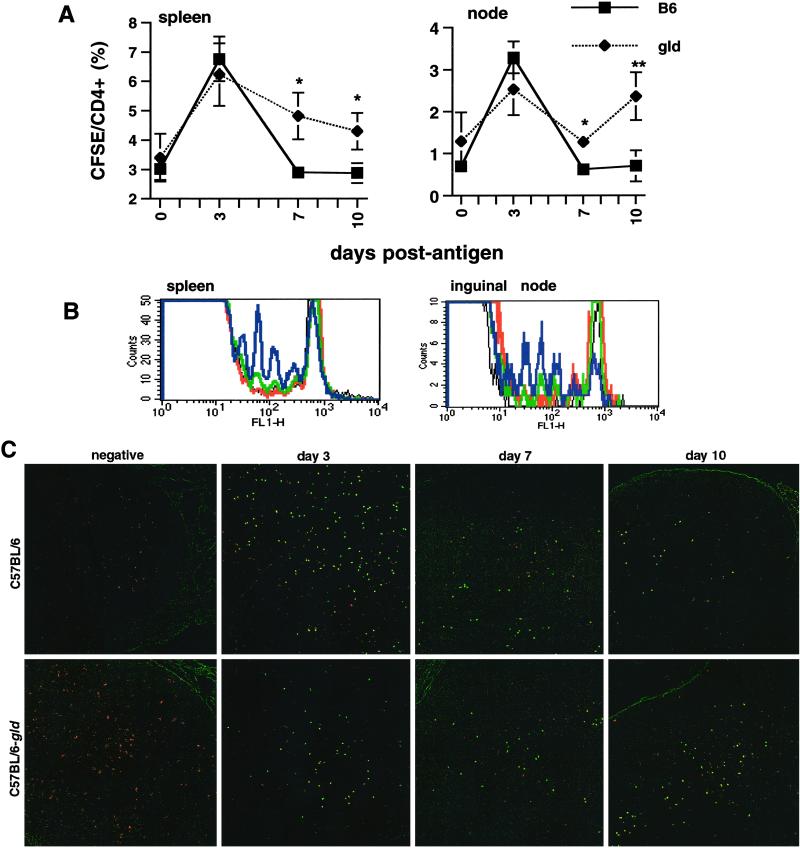
We used TCR transgenic animals that express transgenic TCRs (DO11.10 and OT-2) on CD4+ T cells that recognize OVA peptides presented on I-A (17, 18). To investigate the contribution of nonlymphoid tissues to peripheral lymphocyte deletion, we transferred CFSE-labeled CD4+ T cells from OT-2 TCR transgenic mice into C57BL/6-gld and WT recipients. Twelve hours after transfer, antigen (OVA peptide 323–339) was administered, and fluorescently labeled antigen-specific T cells were tracked in recipient animals. CD45.1+CD4+ cells from B6.SJL were coinjected with OT-2 to control for variation between numbers of antigen-specific cells received by each recipient animal. Whole-lymphocyte preparations were isolated from spleens and lymph nodes at the times indicated, and the number of CFSE-positive cells was measured by flow cytometry relative to the CD4+ populations. Fig. 1A shows the expansion of CFSE-positive OT-2 cells in response to peptide antigen and their progressive decline. Although the lymphoid cells that were transferred were WT for FasL, there was a significant delay in the decline in the numbers of CFSE-positive T cells in C57BL/6-gld recipient animals.
Fig 1.
Nonlymphoid FasL contributes to peripheral lymphocyte deletion. Accumulation of OT-2 lymphocytes in C57BL/6 and C57BL/6-gld recipient mice was measured after transfer of CFSE-labeled CD4+ lymphocytes from OT-2 mice into C57BL/6 and C57BL/6-gld recipient animals. Peripheral lymphocytes were harvested from spleens and nodes at the times indicated, and persistence of antigen-specific T cells was assessed by flow cytometry relative to the total CD4+ population. *, P < 0.003; **, P < 0.01 by paired t test. The data are from a minimum of five animals per data point. (B) CFSE fluorescence profiles of lymphocytes harvested 7 days after antigenic challenge (black, B6 + CFA; red, gld + CFA; green, B6 + OVA; blue, gld + OVA). OT-2 lymphocytes became activated in both recipient strains, and there was an accumulation of activated OT-2 cells in C57BL/6-gld mice. (C) Inguinal nodes from mice treated in A were harvested at the indicated times, fixed, embedded, sectioned, and analyzed by confocal microscopy. (Magnification, ×40.) CFSE-labeled OT-2 cells appear yellow.
We analyzed the CFSE fluorescence profiles from spleen and lymph node lymphocytes and found that OT-2 lymphocytes in both WT and gld recipients became activated and proliferated after injection of antigen, as shown by the sequential dilution of CFSE fluorescence and disappearance of cells in the unstimulated peak highlighted in the vehicle (CFA alone) control animals of each type (Fig. 1B). On day 3 (not shown), proliferation was observed in both groups. By day 7 (Fig. 1B) there was a noticeable difference between WT and gld mice with respect to the number of OT-2 cells in the peripheral lymphocyte pool. The accumulation and subsequent decline of OT-2 cells was also observed when we examined lymphoid tissues by fluorescence microscopy (Fig. 1C).
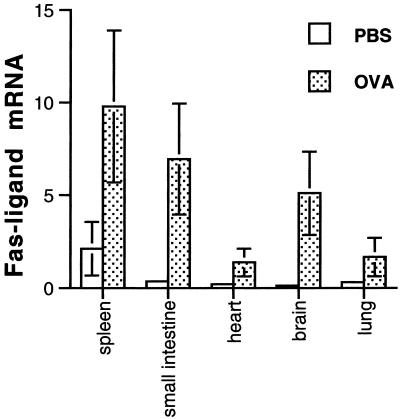
We then set out to identify the tissues responding to antigen-induced T cell expansion with an increase in FasL expression in both OT-2 and DO11.10 TCR transgenic mice. DO11.10 TCR transgenic mice were injected with antigen (OVA peptide 323–339), and FasL mRNA levels were assessed in a number of tissues. Fig. 2 shows the distribution of FasL expression pattern in antigen- and mock-treated DO11.10 transgenic animals. FasL mRNA levels were low in brain, heart, lung, and small intestine in untreated animals but were induced significantly after administration of antigen.
Fig 2.
Tissue distribution and inducible expression profile of nonlymphoid FasL after antigenic challenge in TCR transgenic mice. WT BALB/c and DO11.10 mice were given antigen (OVA 323–339 in PBS) i.v. or PBS alone, and tissues were harvested 24 hours later. FasL mRNA (per microgram total) was quantified by real-time RT-PCR. β-actin was used as an internal control for each respective sample. P < 0.05 by paired t test for all tissues between PBS and OVA treatment. Representative data are shown from triplicate analyses.
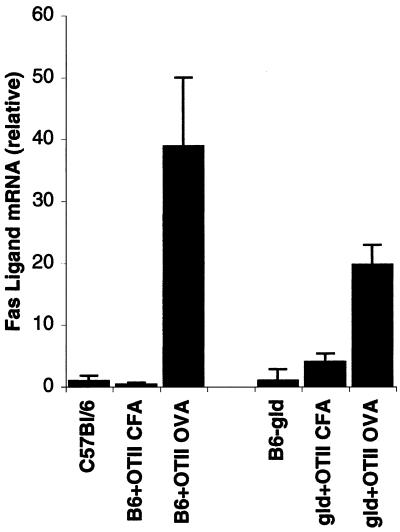
To pursue further the increase in FasL expression in IECs after OVA administration, we transferred OT-2 CD4+ lymphocytes into C57BL/6 and C57BL/6-gld animals and challenged recipient animals with OVA peptide. CFA-treated C57BL/6 recipients were virtually identical to untreated animals, whereas there was a slight increase of IEC FasL in C57BL/6-gld animals that received CFA. However, both C57BL/6 and C57BL/6-gld animals showed a marked increase in FasL mRNA after administration of antigen (Fig. 3). Additional experiments were performed in WT recipients by using both OT-2-C57BL/6 and DO11.10-BALB/c pairs with similar results (data not shown). IEC mRNA was also analyzed for expression of the lymphocyte marker, CD45. This was undetectable in our IEC RNA preparations (data not shown), demonstrating that our IEC preparations were completely devoid of contaminating lymphocytes.
Fig 3.
Both C57BL/6 and C57BL/6-gld recipients of OT-2 cells up-regulate FasL in response to challenge with antigen. CD4+ lymphocytes were isolated from OT-2 mice and injected into C57BL/6 and C57BL/6-gld recipients. Twelve hours after transfer, mice were administered OVA peptide (in CFA) or CFA alone. Three days after antigen, IECs were isolated, and FasL mRNA was measured by real-time RT-PCR. Data from representative mice are shown, and values indicating fold induction are relative to mock-treated animals of each type. IEC RNA samples were analyzed for the CD45 expression. CD45 mRNA was not detected in IEC RNA preparations in our real-time RT-PCR assays (not shown).
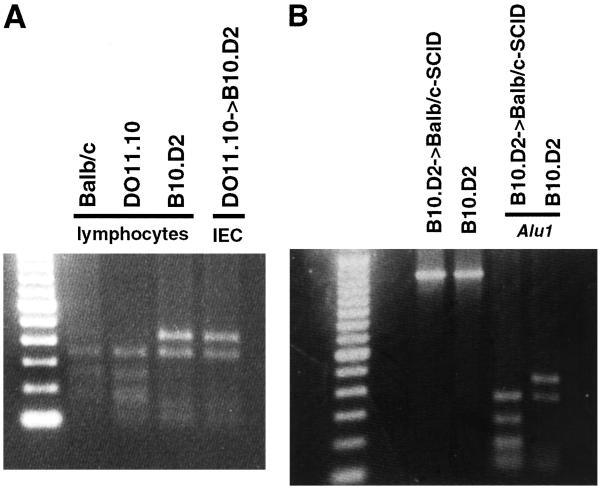
To examine more closely the induction of FasL in IECs, we again used an adoptive transfer approach. We transferred DO11.10 CD4+ cells into B10.D2 recipients. These two strains are the same MHC haplotype (H-2d) but differ in the FasL coding sequence (22), allowing for differentiation of FasL mRNA by an AluI restriction site present in BALB/c but not B10.D2. T cells from DO11.10 were injected i.v. into B10.D2 recipients. Twelve hours later, recipient animals were given OVA peptide. Three days after administration of antigen, IECs were isolated and analyzed for expression of FasL mRNA by RT-PCR. PCR products corresponding to full-length FasL coding sequence were purified, digested with AluI, and analyzed by agarose gel electrophoresis (Fig. 4A). FasL cDNA arising from BALB/c gave rise to the same fragment pattern described earlier (22); however, IECs of B10.D2 mice that received DO11.10 lymphocytes plus antigen expressed FasL with the pattern expected from that of the recipient B10.D2 mouse. Functionality of IEC FasL was confirmed by the assessment of killing activity against Fas-bearing target cells, L1210-Fas (data not shown), as shown for IECs from animals immunized with SEB (23).
Fig 4.
Induction of nonlymphoid FasL mRNA after transfer of TCR transgenic lymphocytes and antigen stimulation. (A) CD4+ lymphocytes from DO11.10 mice were introduced i.v. into B10.D2 recipient animals followed by antigenic stimulation (OVA 323–339 in CFA, s.c.). IECs and splenocytes were harvested 3 days postantigen. cDNA from each respective tissue was generated, and full-length FasL was amplified and digested with AluI to identify the source of FasL RNA. Splenocyte FasL was included from BALB/c, DO11.10, and B10.D2 to demonstrate each restriction-fragment pattern. (B) Further demonstration that induction of IEC FasL is nonlymphoid. B10.D2 mice were given SEB and lymphocytes isolated 3 days later and transferred into BALB/c-SCID recipients. Twenty-four hours after transfer, IECs were isolated and RNA was purified. Full-length cDNA was amplified and subjected to AluI restriction-digest analysis.
To confirm that the FasL we observed was from a nonlymphoid source, we performed the transfer experiment described above in the opposite direction by using SCID recipients. B10.D2 lymphocytes were activated by the superantigen, SEB, for 3 days and isolated, and T cells were transferred into BALB/c-SCID recipients. RNA was isolated from IECs harvested from recipient animals 24 h after transfer. Full-length cDNA was generated and digested with AluI (Fig. 4B). BALB/c-SCID recipients, which do not possess responsive T cells, showed an up-regulation of FasL after transfer of activated B10.D2 lymphocytes, and the source of FasL corresponds to that arising from nonlymphoid IECs of the BALB/c-SCID mouse.
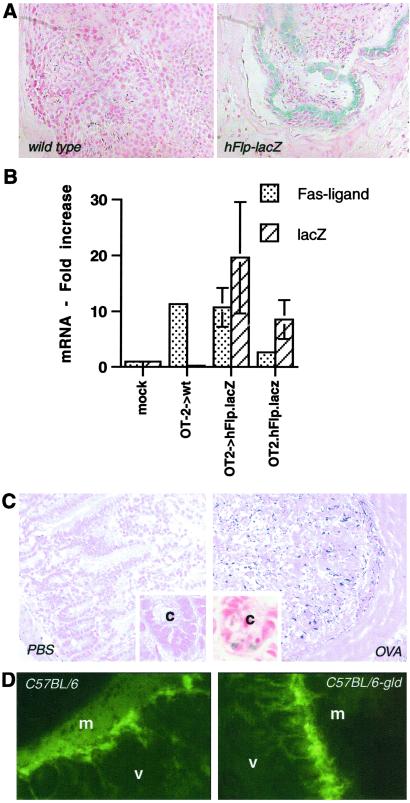
As a means to perform coordinate studies of lymphocyte activation and IEC FasL expression, we generated a transgenic mouse (hFlp-lacZ) expressing a lacZ construct under the control of the proximal promoter of the human FasL gene. As FasL is constitutively expressed in the testes (24, 25), sections of testes were examined for β-galactosidase activity (Fig. 5A). Staining showed cellular specificity only in the transgenic animals. We then transferred CD4+ T cells from OT-2 animals into hFlp-lacZ transgenic recipients, followed by challenge with OVA peptide 12 hours later. After 3 days IECs were isolated, and RNA was analyzed for endogenous FasL and lacZ expression. As shown in Fig. 5B, induced expression of lacZ correlated with production of FasL mRNA, thus demonstrating that activated lymphocyte-induced IEC FasL is controlled by elements in the proximal promoter of the FasL gene. A similar effect was observed when hFlp-lacZxOT-2 double-transgenic animals were immunized with OVA (Fig. 5B).
Fig 5.
In vivo analysis of FasL expression. Transgenic mice were generated that express lacZ under the control of the proximal FasL promoter. (A) Testes of transgenic and WT littermate controls animals were stained for β-galactosidase activity before being fixed, embedded, and sectioned for analysis by light microscopy. (Magnification, ×200.) The transgenic animal (Right) shows a significant number of blue cells that were absent in the negative littermate control (Left). (B) OT-2 CD4+ T cells were transferred into hFlp-lacZ and littermate control animals followed by antigenic challenge. Three days later, RNA from IECs was harvested and analyzed for FasL and lacZ transcript levels. mock, hFlp-lacZ animals that were not challenged with antigen (i.e., CFA alone). OT-2.hFlp-lacZ double-transgenic animals were administered OVA peptide, and IEC RNA was harvested 24 hours later. (C) Small intestines from double-transgenic (OT-2xhFlp-lacZ) animals (24 hours after administration of OVA) were stained for β-galactosidase activity. (Magnification, ×200; Inset ×670.) The intestine of an antigen-challenged animal (Right) and that of a mock-treated (PBS) animal (Left) are shown. The appearance of blue cells corresponds with the induction of endogenous FasL and lacZ mRNA observed above (B). (Inset) Higher magnification of each treatment. c, the lumen of the crypt. (D) CFSE-labeled CD4+ cells from OT-2 mice were transferred into C57BL/6 and C57BL/6-gld recipients as described for Fig. 1. Small intestine sections were examined for the appearance of fluorescent lymphocytes in the regions where we observed induction of FasL promoter activity in the intestinal epithelium. (Magnification, ×200.) In both strains we detected a significant CD4+ infiltrate 3 days after administration of OVA peptide. m and v, the muscularis and villi, respectively.
To study this phenomenon in situ, we stained small intestine samples from antigen-treated hFlp-lacZxOT-2 mice. 5-Bromo-3-indolyl β-D-galactopyranoside-stained tissues were embedded, sectioned, and counterstained with nuclear fast red (Fig. 5C). Few if any blue cells were detectable in PBS-treated animals, but the appearance of blue cells, indicating FasL promoter activity, were readily detectable along the epithelium of the small intestine (Fig. 5C Inset). This pattern is similar to expression of FasL in SEB-treated animals as observed by in situ hybridization (23). To determine whether the localization of induced FasL promoter activity correlated with lymphocyte infiltration, we analyzed small intestine isolated from C57BL/6 and C57BL/6-gld recipient animals that had received CFSE-labeled OT-2 T cells and were subsequently challenged with OVA peptide antigen. Fig. 5D shows the appearance of CFSE-labeled cells in the small intestines of recipient animals 3 days after activation. The T cells infiltrated the same regions that express FasL in response to activated T cells. Fewer lymphocytes were found in the gut of CFA-treated animals at all times points examined (data not shown), suggesting that activated lymphocytes may have a greater propensity to traffic through the small intestine. The trafficking of activated T cells through peripheral tissues has been described in other systems (16, 26). Our results indicate that FasL induced on nonlymphoid tissues by activated T cells contributes to peripheral deletion of the lymphocytes.
Discussion
FasL is expressed in a number of nonlymphoid tissues in response to lymphocyte infiltration and other cellular stresses (see ref. 27). Perhaps the best characterized example is that of immune privilege (14), where certain tissues constitutively express FasL and are cytotoxic for Fas-bearing lymphocytes, thereby suppressing significant lymphocyte infiltration and/or inflammatory responses such as in the eye (28, 29). Similarly, cells of the thyroid express FasL, which is required to prevent cell-mediated autoimmune destruction of thyrocytes observed in Hashimoto's thyroiditis (30, 31), and our data demonstrate that FasL-defective animals cannot perform peripheral deletion equal to that in WT animals even when the lymphocytes themselves have functional FasL.
Huang et al. (32) reported that activated TCR transgenic T cells migrated to the liver where they were disposed and were stimulated to die in a lymphoid-dependent manner. Our results showing reduced peripheral deletion of OT-2 cells in C57BL/6-gld animals do not exclude the involvement of the liver as a site of disposal for T cells triggered to die in the periphery (see ref. 33). It is possible that, rather than a “graveyard,” this is one site of active deletion, because cells of the liver can also express FasL during immune responses (23).
In a recent report (34), deletion of CD4+Vβ8+ cells after SEB challenge was found to be influenced by FasL and tumor necrosis factor but mediated primarily by lymphocyte expression of Bim, a BH3-only member of the Bcl-2 protein family. It is not surprising that more than one mechanism of peripheral deletion may be involved in this process. It is possible that dose of antigen and/or mode of presentation may influence which mechanism(s) of deletion will predominate.
Migration of activated lymphocytes to mucosal epithelia has been proposed for clearance of lymphocytes that have been activated to undergo apoptosis in a strictly lymphoid-dependent fashion (32). However, it has been demonstrated that activated lymphocytes home to the gut before induction of their death programs, and recent evidence suggests that surviving cells may constitute a reservoir of antigen-specific memory cells (16). Activated lymphocytes induce up-regulation of FasL on the cells of the intestinal epithelium (Fig. 5) in a fashion similar to that of inducible immune privilege (23). In the latter model, it is believed that activated lymphocytes enter the small intestine (in addition to the liver and lung) and by a lymphocyte-derived cellular signal induce expression of FasL on epithelial cells, which are then responsible for inducing the activated lymphocytes to undergo apoptosis.
FasL has also been found to induce granulocytosis when its expression is enforced in nonlymphoid tissues in transgenic mice or transplanted tumors (35). However, in none of our experiments did we observe a granulocytosis induced by up-regulation of endogenous FasL (e.g., Fig. 5C, ref. 23, and M.J.P. and D.R.G., unpublished observations). Recently, Janin et al. (36) showed that disseminated destruction of endothelial tissue can be caused by the transfer of FasL-expressing lymphocytes. In tissues or tumors with enforced expression of FasL, endothelia may be contacted, and the resulting damage then may be an underlying cause of neutrophil infiltration. In contrast, it is possible that FasL on the intestinal epithelium is not in sufficient proximity to endothelial tissue to cause its destruction.
The signals responsible for induction of FasL expression in nonlymphoid tissues after T cell activation are only partly understood. Recently we demonstrated that T cell-derived tumor necrosis factor is necessary and sufficient for induction of FasL in IECs in response to SEB (37). Interestingly, tumor necrosis factor has been shown to contribute to peripheral deletion of TCR transgenic T cells (38). However, the role of tumor necrosis factor in FasL expression in other tissues during immune responses is not known.
An emerging model for clonal deletion of activated lymphocytes parallels that of immune privilege (14). Nonlymphoid tissue may act as a monitor of activated T cell numbers, possibly through microenvironment cytokine concentration by the infiltrating T cells, and is then capable of controlling lymphoid expansion through expression of proapoptotic signals such as FasL. In this model, the control of lymphocyte number is not entirely a function of hematopoietic cells but a dynamic interaction between activated T cells and peripheral epithelia.
Acknowledgments
We thank Adam Pinkoski for technical assistance and H. Beere for critical reading of the manuscript. This work was supported by National Institutes of Health Grant AI44828-02. This is La Jolla Institute for Allergy and Immunology article no. 514.
Abbreviations
TCR, T cell receptor
FasL, Fas ligand
SCID, severe combined immunodeficient
CFSE, 5-carboxy-fluorescein diacetate succinimidyl ester
SEB, Staphylococcus enterotoxin B
OVA, ovalbumin
CFA, complete Freund's adjuvant
IEC, intestinal epithelial cell
References
- 1.Lenardo M., Chan, K. M., Hornung, F., McFarland, H., Siegel, R., Wang, J. & Zheng, L. (1999) Annu. Rev. Immunol. 17, 221-253. [DOI] [PubMed] [Google Scholar]
- 2.Mogil R. J., Radvanyi, L., Gonzalez-Quintial, R., Miller, R., Mills, G., Theofilopoulos, A. N. & Green, D. R. (1995) Int. Immunol. 7, 1451-1458. [DOI] [PubMed] [Google Scholar]
- 3.Alderson M. R., Tough, T. W., Davis-Smith, T., Braddy, S., Falk, B., Schooley, K. A., Goodwin, R. G., Smith, C. A., Ramsdell, F. & Lynch, D. H. (1995) J. Exp. Med. 181, 71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju S. T., Panka, D. J., Cui, H., Ettinger, R., el-Khatib, M., Sherr, D. H., Stanger, B. Z. & Marshak-Rothstein, A. (1995) Nature 373, 444-448. [DOI] [PubMed] [Google Scholar]
- 5.Dhein J., Walczak, H., Baumler, C., Debatin, K. M. & Krammer, P. H. (1995) Nature 373, 438-441. [DOI] [PubMed] [Google Scholar]
- 6.Brunner T., Mogil, R. J., LaFace, D., Yoo, N. J., Mahboubi, A., Echeverri, F., Martin, S. J., Force, W. R., Lynch, D. H., Ware, C. F., et al. (1995) Nature 373, 441-444. [DOI] [PubMed] [Google Scholar]
- 7.Kataoka T., Schroter, M., Hahne, M., Schneider, P., Irmler, M., Thome, M., Froelich, C. J. & Tschopp, J. (1998) J. Immunol. 161, 3936-3942. [PubMed] [Google Scholar]
- 8.Kirchhoff S., Muller, W. W., Krueger, A., Schmitz, I. & Krammer, P. H. (2000) J. Immunol. 165, 6293-6300. [DOI] [PubMed] [Google Scholar]
- 9.Kirchhoff S., Muller, W. W., Li-Weber, M. & Krammer, P. H. (2000) Eur. J. Immunol. 30, 2765-2774. [DOI] [PubMed] [Google Scholar]
- 10.Fisher G. H., Rosenberg, F. J., Straus, S. E., Dale, J. K., Middleton, L. A., Lin, A. Y., Strober, W., Lenardo, M. J. & Puck, J. M. (1995) Cell 81, 935-946. [DOI] [PubMed] [Google Scholar]
- 11.Martin D. A., Zheng, L., Siegel, R. M., Huang, B., Fisher, G. H., Wang, J., Jackson, C. E., Puck, J. M., Dale, J., Straus, S. E., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 4552-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe-Fukunaga R., Brannan, C. I., Copeland, N. G., Jenkins, N. A. & Nagata, S. (1992) Nature 356, 314-317. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi T., Tanaka, M., Brannan, C. I., Jenkins, N. A., Copeland, N. G., Suda, T. & Nagata, S. (1994) Cell 76, 969-976. [DOI] [PubMed] [Google Scholar]
- 14.Green D. R. & Ferguson, T. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 917-924. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins M. K., Khoruts, A., Ingulli, E., Mueller, D. L., McSorley, S. J., Reinhardt, R. L., Itano, A. & Pape, K. A. (2001) Annu. Rev. Immunol. 19, 23-45. [DOI] [PubMed] [Google Scholar]
- 16.Reinhardt R. L., Khoruts, A., Merica, R., Zell, T. & Jenkins, M. K. (2001) Nature 410, 101-105. [DOI] [PubMed] [Google Scholar]
- 17.Pape K. A., Kearney, E. R., Khoruts, A., Mondino, A., Merica, R., Chen, Z. M., Ingulli, E., White, J., Johnson, J. G. & Jenkins, M. K. (1997) Immunol. Rev. 156, 67-78. [DOI] [PubMed] [Google Scholar]
- 18.Barnden M. J., Allison, J., Heath, W. R. & Carbone, F. R. (1998) Immunol. Cell Biol. 76, 34-40. [DOI] [PubMed] [Google Scholar]
- 19.Lin T., Brunner, T., Tietz, B., Madsen, J., Bonfoco, E., Reaves, M., Huflejt, M. & Green, D. R. (1998) J. Clin. Invest. 101, 570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Stipdonk M. J., Lemmens, E. E. & Schoenberger, S. P. (2001) Nat. Immunol. 2, 423-429. [DOI] [PubMed] [Google Scholar]
- 21.Kearney E. R., Pape, K. A., Loh, D. Y. & Jenkins, M. K. (1994) Immunity 1, 327-339. [DOI] [PubMed] [Google Scholar]
- 22.Kayagaki N., Yamaguchi, N., Nagao, F., Matsuo, S., Maeda, H., Okumura, K. & Yagita, H. (1997) Proc. Natl. Acad. Sci. USA 94, 3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonfoco E., Stuart, P. M., Brunner, T., Lin, T., Griffith, T. S., Gao, Y., Nakajima, H., Henkart, P. A., Ferguson, T. A. & Green, D. R. (1998) Immunity 9, 711-720. [DOI] [PubMed] [Google Scholar]
- 24.D'Alessio A., Riccioli, A., Lauretti, P., Padula, F., Muciaccia, B., De Cesaris, P., Filippini, A., Nagata, S. & Ziparo, E. (2001) Proc. Natl. Acad. Sci. USA 98, 3316-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.French L. E., Hahne, M., Viard, I., Radlgruber, G., Zanone, R., Becker, K., Muller, C. & Tschopp, J. (1996) J. Cell Biol. 133, 335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masopust D., Vezys, V., Marzo, A. L. & Lefrancois, L. (2001) Science 291, 2413-2417. [DOI] [PubMed] [Google Scholar]
- 27.Pinkoski M. J. & Green, D. R. (2000) Nat. Immunol. 1, 461-462. [DOI] [PubMed] [Google Scholar]
- 28.Griffith T. S., Yu, X., Herndon, J. M., Green, D. R. & Ferguson, T. A. (1996) Immunity 5, 7-16. [DOI] [PubMed] [Google Scholar]
- 29.Griffith T. S., Brunner, T., Fletcher, S. M., Green, D. R. & Ferguson, T. A. (1995) Science 270, 1189-1192. [DOI] [PubMed] [Google Scholar]
- 30.Stassi G., Di Liberto, D., Todaro, M., Zeuner, A., Ricci-Vitiani, L., Stoppacciaro, A., Ruco, L., Farina, F., Zummo, G. & De Maria, R. (2000) Nat. Immunol. 1, 483-488. [DOI] [PubMed] [Google Scholar]
- 31.Stassi G., Zeuner, A., Di Liberto, D., Todaro, M., Ricci-Vitiani, L. & De Maria, R. (2001) J. Clin. Immunol. 21, 19-23. [DOI] [PubMed] [Google Scholar]
- 32.Huang L., Soldevila, G., Leeker, M., Flavell, R. & Crispe, I. N. (1994) Immunity 1, 741-749. [DOI] [PubMed] [Google Scholar]
- 33.Crispe I. N., Dao, T., Klugewitz, K., Mehal, W. Z. & Metz, D. P. (2000) Immunol. Rev. 174, 47-62. [DOI] [PubMed] [Google Scholar]
- 34.Hildeman D. A., Zhu, Y., Mitchell, T. C., Bouillet, P., Strasser, A., Kappler, J. & Marrack, P. (2002) Immunity 16, 759-767. [DOI] [PubMed] [Google Scholar]
- 35.O'Connell J., Bennett, M. W., O'Sullivan, G. C., Collins, J. K. & Shanahan, F. (1999) Immunol. Today 20, 46-52. [DOI] [PubMed] [Google Scholar]
- 36.Janin A., Deschaumes, C., Daneshpouy, M., Estaquier, J., Micic-Polianski, J., Rajagopalan-Levasseur, P., Akarid, K., Mounier, N., Gluckman, E., Socie, G. & Ameisen, J. C. (2002) Blood 99, 2940-2947. [DOI] [PubMed] [Google Scholar]
- 37.Pinkoski M. J., Droin, N. M. & Green, D. R. (2002) J. Biol. Chem. 277, 42380-42385. [DOI] [PubMed] [Google Scholar]
- 38.Sytwu H. K., Liblau, R. S. & McDevitt, H. O. (1996) Immunity 5, 17-30. [DOI] [PubMed] [Google Scholar]