Abstract
We analyzed gene expression patterns in human gastric cancers by using cDNA microarrays representing ≈30,300 genes. Expression of PLA2G2A, a gene previously implicated as a modifier of the ApcMin/+ (multiple intestinal neoplasia 1) mutant phenotype in the mouse, was significantly correlated with patient survival. We confirmed this observation in an independent set of patient samples by using quantitative RT-PCR. Beyond its potential diagnostic and prognostic significance, this result suggests the intriguing possibility that the activity of PLA2G2A may suppress progression or metastasis of human gastric cancer.
Keywords: gene expression profiling, DNA microarray, mucosal immunity, gastritis, Helicobacter
Gastric cancer is the second leading cause of cancer death in the world (1) and is predicted to be the eighth leading cause of all deaths worldwide in the year 2010 (2). Gastric carcinogenesis seems to be a multistep process. Helicobacter pylori infection is the major recognized risk factor for development of gastric cancer (3). Chronic atrophic gastritis, often associated with H. pylori infection, is a common precancerous condition (4). However, other environmental factors, including other bacteria, may also be important in the development of gastric cancer, because ≈25% of gastric cancer patients have no evidence of previous or current H. pylori infection (5).
The prognosis of gastric cancer depends highly on the clinical and pathological stage at diagnosis. Surgical resection, still the mainstay of treatment, is very effective in early-stage cancers. However, most gastric cancer cases are diagnosed at an advanced stage, when the prognosis is extremely poor. Only 20–40% of these patients respond to chemotherapy (6). Currently, the median survival rate for gastric cancer patients in the U.S. is only 6–12 months postdiagnosis (6).
We have carried out a systematic study of gene expression in gastric adenocarcinomas and nonneoplastic gastric mucosa. A full description of these molecular portraits of gastric cancer will be published separately. Here, we show in two independent datasets using different technologies (DNA microarrays and real-time quantitative RT-PCR) that survival in 88 and 59 gastric adenocarcinomas, respectively, is correlated with the expression of a single gene, PLA2G2A. This correlation has potential clinical significance and may provide new insight into the pathogenesis of gastric cancer progression and metastasis.
Phospholipase A2 (PLA2) catalyzes hydrolysis of the sn-2 fatty acyl ester bond of phosphoglycerides, releasing free fatty acids and lysophospholipids. One of the fatty acids that can be released from membrane stores by the activity of PLA2 is arachidonic acid, the critical precursor for biosynthesis of diverse eicosanoids, including prostaglandins, thromboxanes, and leukotrienes (7). At least 15 human genes encode different PLA2 isoenzymes, including both secreted and cytosolic forms (8). PLA2 group IIA (PLA2G2A) is a secreted PLA2. It has been reported to be expressed in human Paneth cells, lacrimal glands, chondrocytes, and amniotic epithelial cells (9–12). PLA2G2A seems to play diverse roles in human diseases, including colon cancer, coronary artery disease, and inflammation (13–15). Induction of PLA2G2A is a frequent feature of inflammatory responses, and elevated expression of PLA2G2A has been reported in several types of malignancies, including pancreatic cancer and prostate cancer (16, 17). The most important and direct link between PLA2G2A and cancer comes from genetic studies in mice; the murine ortholog of this gene, Mom1 (modifier of Min1) has been shown genetically to limit the severity of intestinal neoplasia in the ApcMin/+ mouse, a mouse model of familial adenomatous polyposis (15, 18, 19).
Materials and Methods
Samples and RNA Preparation.
Frozen tumor and normal gastric mucosa were collected from gastrectomy specimens from Queen Mary Hospital, the University of Hong Kong. Ninety primary gastric adenocarcinomas, lymph node metastases from 14 of the 90 primary gastric cancers, and 22 samples of nonneoplastic gastric mucosa were analyzed using DNA microarrays (unpublished data). Another 59 adenocarcinomas were later tested independently by quantitative RT-PCR as described below. This study was approved by the Ethics Committee of the University of Hong Kong and the Internal Review Board of Stanford University.
Tissues were frozen in liquid nitrogen within one-half hour after they were resected. Samples of nonneoplastic mucosa from stomach were dissected free of muscle when fresh and histologically confirmed to be tumor-free by frozen section. The tumors were unselected with respect to grade, but there was, in both cases, selection for specimens judged to have at least 50% tumor cells in the block. The clinical records were surveyed, and the clinical parameters (for this study, overall survival) recovered after the biological experiments for each dataset were complete. Tumors were classified using the Lauren classification into intestinal, diffuse, mixed, and indeterminate types (20). The tumor stage was defined by the General Rules for Gastric Cancer Study of the Japanese Research Society for Gastric Cancer (21). Total RNA was extracted using Trizol (GIBCO/BRL), and mRNA was isolated from total RNA by the FastTrack mRNA isolation kit (Invitrogen).
Microarray Procedure and Data Analysis.
We used a cDNA microarray containing 44,500 cDNA clones, representing ≈30,300 unique genes. The methods for microarray production, hybridization, and data analysis were as described (22–24) and are described in detail elsewhere (unpublished work).
Quantitative RT-PCR.
Quantitative RT-PCR was performed as described (25). In brief, total RNA was further purified with an RNAqueous kit (Ambion, Austin, TX), including DNase I digestion to remove any genomic DNA contamination. Human GAPDH primer and probe reagents (Applied Biosystems) were used as the normalization control in subsequent quantitative analysis. Quantification was performed using the ABI Prism 7900HT sequence detection system (Applied Biosystems) via a two-step nonmultiplexed TaqMan 5′→3′ exonuclease assay (TaqMan reverse transcription reagents kit and TaqMan PCR core reagents kit, Applied Biosystems) according to the relative standard method. Calibration curves were generated for each transcript and validated using linear regression analysis (r2 ≥ 0.99). Transcript quantification was performed in triplicate for every sample and reported relative to GAPDH. The primers and probe used for PLA2G2A in this study are PLA2F, CCGCACTCAGTTATGGCTTCT; PLA2R, AGCGATCCGTTGCATCCTT; and PLA2probe, CACTGTGGCGTGGGTGGCAGA.
In Situ Hybridization.
In situ hybridization for PLA2G2A was performed by using 35S-labeled antisense and sense riboprobes containing nucleotides 14–940 of PLA2G2A, as described (26).
Statistical Analysis.
A nonparametric t test with a P value cutoff of 0.001 based on 10,000 random column permutations was used to identify genes differentially expressed in gastric cancers and nonneoplastic gastric mucosa (27). Before the t test analysis, the missing values in the dataset were estimated with KNNimpute algorithm by using 12 neighbors (28). The false discovery rate was estimated on the basis of the number of genes that passed the P value cutoff of 0.001 in five datasets with randomized columns. Gene-to-gene correlations were calculated using the pairwise correlation function in the statistical package R (http://lib.stat.cmu.edu/R/CRAN) without missing value estimation. The χ2 test with Yates correction was used to analyze relationships between categorical subgroups (29). Kaplan–Meier survival analysis was carried out by using WINSTAT EXCEL plug-in software (www.winstat.com).
Results
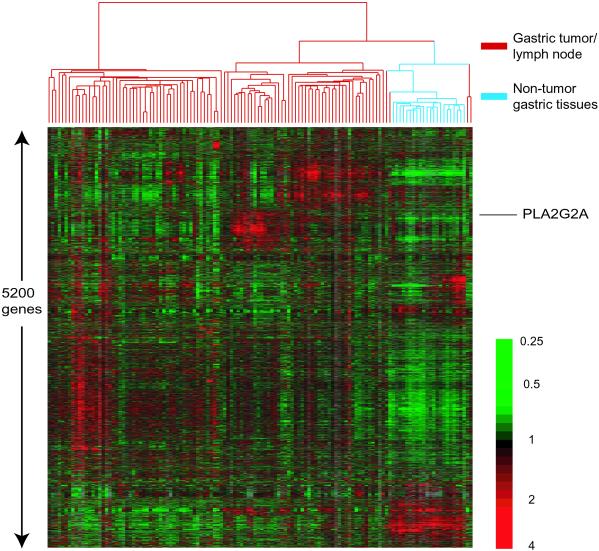
Using a cDNA microarray containing 44,500 clones, corresponding to ≈30,300 different human genes, we profiled mRNA populations from 90 primary gastric adenocarcinomas, 14 lymph node metastases, and 22 samples of nonneoplastic gastric mucosa. We used a hierarchical clustering method to group genes on the basis of similarity in expression across the samples and to group samples based on the similarity in their gene expression patterns. For this analysis, we chose all of the cDNA clones (6,688 altogether, representing ≈5,200 genes) whose expression varied by at least 2.5-fold from their mean in the sample set, in at least three samples (Fig. 1). A detailed analysis of the results displayed in Fig. 1 can be found on our web site, http://genome-www.stanford.edu/Gastric_Cancer.
Fig. 1.
Hierarchical clustering of the patterns of variation in expression of 5,200 genes from 90 primary gastric tumors, 14 lymph node metastases, from 14 of these primary gastric cancers and 22 samples of nonneoplastic gastric mucosa. Each row represents hybridization results from a separate cDNA clone on the microarray (results from a total of 6,688 cDNA clones, representing 5,200 genes, are included in this figure), and each column represents the expression pattern in a separate tumor or tissue sample. The ratio of abundance of transcripts of each gene in each sample to the mean abundance of that transcript across all tissue samples is depicted according to the color scale shown at the bottom. Gray indicates missing or excluded data. The dendrogram at the top provides a representation of the similarity of the gene expression patterns among the samples. The branches in the dendrogram are color coded (red for gastric adenocarcinomas and blue for nonneoplastic tissues). See our web site, http://genome-www.stanford.edu/Gastric_Cancer, for full data.
PLA2G2A Expression and Survival.
Microarray data set.
As a step toward identifying molecular markers for the diagnosis or prognostication of gastric cancer, we used a nonparametric t test to identify genes whose expression levels were significantly different (P ≤ 0.001) between gastric cancer and nonneoplastic gastric mucosa. In total, 2,656 genes satisfied this criterion, with an estimated false discovery rate of 0.13%. A complete list of these genes is given on our web site, http://genome-www.stanford.edu/Gastric_Cancer. Among the genes that were expressed at significantly higher levels in tumors, we searched for genes with especially high variation in their expression levels among the tumors. These genes should be particularly useful for tumor subclassification and prognostication.
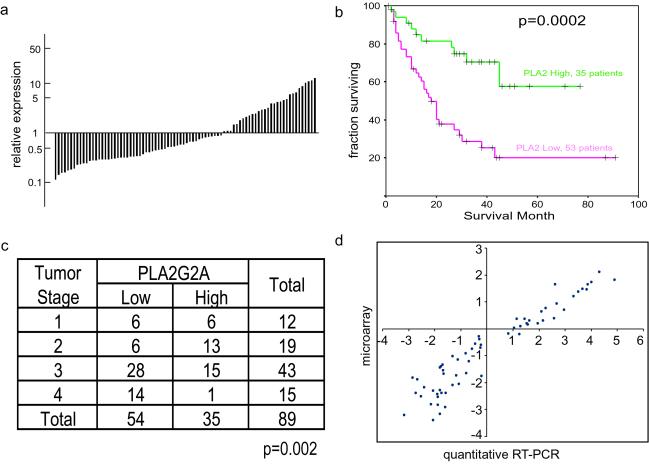
PLA2G2A, the gene that encodes the group II A secreted phospholipase A2, was among the most variably expressed genes in these gastric cancers (Fig. 2a). The range of PLA2G2A transcript levels observed in gastric cancer samples in this microarray analysis exceeded 250-fold. In a subsequent analysis using quantitative RT-PCR, the range exceeded 2,000-fold. Moreover, the pattern of variation in PLA2G2A expression among these gastric cancer samples was not closely related to that of any other genes.
Fig. 2.
Expression of PLA2G2A in gastric cancer and its relationship with survival. (a) Histogram of PLA2G2A expression levels in 89 gastric cancer tissues. (b) Kaplan–Meier plot of survival in 88 gastric cancer patients expressing high or low levels of PLA2G2A. (c) Relationship of PLA2G2A expression with tumor stage. The difference is significant by χ2 test (P = 0.002). (d) Correlation between the PLA2G2A expression levels measured by DNA microarray hybridization and quantitative RT-PCR.
The expression of PLA2G2A was consistent between individual primary tumors and the lymph node metastases from the same patient; the correlation between the expression levels of PLA2G2A in the 14 lymph node metastases and the corresponding primary tumors was 0.884. This suggests that the level of expression of PLA2G2A is an intrinsic characteristic of tumor cells, maintained even when the tumor metastasized to a lymph node.
To test the relationship between the expression of PLA2G2A and survival, we divided the patients into two groups, one with PLA2G2A transcript levels greater than the mean for all tumors tested (Fig. 2a) and the other with levels below the mean. A Kaplan–Meier plot of survival for these two groups of patients is shown in Fig. 2b. The patients with higher levels of PLA2G2A expression had significantly extended survival (P = 0.0002), and an almost 3-fold higher 5-year survival rate. A similar inverse relationship between PLA2G2A expression level and tumor stage at diagnosis (P = 0.002; Fig. 2c) suggests that the expression of PLA2G2A may suppress tumor progression or metastasis.
In the preceding analysis, we relied on expression measurements taken directly from the microarrays. To verify the technical quality of these measurements for PLA2G2A mRNA levels, we analyzed a random subset of 62 of the 90 tumors, using real-time quantitative RT-PCR. We found a complete concordance of the PLA2G2A expression levels as determined by these two different assays (Pearson correlation coefficient = 0.949; Fig. 2d).
Second data set.
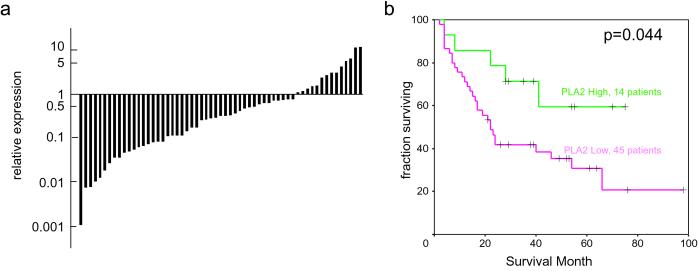
To provide an independent test of the association between PLA2G2A expression and patient outcome, we analyzed a second, completely different set of 59 gastric cancers. We measured the abundance of the PLA2G2A transcripts in these tumors using only quantitative RT-PCR and examined its relationship with patient survival. We segregated the samples into two groups using the same criterion used for microarray analysis (Fig. 2d). The distribution of PLA2G2A mRNA levels and the Kaplan–Meier analysis are shown in Fig. 3. The patients with higher levels of PLA2G2A expression once again survived significantly longer (P = 0.044), with a 3-fold higher 5-year survival rate. We conclude that the expression level of PLA2G2A in gastric cancer is associated with survival.
Fig. 3.
Expression of PLA2G2A in gastric cancer and its relationship with survival assayed in the independent sample set by quantitative RT-PCR. (a) Histogram of the PLA2G2A expression levels in an independent sample set of 59 gastric cancer tissues using quantitative RT-PCR. (b) Kaplan–Meier plot of patient survival for the 59 gastric cancers, stratified into two groups, expressing high or low levels of PLA2G2A, respectively.
Localization of PLA2G2A Expression in Tumors.
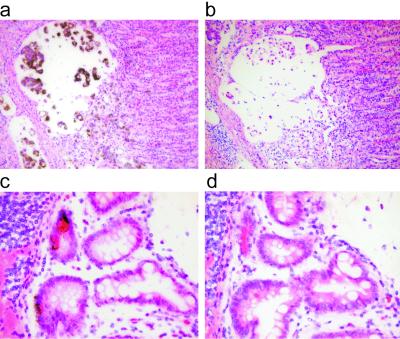
To determine which cells in the tumors expressed PLA2G2A, we used in situ hybridization to examine normal gastric mucosa, metaplastic gastric mucosa, and cancer tissues. We found abundant PLA2G2A mRNA in the cancer cells, whereas none was detectable in the surrounding inflammatory cells and stromal cells (Fig. 4a). PLA2G2A mRNA was also undetectable in the normal or inflamed gastric mucosa but was highly abundant in the Paneth cells found in mucosa with intestinal metaplasia (Fig. 4c). This observation probably accounts for the expression of PLA2G2A in a few of the samples that exhibited intestinal metaplasia, but which were not adjudged to be malignant.
Fig. 4.
In situ hybridization analysis of PLA2G2A expression in gastric tissues. 35S-labeled riboprobe was used, and black grains denote positive signals. (a) PLA2G2A transcripts were detected in gastric cancer cells but not in normal gastric mucosa, stromal cells, and leukocytes. (c) Expression of PLA2G2A is limited to the Paneth cells in gastric mucosa with intestinal metaplasia. (b and d) Control in situ hybridization analysis of tissue sections adjacent to the sections shown in a and c, respectively, using a PLA2G2A sense probe as the negative control.
Discussion
Is PLA2G2A expression simply a fortuitous marker of the propensity of a gastric adenocarcinoma to progress, or might it actually play a role in suppressing progression and metastasis? Several lines of evidence lend credence to such a role.
The strongest evidence comes from studies of the murine PLA2G2A ortholog, the Mom1 gene. Linkage mapping first identified Mom1 as the locus responsible for much of the difference in severity of the ApcMin/+ intestinal polyposis phenotype between mice of the C57BL/6 strain (which were shown to be PLA2G2A-deficient) and AKR, MA, or CAST mice (PLA2G2A positive) (19). Subsequent studies established that expression of PLA2G2A could significantly reduce the number and size of adenomatous intestinal polyps in the ApcMin/+ mouse (15). Although efforts to link mutations or polymorphisms in PLA2G2A to human colon cancers have so far been largely unrewarding, a germ-line frameshift mutation and inactivation of the second allele by deletion of PLA2G2A has been demonstrated in a rare case of sporadic colorectal cancer (30).
Results from studies of strain differences in mice in the response of the gastric mucosa to infection with Helicobacter felis, although not conclusively attributable to differences in PLA2G2A, may provide another clue (31). In C57BL mice, which are genetically deficient in PLA2G2A, H. felis infection results in marked histological changes in the gastric mucosa, as well as increases in both proliferation and apoptosis of mucosal cells. None of these changes were observed in mice of the BALB/c or C3H/HeJ strains, both of which express functional PLA2G2A. Other studies have suggested that PLA2G2A expression may have a protective effect against progression of carcinomas. A previous study of gene expression patterns in hepatocellular carcinoma revealed a reciprocal relationship between PLA2G2A expression and vascular invasion, a pathological indicator of the potential for tumor to spread (32). Expression of PLA2G2A has also been reported to be significantly associated with longer survival after surgery in pancreatic cancer (16). Although the relationship between PLA2G2A and the pathogenesis and progression of these diverse carcinomas has yet to be determined, they each add weight to the hypothesis that it plays a direct role in suppressing gastric cancer progression.
Among the thousands of genes examined in this study, the expression pattern of PLA2G2A was unique (Fig. 1). Only 1 of the 5,200 genes analyzed in this study had an expression pattern with a Pearson correlation coefficient >0.5 to that of PLA2G2A (http://genome-www.stanford.edu/Gastric_Cancer). Such an eccentric expression pattern is fairly unusual. Of the 5,200 genes analyzed, 89% had at least two neighbors with a correlation >0.5, and 68% had at least 10. We therefore suspect that the distinctive pattern of variation in the expression of PLA2G2A may not be simply a secondary consequence of differences among these tumors in a larger program of differentiation or physiological regulation. If that were the case, we would expect to see a PLA2G2A clustered with a group of genes similarly affected by the primary alteration. Instead, we suspect that the distinctive pattern of variation in the expression of PLA2G2A in gastric cancers may point to a primary difference in the PLA2G2A gene itself. It will therefore be important to determine whether the observed variation in expression is related to inherited or acquired variations in the transcriptional control elements, or even somatic mutation or loss of this gene during the genesis or progression of the PLA2G2A-low tumors, as has been found in a case of sporadic colon cancer (25).
If PLA2G2A indeed plays a role in limiting progression of gastric cancer, how might it act? An obvious possibility is that the apparent protective role of PLA2G2A is related to its ability to catalyze release of lysophosphatidic acid and arachidonic acid from membrane phospholipids. The physiological effects of lysophosphatidic acid may include stimulation of cell proliferation, tumor invasion, differentiation, chemotaxis, and platelet aggregation (33). Release of arachidonic acid from membrane phospholipids by PLA2 is the first step, and often a critical regulated step, in synthesis of eicosanoids, including prostaglandins, thromboxanes, hydroxyeicosatetraenoic acids, and leukotrienes (7). Arachidonic acid itself can induce apoptosis in diverse cells, including human colon cancer cells, via an intracellular ceramide-mediated pathway (34). Nonsteroidal antiinflammatory drugs, which inhibit the conversion of arachidonic acid to prostaglandins by cyclooxygenases, seem to confer significantly lower risk of both colorectal cancers and gastric cancers in human populations (35, 36). Moreover, COX-2 inhibitors have been shown to inhibit growth and promote apoptosis in gastric cancer xenografts in nude mice (37). In vitro evidence suggests that the cancer chemopreventative effects of nonsteroidal antiinflammatory drugs may be due to their ability to raise arachidonic acid levels and thereby promote apoptosis in cancer cells (34). Elevated expression of PLA2G2A might similarly inhibit progression of gastric cancer through increased release of arachidonic acid (34).
PLA2s are widespread in nature as both intracellular (cytosolic PLA2) and extracellular (secreted PLA2) enzymes. PLA2G2A is one of at least 15 human genes encoding proteins with phospholipase A2 activity (8). A genetic study of the contrasting effects group IV cytosolic PLA2 and group IIA secreted PLA2 on intestinal neoplasia in mice highlights the complexity of the relationship between PLA2 activity and cancer. Whereas expression of PLA2G2A ameliorates the intestinal neoplasia phenotype in the ApcMin/+ mouse, cytosolic PLA2 expression in the small intestine exacerbates tumor formation (38). Whether and how differences in substrate specificity, regulation, compartmentalization, coupling to downstream metabolic pathways, or other properties might account for the different effects of these two forms of PLA2 in this system remain to be determined. In view of the important role that bacteria can play in the pathogenesis of gastrointestinal malignancies, one intriguing possibility is that the tumor suppressor activity of PLA2G2A might be related to the direct antibacterial activity that distinguishes this enzyme from the other human PLA2 isoenzymes (39–42).
A protective effect of PLA2G2A expression against progression of gastric cancer, if confirmed, could have important therapeutic implications. For example, if PLA2G2A inhibits progression of gastric cancer by raising arachidonic acid levels, treatment of gastric cancer with cyclooxygenase inhibitors might be similarly beneficial. Alternatively, a protective mechanism based on the antibacterial activity of PLA2G2A would suggest that patients with gastric cancer might benefit from antibacterial chemotherapy. Whether and how PLA2G2A might inhibit progression of human gastric cancer, and the corollary possibilities for specific therapy against tumor progression, clearly require further investigation.
Acknowledgments
We thank Wijan Prapong for his help in the preparation of this manuscript and our web site (http://genome-www.stanford.edu/Gastric_Cancer), and the Stanford Functional Genomic Center and Stanford Microarray database for their support. We thank William F. Dove for helpful comments on the manuscript. This work was supported by the National Cancer Institute (P.O.B. and D.B.); the H. M. Lui Foundation (X.C., R.L., and S.S.); the Howard Hughes Medical Institute (P.O.B.); and the Council of the Hong Kong Special Administrative Region Research Grant HKU 7264/01M (to S.Y.L., S.T.Y., and K.M.C.). O.T.G. is a Howard Hughes Medical Institute Predoctoral Fellow and a Stanford Graduate Fellow. P.O.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
PLA2, phospholipase A2
PLA2G2A, PLA2 group IIA
References
- 1.Pisani P., Parkin, D. M., Bray, F. & Ferlay, J. (1999) Int. J. Cancer 83, 18-29. [DOI] [PubMed] [Google Scholar]
- 2.Murray C. J. & Lopez, A. D. (1997) Lancet 349, 1498-1504. [DOI] [PubMed] [Google Scholar]
- 3.Parsonnet J., Friedman, G. D., Vandersteen, D. P., Chang, Y., Vogelman, J. H., Orentreich, N. & Sibley, R. K. (1991) N. Engl. J. Med. 325, 1127-1131. [DOI] [PubMed] [Google Scholar]
- 4.Ming S. C. (1998) Gastric Cancer 1, 31-50. [DOI] [PubMed] [Google Scholar]
- 5.Jonkers D., Houben, P., Hameeteman, W., Stobberingh, E., de Bruine, A., Arends, J. W., Biemond, I., Lundqvist, G. & Stockbrugger, R. (1999) Ital. J. Gastroenterol. Hepatol. 31, 836-841. [PubMed] [Google Scholar]
- 6.Meyerhardt J. A. & Fuchs, C. S. (2002) Semin. Radiat. Oncol. 12, 176-186. [DOI] [PubMed] [Google Scholar]
- 7.Reilly M. P., Lawson, J. A. & FitzGerald, G. A. (1998) J. Nutr. 128, 434S-438S. [DOI] [PubMed] [Google Scholar]
- 8.Six D. A. & Dennis, E. A. (2000) Biochim. Biophys. Acta 1488, 1-19. [DOI] [PubMed] [Google Scholar]
- 9.Nevalainen T. J., Aho, H. J. & Peuravuori, H. (1994) Invest. Ophthalmol. Vis. Sci. 35, 417-421. [PubMed] [Google Scholar]
- 10.Nevalainen T. J., Gronroos, J. M. & Kallajoki, M. (1995) Lab. Invest. 72, 201-208. [PubMed] [Google Scholar]
- 11.Nevalainen T. J., Marki, F., Kortesuo, P. T., Grutter, M. G., Di Marco, S. & Schmitz, A. (1993) J. Rheumatol. 20, 325-330. [PubMed] [Google Scholar]
- 12.Nevalainen T. J., Meri, K. M. & Niemi, M. (1993) Andrologia 25, 355-358. [DOI] [PubMed] [Google Scholar]
- 13.Touqui L. & Alaoui-El-Azher, M. (2001) Curr. Mol. Med. 1, 739-754. [DOI] [PubMed] [Google Scholar]
- 14.Kugiyama K., Ota, Y., Sugiyama, S., Kawano, H., Doi, H., Soejima, H., Miyamoto, S., Ogawa, H., Takazoe, K. & Yasue, H. (2000) Am. J. Cardiol. 86, 718-722. [DOI] [PubMed] [Google Scholar]
- 15.Cormier R. T., Hong, K. H., Halberg, R. B., Hawkins, T. L., Richardson, P., Mulherkar, R., Dove, W. F. & Lander, E. S. (1997) Nat. Genet. 17, 88-91. [DOI] [PubMed] [Google Scholar]
- 16.Kashiwagi M., Friess, H., Uhl, W., Berberat, P., Abou-Shady, M., Martignoni, M., Anghelacopoulos, S. E., Zimmermann, A. & Buchler, M. W. (1999) Gut 45, 605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J., Neubauer, B. L., Graff, J. R., Chedid, M., Thomas, J. E., Roehm, N. W., Zhang, S., Eckert, G. J., Koch, M. O., Eble, J. N. & Cheng, L. (2002) Am. J. Pathol. 160, 667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietrich W. F., Lander, E. S., Smith, J. S., Moser, A. R., Gould, K. A., Luongo, C., Borenstein, N. & Dove, W. (1993) Cell 75, 631-639. [DOI] [PubMed] [Google Scholar]
- 19.MacPhee M., Chepenik, K. P., Liddell, R. A., Nelson, K. K., Siracusa, L. D. & Buchberg, A. M. (1995) Cell 81, 957-966. [DOI] [PubMed] [Google Scholar]
- 20.Lauren P. (1965) Acta Pathol. Microbiol. Scand. 64, 31-49. [DOI] [PubMed] [Google Scholar]
- 21.Japanese Research Society of Gastric Cancer (1995) in Japanese Classification of Gastric Carcinoma, eds. Nishi M. O. Y. & Miwa, K. (Kanehara, Tokyo).
- 22.Alizadeh A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503-511. [DOI] [PubMed] [Google Scholar]
- 23.Perou C. M., Sorlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747-752. [DOI] [PubMed] [Google Scholar]
- 24.Sherlock G., Hernandez-Boussard, T., Kasarskis, A., Binkley, G., Matese, J. C., Dwight, S. S., Kaloper, M., Weng, S., Jin, H., Ball, C. A., et al. (2001) Nucleic Acids Res. 29, 152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin S. A. (2000) J. Mol. Endocrinol. 25, 169-193. [DOI] [PubMed] [Google Scholar]
- 26.Yuen S. T., Chung, L. P., Leung, S. Y., Luk, I. S., Chan, S. Y. & Ho, J. (1994) Am. J. Surg. Pathol. 18, 1158-1163. [DOI] [PubMed] [Google Scholar]
- 27.Troyanskaya O., Garber, M. E., Brown, P. O., Botstein, D. & Altman, R. B. (2002) Bioinformatics 18, 1454-1461. [DOI] [PubMed] [Google Scholar]
- 28.Troyanskaya O., Cantor, M., Sherlock, G., Brown, P., Hastie, T., Tibshirani, R., Botstein, D. & Altman, R. B. (2001) Bioinformatics 17, 520-525. [DOI] [PubMed] [Google Scholar]
- 29.Walpole R. E. & Myers, R. H., (1993) Probablity and Statistics for Engineers and Scientists (Macmillan, New York).
- 30.Nimmrich I., Friedl, W., Kruse, R., Pietsch, S., Hentsch, S., Deuter, R., Winde, G. & Muller, O. (1997) Hum. Genet. 100, 345-349. [DOI] [PubMed] [Google Scholar]
- 31.Wang T. C., Goldenring, J. R., Dangler, C., Ito, S., Mueller, A., Jeon, W. K., Koh, T. J. & Fox, J. G. (1998) Gastroenterology 114, 675-689. [DOI] [PubMed] [Google Scholar]
- 32.Chen X., Cheung, S. T., So, S., Fan, S. T., Barry, C., Higgins, J., Lai, K. M., Ji, J., Dudoit, S., Ng, I. O., et al. (2002) Mol. Biol. Cell 13, 1929-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swarthout J. T. & Walling, H. W. (2000) Cell Mol. Life Sci. 57, 1978-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan T. A., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Proc. Natl. Acad. Sci. USA 95, 681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan T. A. (2002) Lancet Oncol. 3, 166-174. [DOI] [PubMed] [Google Scholar]
- 36.Jolly K., Cheng, K. K. & Langman, M. J. (2002) Drugs 62, 945-956. [DOI] [PubMed] [Google Scholar]
- 37.Sawaoka H., Kawano, S., Tsuji, S., Tsujii, M., Gunawan, E. S., Takei, Y., Nagano, K. & Hori, M. (1998) Am. J. Physiol. 274, G1061-G1067. [DOI] [PubMed] [Google Scholar]
- 38.Hong K. H., Bonventre, J. C., O'Leary, E., Bonventre, J. V. & Lander, E. S. (2001) Proc. Natl. Acad. Sci. USA 98, 3935-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckland A. G. & Wilton, D. C. (2000) Biochim. Biophys. Acta 1488, 71-82. [DOI] [PubMed] [Google Scholar]
- 40.Laine V. J., Grass, D. S. & Nevalainen, T. J. (1999) J. Immunol. 162, 7402-7408. [PubMed] [Google Scholar]
- 41.Ofori-Darko E., Zavros, Y., Rieder, G., Tarle, S. A., Van Antwerp, M. & Merchant, J. L. (2000) Infect. Immun. 68, 3657-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu X. D. & Lehrer, R. I. (1998) Infect. Immun. 66, 2791-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]