Abstract
Acute myelogenous leukemia (AML) is typically a disease of stem/progenitor cell origin. Interestingly, the leukemic stem cell (LSC) shares many characteristics with normal hematopoietic stem cells (HSCs) including the ability to self-renew and a predominantly G0 cell-cycle status. Thus, although conventional chemotherapy regimens often ablate actively cycling leukemic blast cells, the primitive LSC population is likely to be drug-resistant. Moreover, given the quiescent nature of LSCs, current drugs may not effectively distinguish between malignant stem cells and normal HSCs. Nonetheless, based on recent studies of LSC molecular biology, we hypothesized that certain unique properties of leukemic cells could be exploited to induce apoptosis in the LSC population while sparing normal stem cells. In this report we describe a strategy using treatment of primary AML cells with the proteasome inhibitor carbobenzoxyl-l-leucyl-l-leucyl-l-leucinal (MG-132) and the anthracycline idarubicin. Comparison of normal and leukemic specimens using in vitro culture and in vivo xenotransplantation assays shows that the combination of these two agents induces rapid and extensive apoptosis of the LSC population while leaving normal HSCs viable. Molecular genetic studies using a dominant-negative allele of inhibitor of nuclear factor κB (IκBα) demonstrate that inhibition of nuclear factor κB (NF-κB) contributes to apoptosis induction. In addition, gene-expression analyses suggest that activation of p53-regulated genes are also involved in LSC apoptosis. Collectively, these findings demonstrate that malignant stem cells can be preferentially targeted for ablation. Further, the data begin to elucidate the molecular mechanisms that underlie LSC-specific apoptosis and suggest new directions for AML therapy.
Acute myelogenous leukemia (AML) is a serious and often lethal form of hematologic cancer. Although the development of better chemotherapy regimens has improved remission induction and overall survival, relapse remains a common problem, especially among older patients and/or patients with poor prognosis cytogenetics (1). In recent years the clinical characteristics of AML have become better understood in light of studies elucidating the biological origins of the disease. Several lines of evidence clearly indicate that AML is a disease of stem or progenitor cell origin, and that the leukemic stem cell (LSC) stands apart from more mature leukemic cells with its own set of unique biological properties (2–9). Thus, although chemotherapeutic agents effectively ablate leukemic blast cells in a majority of patients, the efficacy of LSC targeting is not known. Indeed, it is attractive to speculate that failure to sustain durable remission may be due to a drug-refractory/resistant malignant stem cell population. Given the potentially critical role of stem cells in both the genesis and perpetuation of AML, recent studies have attempted to better characterize LSC properties (10–12). Notably, although the immunophenotype of LSCs is similar to normal hematopoietic stem cells (HSCs; CD34+, CD38−, HLA-DR−), there are at least three antigens with expression that is known to vary in malignant cells: CD90, CD117, and CD123 (7–9). Thus, it has been possible to prospectively identify and isolate enriched LSC populations, which has allowed subsequent studies to analyze the cell-cycle status and gene-expression characteristics of normal vs. malignant stem cells (13–15). A somewhat counterintuitive finding from cell-cycle studies is that the LSC population seems mostly quiescent, which seems to be true despite the often aggressive characteristics of leukemic disease. Thus, the LSC should not be preferentially susceptible to cycle-active chemotherapeutic agents.
Because standard chemotherapy approaches may not effectively target the LSC population, we have attempted to characterize molecular properties of the LSC that could be useful for apoptosis induction. Data from our laboratory has shown recently that nuclear factor κB (NF-κB) is constitutively activated in the majority of primary AML specimens (15). Although activation of NF-κB is a relatively common feature of many cancers (16–18), a surprising aspect of our studies was the finding that NF-κB is active also in quiescent LSC populations. Thus, strategies to inhibit NF-κB, and thereby block growth and survival pathways regulated by NF-κB, may represent a useful approach to more durable AML therapy. With this concept in mind, we examined several agents that inhibit NF-κB. One such class of drugs that is being widely explored for cancer therapy is proteasome inhibitors (19, 20). Proteasome inhibition blocks degradation of the NF-κB regulator IκBα and results in loss of NF-κB activity. Initial studies of primary AML cells have demonstrated that treatment with the proteasome inhibitor carbobenzoxyl-l-leucyl-l-leucyl-l-leucinal (MG-132) causes rapid inhibition of NF-κB and strongly induces apoptosis (15). Thus, proteasome inhibition seems to be a promising strategy for ablation of leukemic cells.
Another well documented mechanism of apoptosis induction is mediated by activation of specific p53-regulated genes such as Bax, GADD45, and p21WAF/CIP (21–23). This mechanism is typically activated in response to DNA-damaging agents including the anthracycline family of chemotherapy drugs (24, 25), which are widely used for AML-induction therapy. Interestingly, anthracyclines have also been shown to induce NF-κB activity (26, 27), presumably as a cellular survival mechanism to resist toxic effects of the drug. Therefore, we reasoned that anthracycline treatment would increase cellular dependency on NF-κB activity and thus further sensitize cells to the loss of NF-κB activity induced by proteasome inhibitors. Consequently, in the present study an analysis of proteasome inhibitors in combination with anthracyclines has been performed. The results of these studies demonstrate that LSCs are extremely sensitive to a combination of proteasome inhibition and anthracycline treatment and undergo rapid and extensive apoptosis when briefly cultured with these two agents. Importantly, normal HSCs show little to no effect when treated with the same agents. Thus, apoptosis can be induced in noncycling malignant stem cells while sparing the normal HSC population.
Materials and Methods
Cell Isolation and Culture.
AML cells, normal bone marrow, and umbilical cord blood (CB) cells were obtained with informed consent and processed as described (9, 15). In some cases, marrow and CB cells were also obtained from the National Disease Research Interchange (NDRI). Samples were subjected to density-gradient separation to isolate mononuclear cells followed by cryopreservation. As needed, samples were thawed and used immediately. For in vitro studies, cells were cultured in serum-free medium (28) for 1 h before the addition of MG-132 (Calbiochem) and/or idarubicin (IDR, Pharmacia–Upjohn). When IDR and MG-132 where used in combination, cells were incubated with IDR for 15 min prior the addition of MG-132.
Nonobese Diabetic (NOD)/Severe Combined-Immunodeficient (SCID) Mouse Assays.
NOD/SCID (NOD.CB17-Prdkdc Scid/J) mice (The Jackson Laboratory) were exposed to 300 rad of γ irradiation from a 137Cs source 1 day before transplantation. Cells to be assayed were resuspended in 0.2 ml of PBS (GIBCO) with 2% albumin and injected via the tail vein (5–10 million cells per recipient). After 6–8 weeks, animals were killed, and bone marrow was analyzed for the presence of human cells by using flow cytometry.
Flow Cytometry.
Cell-cycle and apoptosis analyses were performed as described (15, 29). To analyze NOD/SCID mice, marrow cells were blocked with the anti-Fc receptor antibody 2.4G2 and 25% human serum followed by labeling with human-specific CD34-FITC and CD45-PE monoclonal antibodies (PharMingen). Control samples consisted of marrow cells from nontransplanted mice. To distinguish HLA-A2+ and HLA-A2− populations, cells were labeled by using FITC-conjugated monoclonal MA2.1 (kindly provided by John Yannelli, Division of Hematology/Oncology, University of Kentucky Medical Center). Adenovirus-infected AML cells were labeled with CD34-PE (PharMingen), and CD34+/GFP+ cells were isolated by fluorescence-activated cell sorting (>94% purity).
Adenovirus Vector Construction and Infection.
Adenovirus vectors used in this study were based on the AdBM5GFP vector from Quantum Biotechnologies (Montreal). Details of the vector construction will be described elsewhere. Briefly, the vector was digested with AflII + BglII to remove the major late promoter, which then was replaced with a cytomegalovirus promoter. A mutant allele of IκBα (serine to alanine at positions 32 and 36) was then cloned immediately downstream of the cytomegalovirus promoter by using the BglII site. Virus was generated by homologous recombination in 293 cells and purified by using standard procedures (30). Infection of primary AML cells was performed exactly as described by Howard et al. (31).
Electrophoretic Mobility-Shift Assay (EMSA), Immunoblot, and RNA Analysis.
EMSA analysis was performed as described (15). For immunoblots, cells were harvested and analyzed as described by Jordan et al. (9). Blots were probed with anti-p53 (clone DO-1) from Santa Cruz Biotechnology and antiactin from Sigma (clone AC-15). RNA was prepared by using Trizol (Invitrogen) per manufacturer instructions. Five micrograms of RNA per sample were analyzed by using the RiboQuant kit and the h-stress-1 multiprobe set (PharMingen) for RNase protection assays. Gels were scanned by PhosphorImager (Molecular Dynamics), and bands were quantitated by using Kodak 1D software.
Results
Proteasome Inhibition Preferentially Ablates Primitive AML Cells in Vitro.
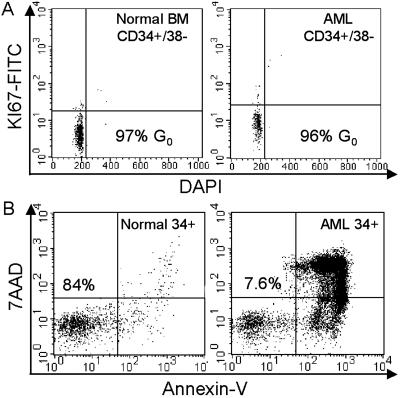
To assess the effects of proteasome inhibition on primitive quiescent cells, studies were performed initially by using multiparameter flow cytometry. In Fig. 1A, cell-cycle analysis shows that primitive cells of both normal and leukemic origin (defined by the immunophenotype CD34+/CD38−/CD123− and CD34+/CD38−/CD123+, respectively) are predominantly in G0. This is evident based on the lack of labeling with nuclear antigen Ki-67, which is a common marker for entry into G1 (32). Based on our studies (15), tests were performed first by using the proteasome inhibitor MG-132 at 1.0 μM for 12 h. This treatment readily induced apoptosis for AML cells, whereas normal CD34+ cells were almost entirely unaffected (as shown by annexin-V labeling, Fig. 1B). Analysis of the 13 AML specimens described in Table 1 showed an average viability of 26 ± 18% after 12 h of culture in MG-132. In contrast, parallel studies of four normal CD34+ cell specimens showed an average viability of 89 ± 5% when analyzed by using the same conditions. All cultures were performed in serum-free medium and in the absence of any exogenous cytokines.
Fig 1.
(A) Cell-cycle profiles for normal and leukemic CD34+/CD38− specimens. BM, bone marrow. (B) Flow-cytometric profile of normal (Left) and leukemic (Right) CD34+ cells stained with annexin-V FITC and 7AAD after 12 h of treatment with 1 μM MG-132. Numbers indicate the percentage of viable cells. DAPI, 4′,6-diamidino-2-phenylindole.
Table 1.
AML specimens
| Case | FAB type | Cytogenetics |
|---|---|---|
| 1 | M4 | 46,XY,del(7) |
| 2 | M2 | 46,XY |
| 3 | M5 | 46,XX |
| 4 | M4 | 48,XY,+8,+13 |
| 5 | M1/M2 | 46,XY |
| 6 | M4 | 46,XX |
| 7 | M5 | 45,X,−Y |
| 8 | M5 | 46,XX |
| 9 | MDS/AML | 46,XY |
| 10 | M1 | 46,XX |
| 11 | M4 | 47,XY,+9 |
| 12 | M2 | 46,XY,t(9;19);t(8;21) |
| 13 | M4 | 46,XY,del(7) |
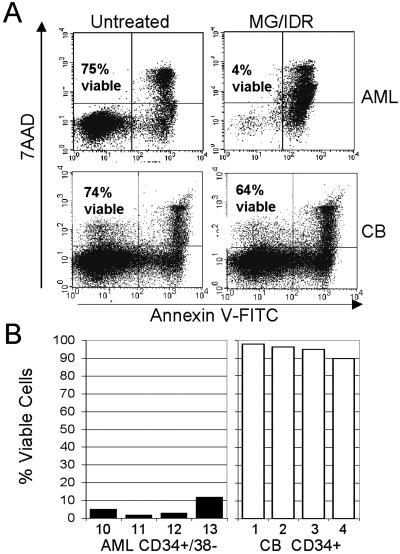
Because MG-132 demonstrated toxicity to phenotypically primitive AML cells in vitro, we sought to further investigate toxicity to the LSC compartment when proteasome inhibition was combined with the commonly used chemotherapeutic anthracycline IDR. Studies have indicated that anthracyclines induce NF-κB in malignant cells (27). Thus, we hypothesized that anthracycline treatment might sensitize leukemic cells to the effects of proteasome inhibition (i.e., loss of NF-κB activity). Moreover, anthracyclines have been shown to increase proteasome activity in leukemic cells, which also could increase sensitivity to proteasome inhibition. To test this theory, both MG-132 and the anthracycline IDR were first titrated to relatively nontoxic doses of 0.25 μM and 15 ng/ml, respectively (data not shown). Each specimen listed in Table 1 then was cultured for 12 h in low-dose MG-132 (0.25 μM) and/or IDR (15 ng/ml). MG-132 treatment alone was only mildly toxic to the cells (mean viability = 73 ± 9%). Similarly, treatment with IDR alone also showed only a small effect on cell viability (mean viability = 88 ± 9%). However, the combination of both drugs synergized strongly to induce robust apoptosis (mean viability = 11 ± 5%). Fig. 2A shows a representative example of an AML specimen (Upper) vs. a normal CB specimen (Lower) treated with the low-dose MG-132/IDR combination. As observed for MG-132 alone, the AML specimen was extremely sensitive to treatment with MG-132/IDR, whereas the normal specimen showed only a slight loss in viability.
Fig 2.
(A) Flow-cytometric analyses of primary AML cells (Upper) compared with normal CB cells (Lower) after 18 h of treatment with or without 0.25 μM MG-132 + 15 ng/ml IDR. Numbers indicate the percentage of viable cells. (B) Percentage of viable CD34+/CD38− cells from primary AML (Left) and normal CB (Right) cell samples after an 18-h treatment with 0.25 μM MG-132 + 15 ng/ml IDR. Viability levels for each sample are normalized to untreated control specimens.
To examine effects on the most primitive cells more specifically, four AML and normal specimens were assayed for apoptosis induction in the CD34+/CD38− population. As shown in Fig. 2B, the primitive AML compartment was also affected dramatically by the combination of MG-132/IDR, whereas the normal CD34+ cells were almost entirely resistant. These data demonstrate that MG-132 in combination with IDR induces apoptosis specifically in primitive AML cells, whereas normal primitive cells seem largely unaffected.
Treatment with MG-132 and IDR Destroys LSCs Able to Engraft NOD/SCID Mice.
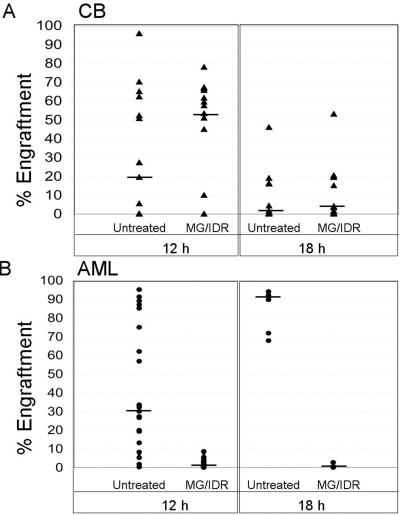
To determine whether stem cells defined by in vivo functional assays were impaired by treatment with MG-132/IDR, normal and leukemic specimens were analyzed by using transplantation into immune-deficient NOD/SCID mice. NOD/SCID xenogeneic models have been used to assess both human HSC and LSC activity (33–35). This model can be used therefore to identify therapies that affect either the LSC or HSC population. Initially, cells from primary CB and AML samples were treated with 0.25 μM MG-132 and 15 ng/ml IDR for 12 h and then injected into NOD/SCID mice. After 6–8 weeks, marrow was analyzed for the presence of donor cells by using a human-specific antibody for CD45. Each triangle in Fig. 3A Left represents the percent engraftment for one animal transplanted with untreated or MG-132/IDR-treated CB cells (15 animals in each group using CB from six independent specimens). The data show that drug treatment does not impair the capacity of normal HSCs to proliferate in NOD/SCID mice. Further, analysis of human cells in animals receiving treated vs. untreated CB cells showed no difference with respect to levels of CD33 (myeloid), CD19 (B lymphoid), and CD34 (primitive) cells, thereby indicating that the differentiation potential of MG-132/IDR-treated CB cells was not impaired (data not shown). In some cases, CB samples lost their ability to engraft after 12 h of culture without drugs. Notably, when the same CB samples were treated with the MG-132/IDR combination, they often retained engraftment ability. Hence, the median engraftment efficiency actually increased for drug-treated specimens (median indicated by horizontal bars). A possible explanation for this surprising result might be that the proteasome inhibitor prevents degradation of proteins involved in homing. Alternatively, the drug treatment may enhance survival or inhibit terminal differentiation of normal stem/progenitor cells. Irrespective of the reason for this observation, the data indicate that engraftment of normal cells is not impaired by MG-132/IDR treatment and supports the in vitro data suggesting the drugs are not toxic to normal stem cells. In contrast, when AML samples were treated in the same fashion and transplanted into NOD/SCID animals, engraftment efficiency was reduced dramatically. Fig. 3B Left shows percent donor engraftment after 12 h of culture ± drug treatment (25 animals in each group using AML cells from four independent specimens). Two AML specimens showed no detectable engraftment, and two specimens displayed only low-level engraftment (3.9–7.5%) after they were exposed to MG-132/IDR. In an attempt to eradicate residual AML cells more completely, we extended the drug exposure to 18 h for the two specimens that showed engraftment at 12 h (Fig. 3B Right). Longer exposure resulted in a further decrease in engraftment but did not eliminate AML cells completely (0.1–2.8% engraftment, seven animals in each group). Finally, to determine whether the few remaining AML cells retained self-renewal potential, marrow from primary animals was transplanted into secondary NOD/SCID recipients (in the absence of any further drug treatment). At 6 weeks posttransplant, no detectable human cells were found in the marrow of secondary recipients, thereby suggesting that the original drug treatment had ablated all NOD/SCID engrafting activity completely. Control experiments using normal CB samples were also performed by using the 18-h MG-132/IDR regimen (Fig. 3A Right). Although overall engraftment of the CB samples was affected by 18 h of culture, treatment with MG-132/IDR did not reduce engraftment efficiency further when compared with untreated controls (15 animals per group).
Fig 3.
(A) Engraftment of NOD/SCID mice with CB cells after 12 or 18 h of culture ± 0.25 μM MG-132 and 15 ng/ml IDR (six independent CB specimens were analyzed at each time point). (B) Engraftment of AML cells after 12 or 18 h of culture ± MG-132 and IDR (four independent AML samples were analyzed after 12 h, and the two showing the highest levels of engraftment were reanalyzed after 18 h of culture). Each triangle or circle represents a single animal analyzed 6–8 weeks posttransplant for the percentage of human cells in marrow. The median level of engraftment is indicated by the horizontal bars.
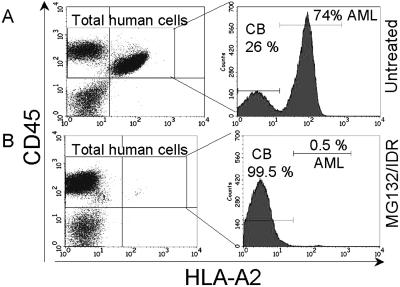
Because treatment with MG-132/IDR strongly induced apoptosis in the total AML cell population, it is possible that LSCs that survive drug treatment might fail to engraft because of the presence of excess dead cells. To address this possibility, an experiment was performed in which CB and AML cells were treated with the combination of MG-132/IDR and coinjected into NOD/SCID animals (n = 6). If apoptotic cells induce an inhibitory effect on the engraftment of viable stem cells, one would predict reduced engraftment when the MG-132/IDR-treated cells are injected together. The specimens used for this experiment expressed disparate HLA-A2 loci such that normal vs. AML cells could be distinguished by flow cytometry. After 6–8 weeks, the animals were killed, and the bone marrow was analyzed for engraftment of human cells. Fig. 4 shows a representative example of bone marrow cells stained with human-specific CD45 and HLA-A2 antibodies. The total percentage of CD45+ cells in both treated and untreated samples was very similar (73–79%). However, labeling with the HLA-A2 antibody shows strong AML cell engraftment in the untreated controls (74% of the CD45+ population) but virtually no leukemic engraftment in MG-132/IDR-treated samples (0.5% of the CD45+ population). These data indicate that the inability of treated AML samples to engraft in the animals is not due to an inhibitory effect caused by apoptotic cells present in the transplanted sample. Taken together, the data in Figs. 2–4 strongly suggest that the combination of MG-132 and IDR effectively eradicates human LSCs while sparing the normal HSC population.
Fig 4.
AML and CB cells were cultured for 12 h ± 0.25 μM MG-132 and 15 ng/ml IDR and injected into sublethally irradiated NOD/SCID mice. Bone marrow was analyzed 6 weeks later with anti-human CD45-PE and HLA-A2-FITC to detect engraftment of human cells. The histograms indicate the percent of human AML vs. CB cells.
Inhibition of NF-κB Contributes to Apoptosis Induced by MG-132/IDR Treatment.
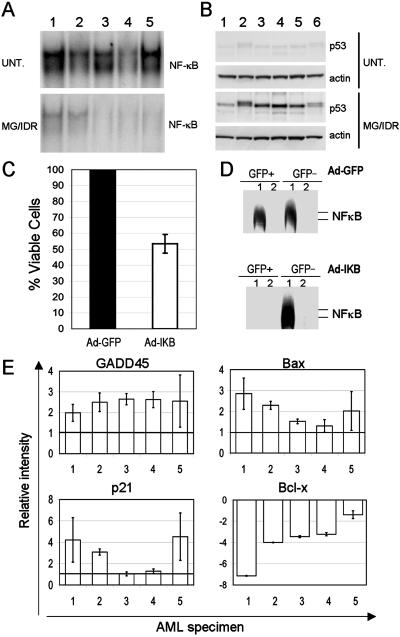
To investigate the mechanisms that underlie LSC-specific apoptosis, a series of molecular studies were performed. Previously we showed that NF-κB is constitutively activated in primary LSC populations but is not detected in normal CD34+ cells (15). Further, these studies showed that treatment with 1.0 μM MG-132 was sufficient to inhibit NF-κB activity. Thus, EMSA analysis was used in the present studies to determine the activity of NF-κB in AML cells treated with 0.25 μM MG-132 and 15 ng/ml IDR. To assess molecular changes occurring at early times posttreatment (i.e., before the onset of overt apoptosis), cells were isolated after 6 h of drug exposure. As shown in Fig. 5A, treatment with MG-132 + IDR induced strong inhibition of NF-κB in comparison to untreated controls in five independent AML specimens. This observation suggests that NF-κB might mediate survival in primary AML cells. To address this possibility directly, an adenovirus vector was constructed that encodes both a mutant version of IκBα and the GFP gene. The IκBα gene used for this vector encodes an allele known as the IκB superrepressor (IκB-SR). This gene is mutated at serines 32 and 36 such that it cannot be phosphorylated and hence is not degraded by normal proteasomal mechanisms. Thus, IκB-SR expression exerts potent inhibition of NF-κB activity as a result of stabilized IκBα protein levels. Three primary AML specimens were transduced with the Ad-IκB-SR vector (Ad-IκB) or a control virus encoding GFP alone (Ad-GFP) as described (31). Infected populations then were monitored over time to assess viability of CD34+ blast cells. The data in Fig. 5C show that although Ad-GFP-infected cells maintained 100% viability over a 36-h period relative to uninfected controls, cells infected with Ad-IκB-SR decreased to ≈50% of control values (52 ± 4% viable cells). EMSA analysis demonstrated that expression of IκB-SR by the vector completely inhibited NF-κB activity in transduced AML cells (Fig. 5D). These data indicate that NF-κB activity is important for survival of primary AML cells, but that inhibition of NF-κB alone is not sufficient to mediate the very rapid and extensive apoptosis observed after treatment with MG-132/IDR.
Fig 5.
(A) NF-κB EMSA of nuclear extracts from five primary AML specimens after 6 h of culture (UNT., untreated; MG/IDR, 0.25 μM MG-132 + 15 ng/ml IDR). Numbers above each lane indicate the specimen number. (B) Immunoblot analysis of p53 for each of the specimens shown in A. Each blot was stripped and reprobed with actin (lower panel of each p53 blot). (C) Three primary AML specimens were infected with Ad-GFP or Ad-IκB-SR and cultured for 36 h. The graph indicates the average percentage of viable GFP+ cells where Ad-GFP is defined as 100%. (D) EMSA analysis of NF-κB in sorted GFP+ or GFP− cells at 12 h postinfection with Ad-GFP or Ad-IκB-SR (lane 1, NF-κB consensus probe; lane 2, probe + 100-fold excess unlabeled consensus probe). (E) Relative change in expression level as determined by RNase protection assay for five primary AML specimens after 6 h of culture in 0.25 μM MG-132 + 15 ng/ml IDR. Untreated specimens were assigned an arbitrary level of 1.0 (shown by the horizontal bar). Analysis of each specimen was performed in triplicate (standard deviation is shown by error bars). Loading was normalized by using L32 ribosomal and glyceraldehyde-3-phosphate dehydrogenase probes as controls.
Activation of a p53-Regulated Mechanism During Apoptosis Induced by MG-132/IDR.
To further characterize molecular mechanisms related to apoptosis induction, pathways commonly associated with cell death were examined. Recent studies by Tergaonkar et al. (36) have shown that treatment with the anthracycline doxorubicin induces a p53-mediated apoptosis response. Moreover, this report showed that loss of NF-κB activity resulted in a stronger induction of apoptosis after activation of p53. Thus, we reasoned that the IDR used in our studies might elicit a similar response. To test this hypothesis, total protein and RNA were prepared from cells cultured for 6 h in the absence or presence of 0.25 μM MG-132 and 15 ng/ml IDR. Immunoblots (Fig. 5B) showed that levels of p53 protein were increased strongly by drug treatment in six independent AML specimens. This observation suggests that p53-mediated changes in gene expression might be involved in apoptosis induced by MG-132/IDR treatment. To examine this question in more detail, RNA was isolated from five primary AML specimens after 6 h of culture and expression of the p53-regulated genes Bax, GADD45, and p21WAF/CIP were analyzed by RNase protection assays. These three genes are strongly associated with p53-induced apoptosis in a variety of different cell types (21–23). As shown in Fig. 5E, RNA levels of Bax, GADD45, and p21WAF/CIP all are increased by treatment with MG-132/IDR. Interestingly, the antiapoptotic protein Bcl-xL was reduced markedly in cells treated with MG-132 + IDR. Bcl-xL is an NF-κB-regulated gene; thus the observed decrease in Bcl-xL mRNA is consistent with the block to NF-κB activity seen in Fig. 5A. Collectively, these data suggest that the combination of MG-132 and IDR induces apoptosis through activation of p53-regulated genes and simultaneous inhibition of NF-κB.
Discussion
Recent studies clearly indicate a central role for LSCs in the pathogenesis of AML and emphasize the need to develop treatment strategies that specifically target this rare population (12). Equally important, it is necessary to identify approaches that minimize toxicity to the normal HSC, which is required to sustain all blood-cell lineages. Consequently, we sought to investigate drug-treatment regimens that would target the LSC population specifically and preferentially. To this end, we have demonstrated that low concentrations of MG-132 (0.25 μM) in combination with IDR (15 ng/ml) are sufficient to induce a strong apoptotic response in primary AML specimens during a short in vitro culture period (≈12 h). More importantly, by using flow-cytometric analysis of phenotypically primitive cells and functional assays in NOD/SCID mice, the data indicate that MG-132 + IDR also effectively ablates the LSC population. In contrast, normal HSCs are almost entirely refractory to the same treatment. These findings formally demonstrate that with the appropriate stimulus quiescent LSCs are preferentially susceptible to apoptosis induction. Thus, it should be possible to develop treatment regimens that specifically target the LSC population. Moreover, recent clinical evaluation of pharmaceutical-grade proteasome inhibitors (37) may soon provide an opportunity to examine the therapeutic potential of such drugs in combination with anthracyclines.
Going forward, it will be important to develop a detailed profile of the molecular events that mediate LSC-specific apoptosis. Studies in this report investigate this phenomenon by analyzing unique molecular characteristics of the LSC population. Previously it was shown that primitive AML cells exhibit substantial NF-κB activity (15). Given the known role of NF-κB in the growth and survival of many tumor types (16), we speculated that inhibition of this factor might be a key step toward inducing LSC apoptosis. Interestingly, although our EMSA data show that NF-κB is inhibited potently by MG-132/IDR treatment, subsequent studies using a dominant-negative allele of IκBα showed that down-regulation of NF-κB activity alone was not sufficient to mediate the same degree of rapid and extensive apoptosis in AML cells as was observed for MG-132/IDR. This finding indicates that other effects of the drug treatment must be contributing to AML cell death. In seeking to characterize LSC apoptosis further, we hypothesized that proteasome inhibition might be stabilizing proteins relevant to apoptosis induction such as p53 (38–40). Moreover, the activity of IDR was expected to activate p53-mediated apoptosis mechanisms. Immunoblot studies confirmed that p53 was increased in drug-treated cells. Further investigation by RNase protection assay showed that several p53-regulated genes known to be mediators of apoptosis were up-regulated in the MG-132/IDR-treated cells. These data indicate that activation of a p53-dependent mechanism is likely to be one component of the overall apoptosis process. Notably, p53 is only mutated in ≈9% of AML specimens (41); thus a strategy that relies on functional p53 for ablation of leukemic cells should be applicable to a majority of patients. Future studies may directly address the role of p53 by expressing a dominant-negative allele of the gene in AML cells and determining whether loss of p53 activity is sufficient to block apoptosis induced by MG-132/IDR.
Another important issue in understanding the biology of LSCs is determining how NF-κB is activated. Our studies indicate that NF-κB plays an important role in the survival of LSCs and represents a potentially useful target for therapeutic intervention. One mechanism for NF-κB activation may be related to mutation of the Flt3 gene. Constitutive activation of Flt3 has emerged recently as the most commonly known aberration in AML (42). Studies have shown that signaling via Flt3 can stimulate Ras (43), which in turn is a known activator of NF-κB (16). Thus, pathways leading from Flt3 to NF-κB seem to exist. Moreover, several recent studies have begun to examine the use of Flt3 inhibitors for AML therapy (44). Consequently, direct analysis of the degree to which Flt3- and NF-κB-regulated pathways overlap are now feasible. Interestingly, the kinetics and degree of primary AML cell death reported for Flt3 inhibitors (45, 46) appear very similar to data in this report using expression of IκB-SR (i.e., 40–50% cell death over 36–72 h of treatment). This similarity may indicate a substantial degree of overlap between strategies that target Flt3 and NF-κB and suggests that Flt3 inhibitors and proteasome inhibitors may be functionally equivalent in the context of leukemic cell biology. If this concept is true, then based on findings in this study, we suggest that combining Flt3 inhibition with IDR treatment may yield a more rapid and robust induction of apoptosis, as was observed for MG-132 + IDR. This approach may be attractive for clinical use, because Flt3 inhibitors are expected to have less nonspecific toxicity than proteasome inhibitors. Further, we suggest that combination of Flt3 inhibitors with agents such as IDR may increase the likelihood that they will target quiescent LSC populations effectively.
Acknowledgments
We thank Drs. Katherine Borden, Gary Van Zant, Hartmut Geiger, and Deborah Echlin for critical evaluation of the manuscript. We also thank Dr. Marty Mayo for helpful discussions and for providing the IκB-SR gene. We gratefully acknowledge the generous support of The Markey Cancer Center Foundation and The Donatina Colachicco Cancer Research Fund. This work was supported by American Cancer Society Grant RPG-99-206-01-LBC (to C.T.J.) and National Institutes of Health Grant R01-CA90446 (to C.T.J.). D.S.H. is a fellow of the Abraham J. and Phyllis Katz Foundation.
Abbreviations
AML, acute myelogenous leukemia
LSC, leukemic stem cell
HSC, hematopoietic stem cell
NF-κB, nuclear factor κB
IκBα, inhibitor of NF-κB
CB, umbilical cord blood
MG-132, carbobenzoxyl-l-leucyl-l-leucyl-l-leucinal
IDR, idarubicin
NOD, nonobese diabetic
SCID, severe combined immunodeficient
EMSA, electrophoretic mobility-shift assay
IκB-SR, IκB superrepressor
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lowenberg B., Downing, J. R. & Burnett, A. (1999) N. Engl. J. Med. 341, 1051-1062. [DOI] [PubMed] [Google Scholar]
- 2.Fialkow P. J., Singer, J. W., Adamson, J. W., Vaidya, K., Dow, L. W., Ochs, J. & Moohr, J. W. (1981) Blood 57, 1068-1073. [PubMed] [Google Scholar]
- 3.Fialkow P. J., Singer, J. W., Raskind, W. H., Adamson, J. W., Jacobson, R. J., Bernstein, I. D., Dow, L. W., Najfeld, V. & Veith, R. (1987) N. Engl. J. Med. 317, 468-473. [DOI] [PubMed] [Google Scholar]
- 4.Lapidot T., Sirard, C., Vormoor, J., Murdoch, B., Hoang, T., Caceres-Cortes, J., Minden, M., Paterson, B., Caligiuri, M. A. & Dick, J. E. (1994) Nature 367, 645-648. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet D. & Dick, J. E. (1997) Nat. Med. 3, 730-737. [DOI] [PubMed] [Google Scholar]
- 6.Blair A., Hogge, D. E. & Sutherland, H. J. (1998) Blood 92, 4325-4335. [PubMed] [Google Scholar]
- 7.Blair A., Hogge, D. E., Ailles, L. E., Lansdorp, P. M. & Sutherland, H. J. (1997) Blood 89, 3104-3112. [PubMed] [Google Scholar]
- 8.Blair A. & Sutherland, H. J. (2000) Exp. Hematol. (Charlottesville, Va) 28, 660-671. [DOI] [PubMed] [Google Scholar]
- 9.Jordan C. T., Upchurch, D., Szilvassy, S. J., Guzman, M. L., Howard, D. S., Pettigrew, A. L., Meyerrose, T., Rossi, R., Grimes, B., Rizzieri, D. A., et al. (2000) Leukemia 14, 1777-1784. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland H. J., Blair, A. & Zapf, R. W. (1996) Blood 87, 4754-4761. [PubMed] [Google Scholar]
- 11.Ailles L. E., Gerhard, B. & Hogge, D. E. (1997) Blood 90, 2555-2564. [PubMed] [Google Scholar]
- 12.Jordan C. T. (2002) Leukemia 16, 559-562. [DOI] [PubMed] [Google Scholar]
- 13.Terpstra W., Ploemacher, R. E., Prins, A., van Lom, K., Pouwels, K., Wognum, A. W., Wagemaker, G., Lowenberg, B. & Wielenga, J. J. (1996) Blood 88, 1944-1950. [PubMed] [Google Scholar]
- 14.Guzman M. L., Upchurch, D., Grimes, B., Howard, D. S., Rizzieri, D. A., Luger, S. M., Phillips, G. L. & Jordan, C. T. (2001) Blood 97, 2177-2179. [DOI] [PubMed] [Google Scholar]
- 15.Guzman M. L., Neering, S. J., Upchurch, D., Grimes, B., Howard, D. S., Rizzieri, D. A., Luger, S. M. & Jordan, C. T. (2001) Blood 98, 2301-2307. [DOI] [PubMed] [Google Scholar]
- 16.Mayo M. W. & Baldwin, A. S. (2000) Biochim. Biophys. Acta 1470, M55-M62. [DOI] [PubMed] [Google Scholar]
- 17.Garg A. & Aggarwal, B. B. (2002) Leukemia 16, 1053-1068. [DOI] [PubMed] [Google Scholar]
- 18.Karin M., Cao, Y., Greten, F. R. & Li, Z. W. (2002) Nat. Rev. Cancer 2, 301-310. [DOI] [PubMed] [Google Scholar]
- 19.Hallahan D. E. & Teng, M. (2000) Int. J. Radiat. Oncol. Biol. Phys. 47, 859-860. [DOI] [PubMed] [Google Scholar]
- 20.Almond J. B. & Cohen, G. M. (2002) Leukemia 16, 433-443. [DOI] [PubMed] [Google Scholar]
- 21.Miyashita T., Krajewski, S., Krajewska, M., Wang, H. G., Lin, H. K., Liebermann, D. A., Hoffman, B. & Reed, J. C. (1994) Oncogene 9, 1799-1805. [PubMed] [Google Scholar]
- 22.Kastan M. B., Zhan, Q., el-Deiry, W. S., Carrier, F., Jacks, T., Walsh, W. V., Plunkett, B. S., Vogelstein, B. & Fornace, A. J., Jr. (1992) Cell 71, 587-597. [DOI] [PubMed] [Google Scholar]
- 23.el-Deiry W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W. & Vogelstein, B. (1993) Cell 75, 817-825. [DOI] [PubMed] [Google Scholar]
- 24.Lowe S. W., Bodis, S., McClatchey, A., Remington, L., Ruley, H. E., Fisher, D. E., Housman, D. E. & Jacks, T. (1994) Science 266, 807-810. [DOI] [PubMed] [Google Scholar]
- 25.Lowe S. W., Ruley, H. E., Jacks, T. & Housman, D. E. (1993) Cell 74, 957-967. [DOI] [PubMed] [Google Scholar]
- 26.Laurent G. & Jaffrezou, J. P. (2001) Blood 98, 913-924. [DOI] [PubMed] [Google Scholar]
- 27.Boland M. P., Foster, S. J. & O'Neill, L. A. (1997) J. Biol. Chem. 272, 12952-12960. [DOI] [PubMed] [Google Scholar]
- 28.Lansdorp P. M. & Dragowska, W. (1992) J. Exp. Med. 175, 1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan C. T., Yamasaki, G. & Minamoto, D. (1996) Exp. Hematol. (Charlottesville, Va) 24, 1347-1355. [PubMed] [Google Scholar]
- 30.Prevec L. & Graham, F. L., (1991) Methods in Molecular Biology: Gene Transfer and Expression Protocols (Humana, Clifton, NJ).
- 31.Howard D. S., Rizzierri, D., Grimes, B., Upchurch, D., Phillips, G., Stewart, A., Yannelli, J. & Jordan, C. T. (1999) Leukemia 13, 1608-1616. [DOI] [PubMed] [Google Scholar]
- 32.Gerdes J., Lemke, H., Baisch, H., Wacker, H. H., Schwab, U. & Stein, H. (1984) J. Immunol. 133, 1710-1715. [PubMed] [Google Scholar]
- 33.Dick J. E. (1996) Semin. Immunol. 8, 197-206. [DOI] [PubMed] [Google Scholar]
- 34.Dick J. E., Bhatia, M., Gan, O., Kapp, U. & Wang, J. C. (1997) Stem Cells (Dayton) 15, 199-203. [DOI] [PubMed] [Google Scholar]
- 35.Lapidot T., Fajerman, Y. & Kollet, O. (1997) J. Mol. Med. 75, 664-673. [DOI] [PubMed] [Google Scholar]
- 36.Tergaonkar V., Pando, M., Vafa, O., Wahl, G. & Verma, I. (2002) Cancer Cell 1, 493-503. [DOI] [PubMed] [Google Scholar]
- 37.Adams J. (2002) Curr. Opin. Chem. Biol. 6, 493-500. [DOI] [PubMed] [Google Scholar]
- 38.Dietrich C., Bartsch, T., Schanz, F., Oesch, F. & Wieser, R. J. (1996) Proc. Natl. Acad. Sci. USA 93, 10815-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopes U. G., Erhardt, P., Yao, R. & Cooper, G. M. (1997) J. Biol. Chem. 272, 12893-12896. [DOI] [PubMed] [Google Scholar]
- 40.Naujokat C., Sezer, O., Zinke, H., Leclere, A., Hauptmann, S. & Possinger, K. (2000) Eur. J. Haematol. 65, 221-236. [DOI] [PubMed] [Google Scholar]
- 41.Stirewalt D. L., Kopecky, K. J., Meshinchi, S., Appelbaum, F. R., Slovak, M. L., Willman, C. L. & Radich, J. P. (2001) Blood 97, 3589-3595. [DOI] [PubMed] [Google Scholar]
- 42.Gilliland D. G. & Griffin, J. D. (2002) Curr. Opin. Hematol. 9, 274-281. [DOI] [PubMed] [Google Scholar]
- 43.Mizuki M., Fenski, R., Halfter, H., Matsumura, I., Schmidt, R., Muller, C., Gruning, W., Kratz-Albers, K., Serve, S., Steur, C., et al. (2000) Blood 96, 3907-3914. [PubMed] [Google Scholar]
- 44.Gilliland D. G. & Griffin, J. D. (2002) Blood 100, 1535-1542. [DOI] [PubMed] [Google Scholar]
- 45.Levis M., Tse, K. F., Smith, B. D., Garrett, E. & Small, D. (2001) Blood 98, 885-887. [DOI] [PubMed] [Google Scholar]
- 46.Levis M., Allebach, J., Tse, K. F., Zheng, R., Baldwin, B. R., Smith, B. D., Jones-Bolin, S., Ruggeri, B., Dionne, C. & Small, D. (2002) Blood 99, 3885-3891. [DOI] [PubMed] [Google Scholar]