Abstract
HLA-G is a nonclassical class I MHC molecule with an unknown function and with unusual characteristics that distinguish it from other class I MHC molecules. Here, we demonstrate that HLA-G forms disulfide-linked dimers that are present on the cell surface. Immunoprecipitation of HLA-G from surface biotinylated transfectants using the anti-β2-microglobulin mAb BBM.1 revealed the presence of an ≈78-kDa form of HLA-G heavy chain that was reduced by using DTT to a 39-kDa form. Mutation of Cys-42 to a serine completely abrogated dimerization of HLA-G, suggesting that the disulfide linkage formed exclusively through this residue. A possible interaction between the HLA-G monomer or dimer and the KIR2DL4 receptor was also investigated, but no interaction between these molecules could be detected through several approaches. The cell-surface expression of dimerized HLA-G molecules may have implications for HLA-G/receptor interactions and for the search for specific receptors that bind HLA-G.
HLA-G is an unusual class I MHC molecule possessing several characteristics that distinguish it from other MHC molecules. Its expression is largely restricted to the invasive extravillous trophoblasts of the decidua (1, 2), it exhibits limited polymorphism (3, 4), and it possesses a truncated cytoplasmic domain (5). Based on its pattern of expression, HLA-G is assumed to play some role at the maternal–fetal interface, but its specific function remains unclear. HLA-G expression has also been reported on some tumors, suggesting a possible role in tumor evasion (6, 7).
Several roles for HLA-G in the decidua are possible. One role may be to provide a signal sequence-derived peptide for binding by HLA-E (also expressed on trophoblasts), a high-affinity ligand for the inhibitory CD94/NKG2A receptor (8). Another role may be to bind KIR2DL4, an activating receptor expressed on NK cells (9–12); lastly, HLA-G may bind the inhibitory ILT2 and ILT4 receptors (13, 14).
Certain aspects of HLA-G suggest that it may function in a manner distinct from other class I MHC molecules. The HLA-G promoter, for example, is divergent from other class I MHC promoters, which may help to explain its pattern of expression (15). HLA-G also has an unusually long half-life on the cell surface, the result of the absence of an endocytosis motif in its truncated cytoplasmic domain (16, 17). Interestingly, this truncation also reveals a dilysine motif that acts to recycle HLA-G from the Golgi back to the endoplasmic reticulum, thus providing an apparent means for the binding by HLA-G of high-affinity peptides (17).
Here, another unusual aspect of HLA-G is demonstrated: its ability to form disulfide-linked dimers both in vitro and on the cell surface. By using site-directed mutagenesis, it is shown that the dimers form via Cys-42 of the HLA-G α1 domain. The formation of a dimerized form of HLA-G may have implications in identifying its functional receptors.
Materials and Methods
Cell Lines and Antibodies.
The 721.221 B-LCLs2 transfected with various class I MHC cDNAs have been described (18, 19). The BW5147.3 mouse thymoma (BW−), and BBM.1 [anti-β2-microglobulin (β2m)] and W6/32 (conformation-sensitive pan-HLA) hybridomas were obtained from American Type Culture Collection. The anti-KIR2DL1 mAb EB6 was obtained from Beckman Coulter. Monoclonal antibodies to HLA-G were prepared by standard procedures from mice immunized with bacterially produced heavy chain, either fully denatured (MEM-G/1, IgG1) or refolded to produce native complexes with β2m and peptide (MEM-G/11, IgG1; see below). KIR2DL4/KIR2DL5-reactive rabbit antiserum was generated by injecting rabbits s.c. with 100 μg of KIR2DL5-Ig fusion protein in complete Freund's adjuvant, followed 14 days later by a second injection s.c. in incomplete Freund's adjuvant (Serasource, Royalston, MA). This polyclonal antibody recognizes both KIR2DL4 and KIR2DL5.
Protein Expression.
Soluble HLA-G was PCR-amplified from JEG-3 choriocarcinoma cell line cDNA and subcloned into the pGMT7 expression vector. The HLA-Cw6 and β2m expression constructs have been described (20). Soluble recombinant proteins were produced in Escherichia coli BL21 (DE3) pLysS (Novagen) and refolded as described (20). HLA-G was refolded with the peptide, VLPKLYVKL, and HLA-Cw6 was refolded with the peptide, YQFTGIKKY.
KIR-Ig fusion constructs were constructed by fusing the signal sequence and extracellular domains of KIRs to the Fc portion of human IgG1 (derived from the CD51neg1, a gift from B. Seed, Massachusetts General Hospital, Boston) and by subcloning into the pCDNA3 expression plasmid (Invitrogen). KIR-Ig plasmids were transfected into COS-7 cells using Fugene 6 (Roche Molecular Biochemicals) and purified over a Protein G/Poros20 column using perfusion chromatography (PerSeptive Biosystems, Framingham, MA). Fusion proteins were eluted in 2% acetic acid and immediately neutralized with 1 M Tris, pH 8.0, followed by dialysis against PBS, pH 7.4.
SDS/PAGE and Western Blotting.
Cells were lysed directly in SDS/PAGE sample buffer containing freshly prepared 50 mM iodoacetamide (Sigma). Purified inclusion bodies in 8 M urea were resuspended in SDS/PAGE sample buffer containing 50 mM DTT. Samples were boiled for 5 min, run on SDS/PAGE, and blotted to nitrocellulose. Heavy chain was detected by using the MEM-G/1 mAb, followed by horseradish peroxidase-conjugated goat anti-mouse IgG mAb (Jackson ImmunoResearch). Blots were visualized by using enhanced chemiluminescence (ECL; Amersham Pharmacia).
Immunohistochemistry.
Decidual tissue was identified macroscopically and washed in PBS, pH 7.2. Decidual pieces were fixed overnight at 4°C in 4% (wt/vol) paraformaldehyde. Fixed tissue was dehydrated in an ethanol/xylene series and embedded in paraffin. Then, 6-μm sections were rehydrated and incubated in 3% (vol/vol) H2O2 in methanol to block endogenous peroxidase activity. MEM-G/1 mAb staining was detected by using a biotinylated secondary mAb and horseradish peroxidase-conjugated streptavidin. Staining was visualized by using diaminobenzidine substrate.
Cell-Surface Labeling and Immunoprecipitation.
Cells were surface biotinylated by first washing with PBS, pH 7.2, supplemented with 0.1 mM CaCl2/1 mM MgCl2 and then incubated for 30 min at 4°C in 0.5 mg/ml sulfo-NHS-biotin (Pierce). Cells then were washed and lysed in lysis buffer [50 mM Tris, pH 7.6/150 mM NaCl/5 mM EDTA/1% (vol/vol) Nonidet P-40 and protease inhibitors (Roche Molecular Biochemicals)]. When iodoacetamide was used, biotinylated cells were split into two aliquots. One aliquot was lysed with lysis buffer alone and one with lysis buffer supplemented with 50 mM iodoacetamide (freshly prepared). Cell lysates were precleared with protein A-Sepharose beads coated with normal rabbit serum and immunoprecipitated overnight at 4°C with BBM.1-coated beads. After washing, the beads were divided into two samples, and one was boiled in SDS/PAGE buffer and one in SDS/PAGE buffer containing 50 mM DTT. Eluted protein was run on SDS/PAGE gels and blotted onto nitrocellulose. Biotinylated proteins were detected by using HRP-conjugated streptavidin (Roche Molecular Biochemicals) and visualized by ECL.
Mutagenesis.
The HLA-A2 crystal structure was visualized by using RASMOL. Cys-42 was mutagenized to serine by using site-directed mutagenesis according to the manufacturer's instructions (Stratagene). The PAGE-purified primer Gmut1: 5′-GACAGCGACTCGGCGAGTCCGAGGATGGAGCCG-3′ and its reverse complement were used for mutagenesis.
KIR-ζ Fusion Constructs and Transfectants.
A KIR2DL4-ζ fusion protein in which the extracellular domain of KIR2DL4 was fused to the transmembrane and cytoplasmic domains of CD3-ζ was generated by overlapping PCR. The KIR2DL4 extracellular domain was amplified by using 5′K103Kpn, 5′-GGCGGTACCATGTCCATGTCACCCACG-3′, and 3′K103-ζ, 5′-CAGGCCAAAGCTCTGATGCAGGTGTCTGGC-3′. ζ-chain was amplified by using 5′K103-ζ 5′-GCCAGACACCTGCATCAGAGCTTTGGCCTG-3′ and 3′ζ, 5′-CGGAATTCCGTTAGCGAGGGGCAGG-3′. The gel-purified templates were reamplified in a second PCR by using 5′K103Kpn and 3′ζ and cloned into the pEF6 expression vector (Invitrogen). KIR-ζ constructs were electroporated into mouse BW cells (250 V, 500 μF), and stable transfectants were established.
BW/KIR-ζ transfectants were cocultured in 96-well round- bottom plates with γ-irradiated (3,000 rad) stimulators for 72 h. As positive controls, BW/KIR-ζ transfectants were incubated in wells coated with either 10 μg/ml EB6 (for KIR2DL1-ζ) or with 20 μg/ml purified anti-KIR2DL4/KIR2DL5 antiserum (for KIR2DL4-ζ). Supernatants were tested for mouse IL-2 by using ELISA (BioSource International, Camarillo, CA).
Surface Plasmon Resonance.
Goat anti-human Fc receptor antibody (Jackson ImmunoResearch) was coupled to CM5 chips by using standard amine coupling according to the manufacturer's instructions (Biacore, Piscataway, NJ). KIR2DL1-Ig and KIR2DL4-Ig fusion proteins (30 μg/ml) were then bound to the surface at a flow rate of 5 μl/min. Recombinant class I MHC molecules were purified by gel filtration just before use, diluted in HBS-EP running buffer, and flowed over the chip at a flow rate of 5 μl/min. Both mock-coupled and anti-human Fc-coupled surfaces were used as controls. Binding data were evaluated by using BIAEVALUATION V.3.0 (Biacore).
Results
Generation and Characterization of Anti-HLA-G mAbs.
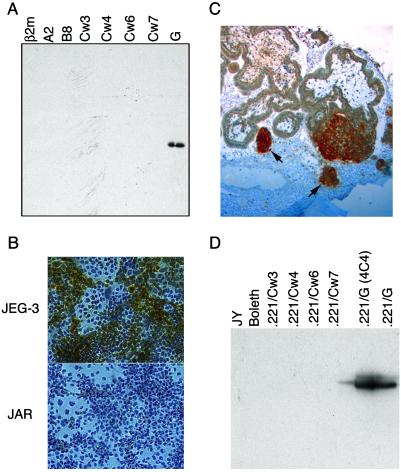
To facilitate the examination of HLA-G in vitro and in vivo, HLA-G-specific mAbs were generated against both denatured heavy chain (MEM-G/1 mAb) and HLA-G that had been refolded with β2m and peptide (MEM-G/11 mAb). MEM-G/1 was specific for HLA-G heavy chain in Western blots (Fig. 1A), and it stained cytospin preparations of JEG-3 (HLA-G+) but not JAR (HLA-G−) trophoblast cell lines (Fig. 1B). Furthermore, MEM-G/1 was specific for the extravillous trophoblast population when used in immunohistochemical staining of paraffin-embedded placental sections (Fig. 1C). Immunoprecipitation of a panel of cell lines and HLA transfectants demonstrated that MEM-G/11 recognized HLA-G specifically (Fig. 1D). The HLA-G specificity of MEM-G/11 was confirmed by using fluorescence activated cell sorter (FACS) analysis of a panel of HLA-transfected cell lines (data not shown).
Fig 1.
Characterization of HLA-G-specific mAbs. (A) Western blot of bacterially produced solubilized inclusion bodies from various soluble recombinant HLA molecules detected by MEM-G/1 mAb. (B) Cytospin preparations of the HLA-G+ choriocarcinoma cell line JEG-3 and the HLA-G− cell line JAR were stained with the MEM-G/1 mAb. Whereas JAR is completely negative, HLA-G staining (brown) can be seen clearly in the JEG-3 sample. Note that not all of the JEG-3 cells stain positively, which is consistent with the heterogeneous nature of this tumor line. (C) Immunohistochemistry of paraffin-embedded placental sections using the MEM-G/1 mAb. HLA-G staining (dark brown) was detected on the cell islands (denoted by arrows) that had migrated from the villi. (D) The MEM-G/11 mAb was used to immunoprecipitate HLA-G from surface-biotinylated B-lymphoblastoid cell lines and from various class I MHC transfected into the MHC-deficient 721.221 B-lymphoblastoid cell line. ECL visualization demonstrated that only the two different clones of HLA-G+ transfectants were recognized.
In Vitro Dimerization of HLA-G.
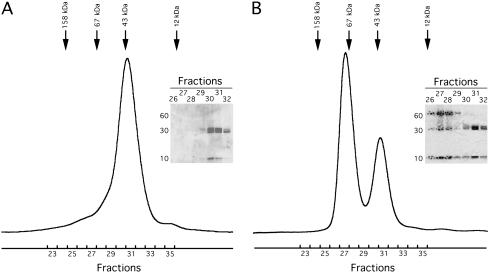
Repurification by gel filtration of refolded HLA-G/β2m/peptide heterotrimers (designated throughout as HLA-G monomer), resulted in the serendipitous discovery of two peaks, one corresponding to the expected molecular mass (MM) of 43 kDa and one approximately twice that. These data suggested that HLA-G dimerization had occurred. To confirm this, HLA-G was refolded with β2m and peptide and purified to homogeneity by using gel filtration (Fig. 2A). The purified HLA-G monomer was kept at 4°C for 21 days, and the sample was analyzed again by gel filtration. After the incubation, the majority (≈75%) of the refolded HLA-G eluted earlier than the HLA-G monomer. The early eluting peak corresponded to a MM of approximately twice that of the monomeric form of HLA-G (Fig. 2B). Analysis of the fractions by SDS/PAGE under nonreducing conditions confirmed that this new peak contained dimerized (≈66 kDa) and monomeric (≈33 kDa) HLA-G heavy chain. The HLA-G dimers could be reduced to monomers under reducing conditions, suggesting that the dimers were disulfide-linked. Thus, HLA-G was capable of forming disulfide-linked dimers in vitro.
Fig 2.
Soluble HLA-G dimerizes in vitro. (A) Chromatogram of recombinant soluble HLA-G analyzed by size-exclusion chromatography using a Superdex 200 10/30. Arrows depict MM calibration standards. (Inset) A nonreducing SDS/PAGE gel of the fractions before pooling. Because it is a soluble truncated form of HLA-G, the heavy chain has an expected MM of 32 kDa, and the refolded HLA-G monomer has an expected MM of ≈44 kDa. (B) Chromatogram of HLA-G after a 21-day incubation at 4°C run on a Superdex 200 10/30. Approximately 25% of the sample eluted in a volume corresponding to a MM of ≈43 kDa and 75% of the sample eluted in a volume corresponding to roughly twice the MM of the monomer. (Inset) A nonreducing SDS/PAGE gel of the fractions from both peaks.
HLA-G Dimerization on the Cell Surface.
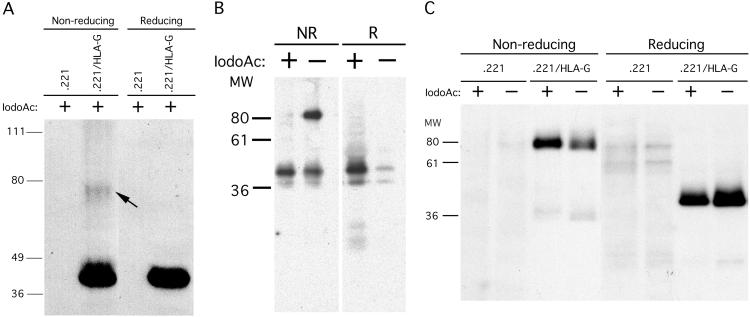
To investigate whether HLA-G dimerization occurred naturally, HLA-G transfectants of 721.221 B-LCLs were lysed, run on SDS/PAGE gels under both nonreducing and reducing conditions, and blotted to nitrocellulose. Detection of HLA-G heavy chains using the HLA-G-specific mAb MEM-G/1 demonstrated the presence of two bands under nonreducing conditions, one of ≈39 kDa, the expected MM of the HLA-G heavy chain, and the other of ≈78 kDa, the expected MM of dimerized HLA-G heavy chain (Fig. 3A). Under reducing conditions, this higher MM band was not present, suggesting that it was disulfide-bonded HLA-G heavy chain. However, only a small fraction of the entire pool of HLA-G heavy chains existed in the dimerized form.
Fig 3.
Cell-surface dimerization of HLA-G. (A) 721.221/HLA-G transfectants and the 721.221 parental cell line were lysed in SDS/PAGE buffer containing iodoacetamide and run under nonreducing and reducing conditions. After blotting to nitrocellulose, HLA-G heavy chains were detected by using the HLA-G-specific mAb, MEM-G/1. (B) Immunoprecipitation of cell surface-biotinylated HLA-A2 molecules from 721.221/HLA-A2 transfectants. Inclusion of iodoacetamide abrogated the formation of HLA-A2 dimer artifacts, because under nonreducing conditions the ≈85-kDa MM band does not appear in samples lysed in the presence of iodoacetamide. (C) Cell-surface molecules of 721.221/HLA-G transfectants and the 721.221 parental cell line were biotinylated, and cells were lysed in the presence or absence of iodoacetamide. Class I MHC molecules were immunoprecipitated with BBM.1 and run under both nonreducing and reducing conditions. Even in the presence of iodoacetamide, HLA-G dimers were detected.
To determine whether HLA-G/β2m/peptide heterotrimers existed in a dimerized form before cell lysis, cells were lysed in the presence and absence of iodoacetamide, which carboxymethylates free cysteines and blocks formation of disulfide bonds. First, in a control experiment, HLA-A2 was immunoprecipitated by using anti-β2m mAb (BBM.1) beads from surface-biotinylated HLA-A2 transfectants in the presence and absence of iodoacetamide. As reported (21), dimerized HLA-A2 heavy chains were observed under nonreducing conditions in the absence of iodoacetamide, but this dimerization was completely abrogated by the addition of iodoacetamide (Fig. 3B). Thus, the observed disulfide-bonded HLA-A2 dimers were an artifact of cell lysis and immunoprecipitation, and the inclusion of iodoacetamide in the lysis buffer allowed us to distinguish between preexisting and artifactual class I MHC dimers. Similarly, when HLA-G was immunoprecipitated from surface-biotinylated HLA-G transfectants, only HLA-G dimers were detected under nonreducing conditions even in the presence of iodoacetamide (Fig. 3C), suggesting that they were preexisting and did not form after cell lysis, and that HLA-G is present on the surface of these transfectants only as the dimer. Immunoprecipitations with the conformation-specific W6/32 mAb and the HLA-G-specific MEM-G/11 mAb yielded similar results (data not shown). Thus, disulfide-linked dimers of HLA-G/β2m/peptide are expressed on the cell surface of 721.221/HLA-G transfectants. When this experiment was repeated with the HLA-G+ JEG-3 choriocarcinoma cell line, no dimers were observed (data not shown). However, the level of HLA-G expression was much (one log) lower than on the transfectant (data not shown), suggesting that dimerization may depend on the cell-surface density of HLA-G.
Cell-Surface Dimerization of HLA-G Is Mediated Through Disulfide Bonds of Cys-42 of the Heavy Chain.
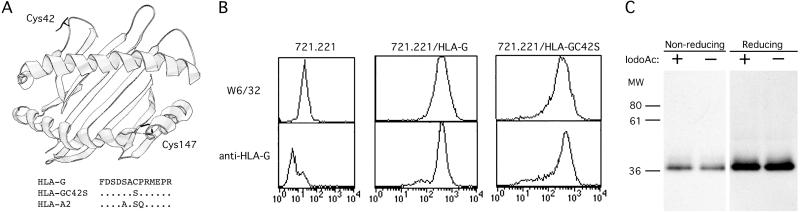
Examination of the predicted amino acid sequence of HLA-G revealed the presence of two free cysteines, Cys-42 and Cys-147, not normally found in class I MHC molecules. Superimposition of these residues onto the HLA-A2 crystal structure indicated that Cys-42 resides on a loop between the third and fourth β strands of the α1 domain, projecting out from the class I MHC molecule (Fig. 4A). Cys-147, in contrast, was predicted to be located on the α2 helix, a position normally occupied in HLA-A2 by a tryptophan pointing into the groove. Therefore, because of its accessibility, Cys-42 was the most likely participant in an intermolecular disulfide linkage. Site-directed mutagenesis was performed, mutating Cys-42 to Ser-42 (the amino acid occurring at position 42 in HLA-A2 and in most other class I MHC molecules; Fig. 4A), and the resulting HLA-G/C42S construct was transfected into 721.221 B-LCLs.
Fig 4.
Identification and mutagenesis of extracellular cysteines in HLA-G. (A) A ribbon diagram of the crystal structure of HLA-A2 with Cys-42 and Cys-147 residues superimposed. Shown below is a portion of the α1 domain HLA-G sequence containing Cys-42 which was chosen for mutagenesis to a serine. Dots (.) indicate the residue identity with the HLA-G sequence. (B) Cell-surface expression of HLA-G/C42S. 721.221 B-LCLs transfected with HLA-G/C42S were stained with the conformation-specific mAb W6/32 and the HLA-G-specific mAb, MEM-G/11. After FACS-sorting, HLA-G/C42S and the wild-type HLA-G transfectants expressed similar levels of protein. (C) Mutagenesis of Cys-42 to Ser-42 completely abrogates HLA-G dimerization. HLA-G/C42S transfectants were cell surface-biotinylated and lysed in the presence or absence of iodoacetamide. Class I MHC molecules were immunoprecipitated and run under both nonreducing and reducing conditions. Even in the absence of iodoacetamide, HLA-G dimers could not be detected.
FACS analysis demonstrated that HLA-G/C42S was expressed at the cell surface and could be detected by using both the conformation-specific pan-class I MHC mAb W6/32 and the HLA-G-specific mAb MEM-G/11 (Fig. 4B). HLA-G/C42S transfectants were FACS-sorted to ensure that they expressed similar levels of class I MHC at the cell surface as the HLA-G transfectants used in the previous experiments. Interestingly, mutation of Cys-42 to Ser-42 completely abrogated dimerization of HLA-G in the absence of iodoacetamide (Fig. 4C). Thus, dimerized HLA-G exists on the cell surface linked by means of a Cys-42-mediated disulfide bond.
HLA-G–KIR2DL4 Interactions.
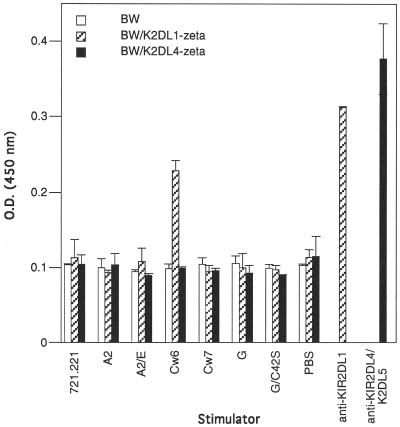
The existence of a dimerized form of HLA-G on the cell surface may have implications for the identification of HLA-G receptors. HLA-G has been reported to bind ILT-2 and KIR2DL4. Attempts by us, however, to detect an interaction between HLA-G and KIR2DL4-Ig fusion proteins as reported (9, 10) have been unsuccessful (unpublished results). To explore the HLA-G–KIR2DL4 interaction further, two additional approaches were taken. First, KIR2DL4-ζ and KIR2DL1-ζ chain fusion proteins were constructed in which the extracellular domains of the KIRs were fused to the transmembrane and cytoplasmic domain of the CD3-ζ chain. These constructs then were stably transfected into the BW− mouse thymoma line. Anti-KIR2DL1 (mAb EB6)–coated and anti-KIR2DL4/KIR2DL5-coated wells were used as positive controls for the ability of the BW transfectants to signal. Whereas BW−/KIR2DL1 cells secreted IL-2 upon coculture with HLA-Cw6 but not –Cw7 transfectants, BW−/KIR2DL4 transfectants did not signal upon coculture with the HLA-G transfectant or any of the other class I MHC protein-expressing targets (Fig. 5).
Fig 5.
Cell surface-expressed KIR2DL4-ζ is not crosslinked by HLA-G. BW cells stably transfected with KIR-ζ constructs were cocultured with various class I MHC transfectants. After 72 h, supernatants were collected and IL-2 secretion was measured by ELISA. As positive controls, some wells were coated with anti-KIR2DL1-specific mAb EB6 and anti-KIR2DL4/KIR2DL5 polyclonal antibody.
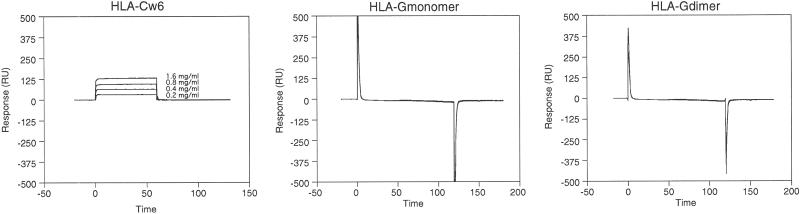
A direct examination of HLA-G–KIR2DL4 interaction was performed with surface plasmon resonance. KIR2DL4-Ig and KIR2DL1-Ig were bound to CM5 sensor chips coupled with goat anti-human Fc. Whereas HLA-Cw6 bound the KIR2DL1-Ig fusion protein with the expected affinity, neither HLA-G monomer nor HLA-G dimer bound KIR2DL4-Ig fusion proteins at the highest concentrations used (5 mg/ml; Fig. 6).
Fig 6.
SPR analysis of HLA-G–KIR2DL4 interaction. KIR-Ig coupled to sensor chips by means of a goat anti-human IgG antibody were used to detect interactions between HLA-Cw6 and KIR2DL1-Ig (A), HLA-G monomer and KIR2DL4- and KIR2DL1-Ig (B), and HLA-G dimer and KIR2DL4- and KIR2DL1-Ig (C). The varying concentrations of HLA-Cw6 that flowed over the surface are indicated. Neither HLA-G monomer nor HLA-G dimer bound to the KIR-Ig fusion proteins when run at a concentration of 5 mg/ml (>100 μM).
Discussion
HLA-G is an unusual class I MHC molecule whose function is unknown. The data presented here indicate an additional unique aspect of HLA-G: its ability to form disulfide-linked cell surface-expressed dimers. Numerous other class I MHC molecules possess unpaired cysteines, but the majority of these are located in the transmembrane and cytoplasmic domains. However, consistent with the reducing environment inside the cell, these cysteines do not form disulfide linkages in vivo (21). Only a few HLA molecules possess unpaired free cysteines in their extracellular domains, (i.e., exposed to the oxidizing extracellular environment), and even fewer possess unpaired free cysteines in regions capable of intermolecular contact. An example of such an unpaired free cysteine is Cys-67 that exists in some HLA-B27, HLA-B15, and HLA-B39 allotypes. The question of whether Cys-67 can mediate formation of disulfide-linked HLA-B27 heavy chain homodimers is controversial (22, 23).
HLA-G dimers could be formed either through oxidation at the cell surface or through an intracellular enzymatic redox reaction. The bulk of the evidence, however, points to the former hypothesis as the more likely one. First, HLA-G heavy chain dimers comprised only a relatively small proportion of the total cellular pool of HLA-G heavy chain, yet they comprised a large proportion of the cell-surface HLA-G. This finding suggests that most of the dimer is on the cell surface. Second, soluble recombinant HLA-G was able to spontaneously dimerize. The presence of monomeric heavy chains in fractions (Fig. 2B, fractions 27 and 28) in the peak that also contains disulfide-linked dimers suggests that HLA-G dimer formation is a two-step process: first, a stable, noncovalent dimer is formed, followed afterward by Cys-42-mediated disulfide bond formation. Lastly, the very long half-life of cell-surface HLA-G (16, 17), may promote dimer formation on the cell surface. Therefore, a likely hypothesis is that HLA-G arrives at the cell surface as a monomer, where it subsequently dimerizes. HLA-G dimerization may depend on a high cell-surface density of HLA-G because dimers were detected on high-expressing HLA-G transfectants but not on the low-expressing JEG-3 choriocarcinoma cell line. Thus, conditions favoring high local concentrations of class I MHC protein such as lipid raft formation, or clustering with a ligand at a cell–cell synapse, may promote HLA-G dimer formation.
The existence of class I MHC protein dimers is especially interesting in light of the fact that KIR inhibitory receptors also dimerize by means of divalent metal cations such as cobalt and zinc (24, 25). KIR dimerization has been demonstrated to increase the affinity of KIRs for their MHC ligands (25, 26), which helps to explain the initial observation that KIR-mediated inhibition of NK cells is zinc-dependent (27, 28). The data presented here suggest that the converse may also be true; HLA-G dimerization could influence its interactions with putative receptors such as KIR2DL4 or ILT-2 and -4. However, an interaction between KIR2DL4 and HLA-G (monomer or dimer) could not be detected by using either a cell-based system or surface plasmon resonance. The failure to detect a HLA-G–KIR2DL4 interaction suggests that a specific interaction between these two proteins does not occur. Alternatively, it could point to the existence of an additional NK-specific (or human) protein critical for the interaction, or it may reflect an inherent low-affinity interaction as is seen for other KIR-activating receptors with their MHC ligands (29). Thus, the nature of the HLA-G–KIR2DL4 interaction needs to be explored in more detail.
The expression of HLA-G in a dimerized form on the cell surface may have implications for its interaction with putative receptors. Dimer formation may affect the specificity of HLA-G for its receptors, either positively (in which the dimerized HLA-G is the functional receptor ligand) or negatively (in which the dimerized HLA-G cannot function as a receptor ligand). Alternatively, dimer formation may have more subtle effects, such as modulating the affinity (again, either positively or negatively) of HLA-G for its receptor. Future evaluation of these possibilities may yield insights into the function of HLA-G.
Acknowledgments
We thank Dr. Hugh Reyburn for providing the KIR2DL1-ζ construct, and we thank Drs. Sumati Rajagopalan and Eric Long for providing their KIR2DL4-Ig fusion construct. This work was supported by a National Research Service Award (to J.E.B.), by National Institutes of Health Grants CA-47554 and AI-50207-02 (to J.L.S.), and by Center of Molecular and Cellular Immunology Grant LN00A026 (to P.A.).
Abbreviations
β2m, β2-microglobulin
MM, molecular mass
References
- 1.Kovats S., Main, E. K., Librach, C., Stubblebine, M., Fisher, S. J. & DeMars, R. (1990) Science 248, 220-223. [DOI] [PubMed] [Google Scholar]
- 2.McMaster M. T., Librach, C. L., Zhou, Y., Lim, K. H., Janatpour, M. J., DeMars, R., Kovats, S., Damsky, C. & Fisher, S. J. (1995) J. Immunol. 154, 3771-3778. [PubMed] [Google Scholar]
- 3.Alizadeh M., Legras, C., Semana, G., Le Bouteiller, P., Genetet, B. & Fauchet, R. (1993) Hum. Immunol. 38, 206-212. [DOI] [PubMed] [Google Scholar]
- 4.Kirszenbaum M., Djoulah, S., Hors, J., Le Gall, I., de Oliveira, E. B., Prost, S., Dausset, J. & Carosella, E. D. (1997) Hum. Immunol. 53, 140-147. [DOI] [PubMed] [Google Scholar]
- 5.Geraghty D. E., Koller, B. H. & Orr, H. T. (1987) Proc. Natl. Acad. Sci. USA 84, 9145-9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefebvre S., Antoine, M., Uzan, S., McMaster, M., Dausset, J., Carosella, E. D. & Paul, P. (2002) J. Pathol. 196, 266-274. [DOI] [PubMed] [Google Scholar]
- 7.Davies B., Hiby, S., Gardner, L., Loke, Y. W. & King, A. (2001) Am. J. Reprod. Immunol. 45, 103-107. [DOI] [PubMed] [Google Scholar]
- 8.King A., Allan, D. S., Bowen, M., Powis, S. J., Joseph, S., Verma, S., Hiby, S. E., McMichael, A. J., Loke, Y. W. & Braud, V. M. (2000) Eur. J. Immunol. 30, 1623-1631. [DOI] [PubMed] [Google Scholar]
- 9.Cantoni C., Verdiani, S., Falco, M., Pessino, A., Cilli, M., Conte, R., Pende, D., Ponte, M., Mikaelsson, M. S., Moretta, L. & Biassoni, R. (1998) Eur. J. Immunol. 28, 1980-1990. [DOI] [PubMed] [Google Scholar]
- 10.Rajagopalan S. & Long, E. O. (1999) J. Exp. Med. 189, 1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan S., Fu, J. & Long, E. O. (2001) J. Immunol. 167, 1877-1881. [DOI] [PubMed] [Google Scholar]
- 12.Faure M. & Long, E. O. (2002) J. Immunol. 168, 6208-6214. [DOI] [PubMed] [Google Scholar]
- 13.Navarro F., Llano, M., Bellon, T., Colonna, M., Geraghty, D. E. & Lopez-Botet, M. (1999) Eur. J. Immunol. 29, 277-283. [DOI] [PubMed] [Google Scholar]
- 14.Allan D. S., Colonna, M., Lanier, L. L., Churakova, T. D., Abrams, J. S., Ellis, S. A., McMichael, A. J. & Braud, V. M. (1999) J. Exp. Med. 189, 1149-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gobin S. J., Biesta, P., De Steenwinkel, J. E., Datema, G. & Van Den Elsen, P. J. (2002) J. Biol. Chem. 277, 39525-39531. [DOI] [PubMed] [Google Scholar]
- 16.Davis D. M., Reyburn, H. T., Pazmany, L., Chiu, I., Mandelboim, O. & Strominger, J. L. (1997) Eur. J. Immunol. 27, 2714-2719. [DOI] [PubMed] [Google Scholar]
- 17.Park B., Lee, S., Kim, E., Chang, S., Jin, M. & Ahn, K. (2001) Immunity 15, 213-224. [DOI] [PubMed] [Google Scholar]
- 18.Colonna M., Borsellino, G., Falco, M., Ferrara, G. B. & Strominger, J. L. (1993) Proc. Natl. Acad. Sci. USA 90, 12000-12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandelboim O., Pazmany, L., Davis, D. M., Vales-Gomez, M., Reyburn, H. T., Rybalov, B. & Strominger, J. L. (1997) Proc. Natl. Acad. Sci. USA 94, 14666-14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vales-Gomez M., Reyburn, H. T., Mandelboim, M. & Strominger, J. L. (1998) Immunity 9, 337-344. [DOI] [PubMed] [Google Scholar]
- 21.Springer T. A., Robb, R. J., Terhorst, C. & Strominger, J. L. (1977) J. Biol. Chem. 252, 4694-4700. [PubMed] [Google Scholar]
- 22.Allen R. L., O'Callaghan, C. A., McMichael, A. J. & Bowness, P. (1999) J. Immunol. 162, 5045-5048. [PubMed] [Google Scholar]
- 23.Malik P., Klimovitsky, P., Deng, L., Boyson, J. E. & Strominger, J. L. (2002) J. Immunol. 169, 4379-4387. [DOI] [PubMed] [Google Scholar]
- 24.Fan Q. R., Long, E. O. & Wiley, D. C. (2000) J. Biol. Chem. 275, 23700-23706. [DOI] [PubMed] [Google Scholar]
- 25.Vales-Gomez M., Erskine, R. A., Deacon, M. P., Strominger, J. L. & Reyburn, H. T. (2001) Proc. Natl. Acad. Sci. USA 98, 1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Q. R., Long, E. O. & Wiley, D. C. (2000) Eur. J. Immunol. 30, 2692-2697. [DOI] [PubMed] [Google Scholar]
- 27.Rajagopalan S., Winter, C. C., Wagtmann, N. & Long, E. O. (1995) J. Immunol. 155, 4143-4146. [PubMed] [Google Scholar]
- 28.Rajagopalan S. & Long, E. O. (1998) J. Immunol. 161, 1299-1305. [PubMed] [Google Scholar]
- 29.Vales-Gomez M., Reyburn, H. T., Erskine, R. A. & Strominger, J. L. (1998) Proc. Natl. Acad. Sci. USA 95, 14326-14331. [DOI] [PMC free article] [PubMed] [Google Scholar]