Abstract
Long Interspersed Elements (LINE-1s, L1s) are the most active mobile elements in the human genome and account for a significant fraction of its mass. The propagation of L1 in the human genome requires disruption and repair of DNA at the site of integration. As Barbara McClintock first hypothesized, genotoxic stress may contribute to the mobilization of transposable elements, and conversely, element mobility may contribute to genotoxic stress. We tested the ability of genotoxic agents to increase L1 retrotransposition in a cultured cell assay. We observed that cells exposed to gamma radiation exhibited increased levels of L1 retrotransposition. The L1 retrotransposition frequency was proportional to the number of phosphorylated H2AX foci, an indicator of genotoxic stress. To explore the role of the L1 endonuclease in this context, endonuclease-deficient tagged L1 constructs were produced and tested for their activity in irradiated cells. The activity of the endonuclease-deficient L1 was very low in irradiated cells, suggesting that most L1 insertions in irradiated cells still use the L1 endonuclease. Consistent with this interpretation, DNA sequences that flank L1 insertions in irradiated cells harbored target site duplications. These results suggest that increased L1 retrotransposition in irradiated cells is endonuclease dependent. The mobilization of L1 in irradiated cells potentially contributes to genomic instability and could be a driving force for secondary mutations in patients undergoing radiation therapy.
INTRODUCTION
L1s are autonomous mobile elements that have proliferated throughout eukaryotic genomes for hundreds of millions of years (1). In humans, L1s comprise ∼17% of the mass of the genome (2). An active L1 propagates via an RNA intermediate. L1 RNA not only serves as a cDNA template but also encodes the two L1 proteins, ORF1 and ORF2, both of which are required for retrotransposition (3,4). Because L1 retrotransposition involves the insertion of DNA into random sites in the genome, L1 has the capacity to disrupt genes (5) and contribute to genetic instability (6,7).
The generation of a new genomic L1 copy requires the disruption and repair of DNA. It is likely that most L1s enter the genome via a pathway that uses the element-encoded endonuclease to create a nick in the target DNA that produces a free DNA end that can be used as a cDNA primer. This process of target primed reverse transcription is based mainly on elegant studies with the non-LTR retrotransposon R2Bm (8,9). The R2Bm element-encoded endonuclease creates sequential staggered DNA breaks at the genomic target site (10). A similar mechanism likely operates for L1 insertion, albeit at a less specific target site (5′-TTTT/A-3′) (6,7,11–14). It is less clear how, after first-strand L1 cDNA synthesis, the second strand is created and the insertion site is repaired. Non-homologous recombination machinery may use short stretches of sequence homology between the L1 cDNA and the genomic insertion flank to patch the 5′ end of a nascent L1 insertion and/or prime second strand cDNA synthesis (7,15–18). Using a bioinformatic analysis, it has recently been reported that the 5′ ends of truncated L1 insertions contain short stretches of microhomology to the target site, whereas longer L1 insertions and Alu elements do not show this pattern (19). These and other findings (see below) suggest that there is more than one pathway by which L1s can breach genomic DNA.
L1s can also enter the genome in a manner that does not require L1 endonuclease. An endonuclease-deficient tagged L1 element can retrotranspose, albeit with greatly diminished activity compared to the wild type tagged L1 element (20). Presumably, this endonuclease-deficient L1 inserts into pre-existing DNA breaks. This hypothesis is supported by studies that document that other mobile elements, including retrotransposons, can insert into DNA breaks (21–23). Endonuclease-deficient L1s can retrotranspose at high frequencies in cells deficient in non-homologous end joining (NHEJ) DNA repair, lending further credence to the idea that L1 insertions can integrate and possibly repair double strand DNA breaks (24,25).
To determine if L1s insert into pre-formed DNA breaks in cells with intact DNA repair machinery, L1 retrotransposition was analyzed in tissue culture cells that were subjected to different forms of DNA damage. Here, we report that retrotransposition of a tagged L1 element increases in cultured cells that are subjected to gamma irradiation, a source of genotoxic stress that produces double strand DNA breaks. We document a dose-dependent increase in phosphorylated H2AX foci, an indicator of genotoxic stress, under the conditions of the L1 retrotransposition assay. To determine whether L1 insertions in gamma irradiated cells require the L1 endonuclease, genomic DNA flanking the L1 insertions was isolated and sequenced. The sequences harbored evidence of conventional, endonuclease-dependent L1 insertion. Furthermore, the retrotransposition activity of an endonuclease-deficient L1 element did not significantly increase when cells were subjected to gamma irradiation. Taken together, these findings suggest that L1 usually enters the genome via an endonuclease-dependent pathway in cells subjected to gamma irradiation.
MATERIALS AND METHODS
Recombinant DNA plasmids
All plasmids consist of the L1RP element tagged with an enhanced green fluorescent protein (EGFP) cassette (pL1RP-EGFP) in a pCEP4 backbone (Invitrogen) as described previously (26). The L1 element is driven by its 5′-UTR and an upstream CMV MIE promoter. L1-EGFP (EF06R) was derived from L1RP-EGFP by blunt ligation of 1–1392 nt of pPur (BD/Clontech) into the NruI site of pCEP4. A negative control ‘dead’ L1-EGFP (EF05J) was similarly derived from pL1RP(JM111)-EGFP, which contains disabling mutations in ORF1 (3). A positive control with constitutive EGFP expression (EF03N) was derived from pEGFP-N1 (BD/Clontech) by cloning pPur into the EcoO191 site of pEGFP-N1. An EN− plasmid (EF13E) was created by swapping 1927–3708 nt (Age I–Bcl I) from JM102D205A (24), which contains a point mutation in the EN domain, into EF06R. A control plasmid (EF12J) was similarly derived from JM102 (3). JM102 and JM102D205A are derived from L1.3, which differs from L1RP by 10 base changes in the region swapped.
Cell lines
The 143B human osteosarcoma cells were a gift from H. Kazazian (University of Pennsylvania) and CHO-K1 cells from J. Moran (University of Michigan). Both 143B and CHO-K1 cells were cultured at 37°C, 5% CO2 and 100% humidity in DMEM (Gibco BRL) supplemented with 10% fetal calf serum (US Biotechnologies) and 100 U/ml of penicillin and streptomycin.
Gamma irradiation
On day 0, 1 × 105 adherent 143B cells were transfected with 1 µg of L1 plasmid and 0.2 µg of DsRed-Express-N1 (Clontech) in 6 µl FuGENE6 (Roche) and 100 µl OptiMEM (Gibco BRL). On day 2, cells were harvested with Versene [2% EDTA in phosphate-buffered saline (PBS)] and irradiated with 0–4 Gy using a 137Cesium source. Cells were returned to culture, harvested on day 8, and the percentage of EGFP+ cells analyzed by flow cytometry (BD FACSCaliber), gating on live cells by forward/side scatter and ToPro3 exclusion. Transfection efficiency was normalized using DsRed (BD Biosciences). Alternately, cells were selected for transfection with 0.6 µg/ml of puromycin on days 4–6 and analyzed by flow cytometry on day 12.
Other chemotherapeutic agents
The 143B cells were transfected as above. Cells were exposed to genotoxic agents for 1 h on day 2 in standard culture conditions, washed twice with PBS and returned to culture. Agents tested were arsenite (Sigma, in PBS), calicheamicin γ1 [gift from Wyeth, in dimethyl sulfoxide (DMSO)], camptothecin (Sigma, in DMSO) and cisplatin (Sigma, in PBS). Etoposide (Sigma, in DMSO) exposure was for 10 min. Ultraviolet irradiation was performed in a Stratalinker (Stratagene) with cells in 3 mm depth of PBS.
DNA damage quantification
Cells grown on coverslips were fixed and permeabilized with 3.5% paraformaldehyde + 1% TritonX-100, washed with PBS and blocked with KB (0.01 M Tris pH 7.5, 0.15 M NaCl, 0.1% BSA, 0.1% Na Azide). Coverslips were stained with mouse anti-phosphorylated H2AX (Upstate, Charlottesville) and washed first 1× with 0.1% Triton in PBS and then 2× with KB. Secondary staining was with anti-mouse AlexaFluor 594 (Molecular Probes, Eugene), and nuclei were visualized by DAPI staining (Sigma). Stained cells were examined with a 100X PlanNeofluor objective mounted on a Nikon TE-200 microscope. For assessments of H2AX foci, the number of foci detectable in each nucleus was counted by focusing through the entire thickness of the nucleus and recording the number of foci from all levels. At least 300 cells were counted per time point/treatment. Images were captured with a Hammamatsu CCD camera.
Quantitative RT–PCR
The 143B cells were disrupted in a QiaShredder column (Qiagen) and RNA was extracted using RNeasy columns (Qiagen) per the manufacturer's instructions. A total of 100 ng of RNA was reverse transcribed from a primer in the L1 3′-UTR (SV40rev-TCCAAACTCATCAATGTATCTTATCAT) using Taqman Multiscribe (ABI). Quantitative real-time PCR was performed using High Capacity cDNA archive kit (ABI) and detected on a PRISMR 7900HT (ABI) using a custom designed TaqmanL1 probe/primer (ABI). GAPDH was similarly quantified using random hexamer reverse transcription and human GAPDH Taqman probe/primers (ABI). Data were analyzed using SDS 2.2.1 (ABI) and Excel.
Cloning and sequencing of genomic flanks
EGFP+ clones were handpicked or isolated by flow cytometry (FACSDiva). Clones were expanded in culture for 2 weeks, and DNA purified using a DNeasy kit (Qiagen). Amplification was either by inverse PCR as previously described (27) or by modified suppression PCR (28). For suppression PCR, genomic DNA was digested by Ase1, EcoR1 or HindIII (New England Biolabs). A linker/dummy combination was annealed and ligated (New England Biolabs) to the digested genomic DNA: ATlink-GTGGCGGCCAGTATTCGTAGGAGGGCGCGTAGATAGAACG. Excess linkers were removed with Centricon-40 columns (Princeton Separations). Genomic flanks were amplified using a primer in the linker ATX-GTGGCGGCCAGTATTC and seminested primers in L1-EGFP: SV40for1-ATGATAAGATACATTGATGAGTTTGGA and SV40for2-GGACAAACCACAACTAGAATGC. The primary PCR was done in a reaction volume of 25 µl containing 1 µM SV40for1, 50 nM ATX, 1× FailSafe buffer D, 4% DMSO, 1.25 U FailSafe Taq (Epicentre) and 8 µl linked DNA. Amplification used the following cycling conditions: 68° for 5 s (extension of dummy), 94° for 4 min (primary denaturation), followed by 40 cycles at 94° for 30 s, 85° for 10 s, 75° for 10 s, 63° for 30 s and 68° for 120 s, with a final extension at 68° for 10 min. The secondary PCR was done in a reaction volume of 20 µl containing 1 µM SV40for2, 50 nM ATX, 50 µM dNTPs, 1× PCR buffer D, 4% DMSO, 1.25 U FailSafe Taq and 1 µl primary PCR product. Amplification used the following cycling conditions: 94° for 4 min, followed by 40 cycles at 94° for 30 s, 63° for 30 s and 68° for 120 s, with a final extension at 68° for 10 min. PCR products were run on a 0.8% agarose gel and extracted by QIAQuick (Qiagen), TA cloned into pCR2.1 (Invitrogen), and transformed into One-Shot bacteria (Invitrogen). Plasmid DNA was extracted from expanded clones by QiaPrep (Qiagen) and sequenced. The 3′ flank DNA was localized in the human genome using MegaBLAST against the NCBI database and 5′ primers were generated from upstream sequence (IDT DNA). The 5′ flank was amplified using insertion-specific primers as well as primers in L1 (L1ORF1AS: GGTTGTTCCTTTCCATGTTTAGC, L1RP3435AS: GCTGTGAATCCATCTGGTCC, and HSVtk: CCGATTCGCAGCGCATCGCCTT). The PCR was performed in a reaction volume of 20 µl containing 300 nM of each primer, 50 µM dNTPs, 1× PCR buffer I (Roche), 1.5 U TaqGold (Roche) and 200 ng genomic DNA and used the following cycling conditions: 95° for 10 min, followed by 40 cycles at 95° for 25 s, 58° for 30 s and 72° for 210 s, with a final extension at 72° for 10 min. PCR products were isolated and sequenced as above.
RESULTS
Gamma radiation increases L1 retrotransposition in a cultured cell assay
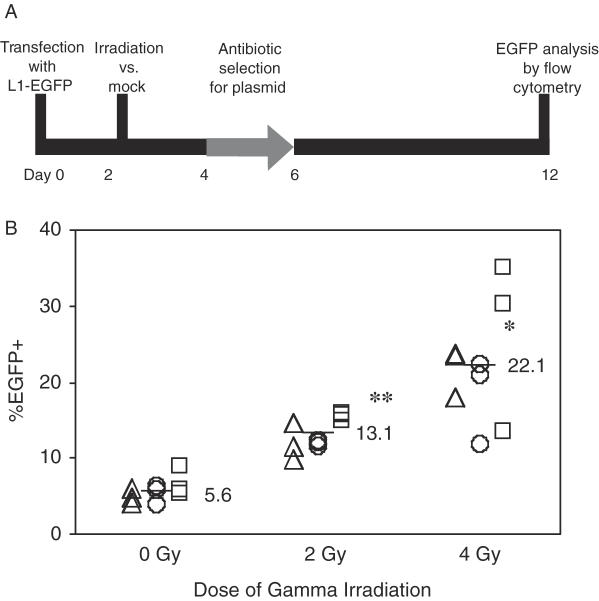
The use of a transient plasmid-based assay offers unique advantages for determining the mechanism of genotoxic-induced L1 mobility. A plasmid-based assay measures retrotransposition directly rather than as a by-product of L1 activity such as L1 RNA or protein levels. By studying the activity of a single active element, the retrotransposition assay is not confounded by the genomic context of different elements. To determine if DNA damage affects L1 mobility, L1 retrotransposition frequency was measured in a cultured cell assay in the presence and absence of gamma radiation (Figure 1A). This assay made use of an active human element, L1RP, tagged with an EGFP reporter as described previously (26). An L1 insertion of sufficient length into a transcriptionally permissive location in the genome will express EGFP. In human 143B osteosarcoma cells selected for the presence of the L1-EGFP plasmid with puromycin, 2 and 4 Gy of gamma radiation increased the percentage of EGFP+ cells (Figure 1B). At these doses, DSBs generated were also proportional to radiation exposure, suggesting a correlation between DNA damage and retrotransposition (Figure 3A). A functionally inactive L1 with two missense mutations in ORF1 (3) showed no detectable retrotransposition with or without irradiation (data not shown), indicating that the radiation-induced increase in EGFP+ cells was specific to the L1 retrotransposition mechanism. Because inhibition of protein synthesis by puromycin may act independently or in concert with radiation to increase retrotransposition (29), the experiment was also performed without antibiotic selection. Without antibiotic selection, gamma radiation again increased the percentage of EGFP+ cells up to 3-fold over mock treated cells (Supplementary Figure 1). A further consideration is that the L1-EGFP expression plasmids used in this study may be themselves sensitive to the genotoxic treatments. To test this, we measured the transcriptional activity of the L1 plasmid by quantifying L1-EGFP RNA via quantitative strand specific real-time PCR. L1-EGFP RNA levels were indistinguishable between irradiated and unirradiated cells at 3 and 24 h post-radiation (Supplementary Figure 2). This suggests that plasmid transcription and copy number remain constant in the face of gamma radiation.
Figure 1.
Gamma radiation increases the L1 retrotransposition frequency. (A) The 143B human osteosarcoma cells transfected with L1-EGFP were exposed to gamma radiation and subjected to puromycin selection for the presence of plasmid. The percentage of cells expressing EGFP was measured by flow cytometry. (B) Percentage of EGFP+ cells on day 12 from three independent L1-EGFP transient transfections (triangles, circles and squares) following 0, 2 or 4 Gy of gamma irradiation and puromycin selection. *P < 0.0005, **P < 0.0001, as compared to 0 Gy by two tailed Student's t-test. A functionally inactive L1 was also tested and had undetectable activity at each dose of irradiation (data not shown).
Other genotoxic agents do not increase L1 retrotransposition in 143B cells
If damaged DNA serves as a preferred substrate for L1 insertion (Figure 2), then other DNA damaging agents besides gamma radiation should increase the L1 retrotransposition frequency. Therefore, L1 retrotransposition was monitored in cells subjected to a variety of DNA damage agents. These agents had various mechanisms of action, including oxidative damage, topoisomerase inhibition and adduct formation. Only gamma radiation resulted in increased L1 retrotransposition (Table 1).
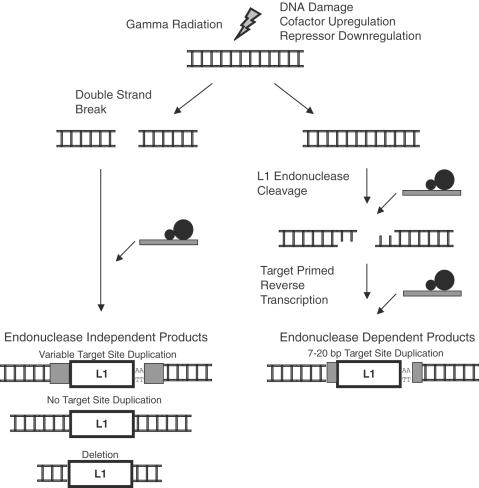
Figure 2.
Potential mechanisms of gamma radiation-induced retrotransposition. Left arrow: gamma radiation-induced double strand breaks could serve as substrates for L1 insertion. Insertions at pre-formed DNA breaks may not require L1 endonuclease; genomic DNA flanking such insertions is expected to lack target site duplications and contain deletions. Right arrow: gamma irradiation may make the host environment more amenable to retrotransposition by upregulating cofactors or downregulating repressors for endonuclease-dependent retrotransposition. An alternative pathway (not shown) is that L1 inserts near DSBs in irradiated cells, but still uses its endonuclease to cleave and/or process DNA at the integration site. This alternative is discussed in the text.
Table 1.
Influence of various genotoxic agents on L1 retrotransposition in a cultured cell assay
| Agent | Dose | Break type | Mechanism | Retrotransposition |
|---|---|---|---|---|
| Gamma Irradiation | 1–10 Gy | ds | Radical attack | + |
| Calicheamicin γ1 | 5–20 pM | ds | Radical attack | − |
| Etoposide | 0.25–100 µg/ml | ds/ss | Topoisomerase II inhibition | − |
| Cisplatin | 0.25–10 µg/ml | ds/ss | Strand crosslinking | N/S |
| Camptothecin | 1–5 uM | ds/ss | Topoisomerase I inhibition | N/S |
| Arsenite | 100–500 uM | ds/ss | Oxidative | N/S |
| Ultraviolet | 25–200 J/m2 | ss | TT dimers | N/S |
143B cells were transiently transfected with L1-EGFP and exposed to DNA damaging agents at various doses up to and including a toxic dose. The agent, dose range tested, spectrum of single stranded (ss) versus double strand (ds) break generated, the major mechanism of damage and effect on L1-EGFP mobility are summarized. +, increase in L1 mobility; −, decrease in L1 mobility; N/S, no significant change.
Gamma radiation and calicheamicin γ1 generate similar numbers of H2AX foci
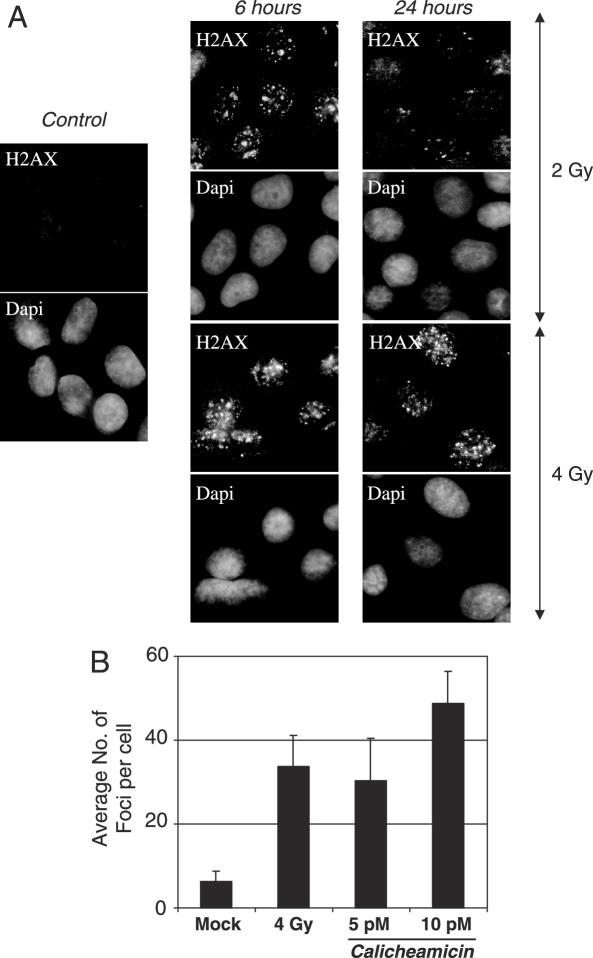
One possible interpretation of the chemotherapy experiments is that toxicity was reached before a comparable level of genotoxic stress was achieved. To investigate this issue, we focused on the capacity of gamma irradiation and calicheamicin γ1 treatment to induce γH2AX foci. Calicheamicin γ1 was chosen for this more detailed analysis because of its higher activity in producing DSBs than the other chemotherapeutic agents (30). To monitor DNA damage, cells were subjected to γ irradiation or calicheamicin γ1 treatment and stained for phosphorylated H2AX foci (γH2AX). γH2AX foci appear rapidly, persist for hours and are proportional to the number of DSBs [Figure 3A, (31,32)]. When γH2AX foci were measured 6 h after treatment, 5–10 pM calicheamicin γ1 elicited a comparable number of foci to 4 Gy of gamma radiation (Figure 3B). Cell viability was also comparable between calicheamicin γ1 and gamma radiation in this dose range (data not shown). One possible reason for this result is that DSBs per se are not responsible for increasing the retrotransposition frequency in irradiated cells. Alternatively, DSBs are still the relevant lesion, but gamma radiation differs from other forms of DNA damage in the structure or kinetics of DSB production. According to this latter alternative, even though the number of H2AX foci is similar, the type of DNA damage and/or longevity of the damage may differ between radiation and calicheamicin γ1.
Figure 3.
Gamma irradiation and calicheamicin γ1 generate comparable numbers of γH2AX foci. 143B cells were permeabilized and fixed 6 and 24 h after exposure to gamma irradiation (54 and 72 h post-transfection). Double strand breaks were detected by staining for γH2AX. Nuclei were visualized by DAPI staining. (A) Fluorescent images showing γH2AX foci at 6 and 24 h post-irradiation. All images are 400×. (B) Average numbers of repair foci per cell 6 h after exposure to mock, 4 Gy gamma radiation or 5 and 10 pM of calicheamicin γ1. The bar graphs show the mean number of γH2AX foci per cell determined from three separate assessments of at least 100 cells in different regions of the coverslip. Error bars indicate standard deviation.
Endonuclease-deficient L1 retrotransposition in irradiated cells
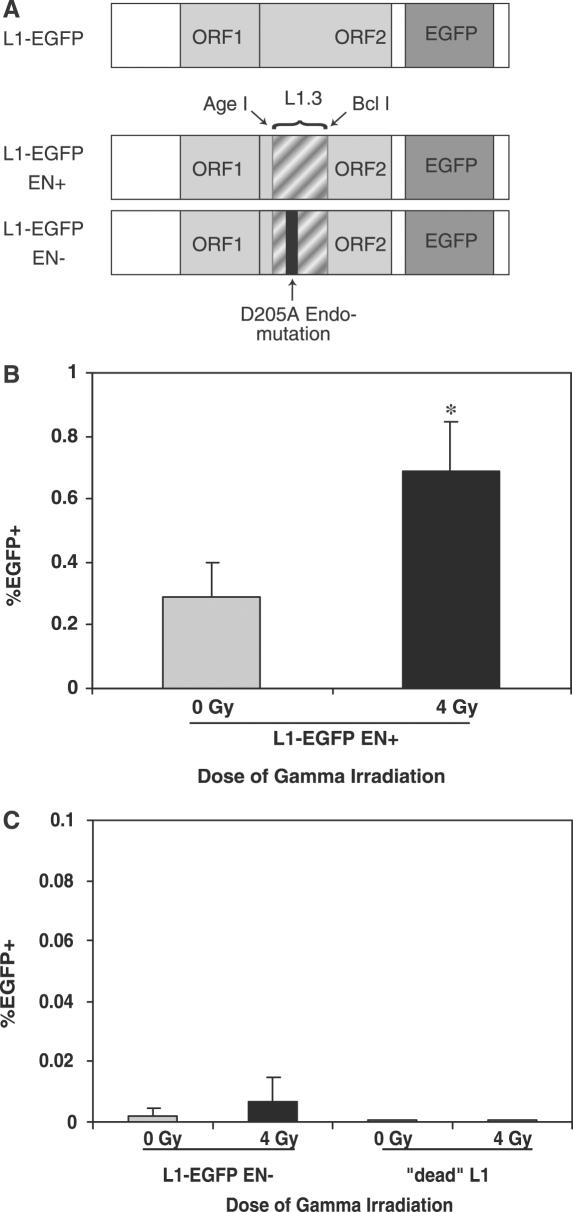
To further investigate the potential role of DSBs in activating L1 retrotransposition, we set out to determine whether the increased retrotransposition frequency in gamma irradiated cells required L1 endonuclease. If radiation-induced DNA breaks were preferred substrates for insertion, then an endonuclease-deficient L1 element should exhibit increased retrotransposition in the presence of radiation-induced DNA damage. A hybrid L1RP/L1.3 element was generated that uses the EGFP reporter and harbors a mutation abrogating endonuclease function (Figure 4A) (24). L1RP has greater activity than L1.3 in a retrotransposition assay (26,33). There are 10 nt differences causing three changed amino acids in the swapped region between L1.3 and L1RP (34,35). Because these sequence differences have uncharacterized effects on retrotransposition rates, the parental L1.3 was also swapped for use as an endonuclease-competent control (Figure 4A). Retrotransposition activity of these elements was tested in CHO-K1 cells, previously shown to support high levels of endonuclease-independent retrotransposition (24). Following irradiation, an increased fraction of CHO-K1 cells transfected with the endonuclease-competent L1-EGFP expressed EGFP (Figure 4B). Gamma radiation is therefore able to increase retrotransposition in an additional mammalian cell line. In contrast, cells transfected with an endonuclease-deficient L1 did not exhibit a statistically significant increase in retrotransposition following irradiation (Figure 4C). Therefore, most of the increased retrotransposition in the setting of gamma radiation probably occurs via an endonuclease-dependent pathway.
Figure 4.
Endonuclease-independent L1 retrotransposition in CHO-K1 cells subjected to gamma radiation. (A) An endonuclease-deficient L1 was generated by swapping in a portion of ORF2 from L1.3 containing a point mutation at the endonuclease active site (see Materials and Methods). A control endonuclease-competent chimeric L1 element was also generated. (B) CHO-K1 cells were transfected with endonuclease-competent L1-EGFP and retrotransposition measured as shown in Figure 1. n = 9 (three independent transfections measured in triplicate) for each bar. *P < 0.001 by two tailed Student's t-test. (C) CHO-K1 cells were transfected with endonuclease-deficient L1-EGFP and retrotransposition was measured. Total EGFP+ events per gated (live) events: 0 Gy: 43/2.3 × 106, 4 Gy: 154/2.2 × 106. The difference in retrotransposition frequency between the irradiated and unirradiated endonuclease-deficient L1s is not significant. A retrotransposition-incompetent L1 did not exhibit detectable retrotransposition with or without irradiation.
Genomic flanks of novel insertions have endonuclease-dependent features
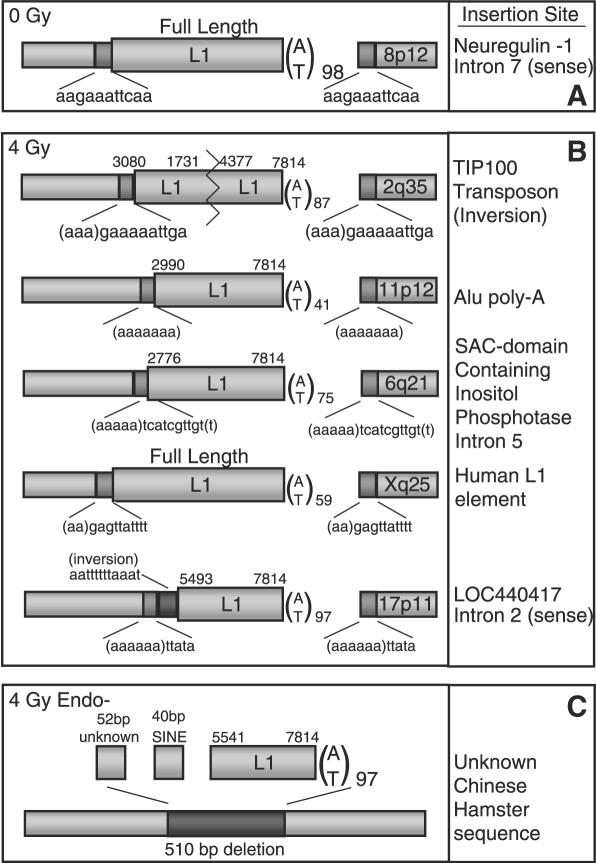
To confirm that L1 integration in the setting of gamma radiation occurs mainly via an endonuclease-dependent pathway, genomic sequences flanking L1 insertions were cloned. Irradiated cells that expressed EGFP were isolated by flow cytometry and expanded in tissue culture. Suppression PCR and inverse PCR were used to characterize the genomic flanks of new L1 insertions (see Materials and Methods). Typical endonuclease-dependent insertions have 7–20 bp target site duplications, an AT rich target site and a poly-A tail, as evidenced by an insertion cloned from an unirradiated 143B cell (Figure 5A) (36). In contrast, endonuclease-independent insertions lack these features and may instead contain large deletions, cDNA transduction or 3′ element truncation (13,14,24). Insertion flanks sequenced from irradiated cells had features consistent with endonuclease-dependent retrotransposition (Figure 5B). All had target site duplications, AT rich target sites, and three resided within older mobile elements.
Figure 5.
L1 insertions from irradiated clones have endonuclease-dependent features. Single L1-EGFP transfected 143B cells expressing EGFP were isolated by flow cytometry and expanded. DNA was extracted and the 3′ genomic flank amplified and sequenced by suppression PCR or inverse PCR (see Materials and Methods). The 5′ flank was then amplified and sequenced using primers designed from the human genome database. Hallmarks of endonuclease-dependent L1 insertion include 7–20 bp target site duplications (TSDs), AT rich consensus target sites and poly-A tails. Dark gray boxes denote TSDs. TSD sequences are displayed beneath each dark gray box. Numbers represent map positions in L1-EGFP; a full length insertion (including the spliced EGFP cassette) is 7814 bp long. Poly-A tail length is given as the subscripted number next to A/T. Chromosome insertion location is given in the 3′ flank. (A) L1 insertion flanks from an unirradiated cell. (B) Insertion flanks recovered following 4 Gy irradiation resemble most endonuclease-dependent genomic L1 insertions. (C) An endonuclease-deficient L1 insertion in a CHO-K1 cell has a deletion at the site of insertion, lacks target site duplications and has 5′ transduced sequence.
To ensure that the method of recovering L1 insertion flanks could recover endonuclease-independent insertions, an insertion from an endonuclease-deficient L1 was cloned. This insertion lacked target site duplications and had a large deletion at the site of insertion when compared with the corresponding genomic empty site (Figure 5C). In addition, SINE cDNA capture occurred at the 5′ end of this L1 insertion. SINE capture has been described previously for endonuclease-independent L1 insertions (23). These data suggest that, despite large numbers of gamma radiation-induced DNA breaks, L1 insertion usually occurs via an endonuclease-dependent pathway. Exposure to gamma radiation therefore probably changes the cellular environment in a manner that is favorable for endonuclease-dependent retrotransposition.
The analysis of L1 insertion flanks revealed some additional interesting features. A 5′ inversion, a feature common to genomic L1 insertions, was identified in one of the insertions (17). This inversion was unusually long, as were many of the insertions sequenced. Long insertions have been associated with highly active elements and may occur naturally in 143B cells. In addition, two of the insertions were in introns. Insertion of L1 sequences near or in genes has been noted to alter transcription, disrupt coding sequences and interfere with transcriptional elongation (5,37,38).
DISCUSSION
Gamma radiation increases L1 retrotransposition
L1 retrotransposition increases when cultured cells are subjected to gamma radiation. Using different assay conditions, gamma radiation increased L1 retrotransposition up to 4-fold. The increase is most evident in the presence of antibiotic selection for the L1-EGFP plasmid. L1 retrotransposition is also increased following irradiation without antibiotic selection. The lower retrotransposition frequencies in assays without antibiotic selection are due in part to measurement of EGFP at an earlier time point. The frequency of EGFP+ cells is known to increase for at least the first week of the assay (26). Antibiotic exposure may also help drive retrotransposition by increasing the copy number of the L1 plasmid or by acting as a stressor in concert with irradiation. The increase in L1 retrotransposition is proportional to the dose of gamma radiation in the 0–4 Gy range. These doses induce many DNA breaks per cell, as measured by staining for γH2AX foci. Increased retrotransposition following irradiation was observed in two different immortalized cell lines, 143B human osteosarcoma cells and CHO-K1 Chinese hamster ovary cells. These cell lines were chosen for this analysis because they were known to support high levels of L1 retrotransposition (24,39). Increased retrotransposition following irradiation in both cell types suggests that the increase in L1 activity is not a unique peculiarity of a single species or cell line.
Endonuclease-dependent retrotransposition likely predominates following gamma radiation
We considered whether gamma radiation promoted integration of L1 into DSBs. To address this possibility, we compared the retrotransposition frequency of an endonuclease-deficient L1 in irradiated and unirradiated cells. Despite inducing a significant number of γH2AX foci, no significant increase in endonuclease-deficient retrotransposition was observed. In addition, calicheamicin γ1, while creating comparable numbers of H2AX foci to gamma irradiation, did not increase L1 mobility.
The analysis of genomic sequences flanking insertions also favors the predominant use of an endonuclease-dependent pathway in irradiated cells. All five genomic flanks had target site duplications, used A/T rich target sites and had poly-A tails. In four out of the five insertions cloned from irradiated cells, the flanking DNA sequences were perfectly intact (data not shown). In contrast, the genomic DNA flanking an endonuclease-deficient L1 insertion harbored a deletion and lacked target site duplications. These data are consistent with an endonuclease-dependent pathway of L1 entry in irradiated cells. The deletions in the endonuclease-deficient L1 insertion are consistent with L1 entry into a DSB that is repaired by an error-prone form of NHEJ. However, one of the insertions in irradiated cells did have unusual features, including the loss of 3 nt at the target site and inversions of both the genomic flanking sequence and the L1 sequence. Specifically, 10 bases of the 5′ genomic flank were inverted, as well as 7 bases of the L1 proximal to the point of 5′ truncation. These features are similar to recently described L1 insertions containing palindromic sequences (14). This insertion may reveal unusual enzymatic properties of the L1 machinery or could be due to the minority of L1s entering into a DSB that is repaired by error-prone NHEJ. We also note that the absence of prominent deletions does not rule out the possibility that L1s may have inserted near DSBs (but outside of the sequenced region which in some clones extends several hundred bases). An alternative possibility is that many DSBs in irradiated 143B cells are repaired by a non-error-prone form of NHEJ, in which case a repaired break near the L1 insertion would not look different from the uninterrupted genomic flanking sequence.
Overall, the simplest interpretation of the endonuclease-deficient L1 experiment and the flanking sequence analysis is that L1 usually retrotransposes via an endonuclease-dependent pathway in irradiated cells. At first glance, these results may seem inconsistent with earlier studies that document the insertion of L1 elements into pre-formed DNA breaks. However, in those earlier studies L1 integration events into DNA breaks either conferred a selective advantage or were assayed using methods designed to specifically recover insertions into breaks (21–24). Interestingly, XRCC4 deficient cells support increased absolute numbers of endonuclease-independent insertions, while DNA-PKcs deficient cells have fewer insertions relative to parental lines (24). One possibility is that endonuclease-independent insertion is favored at the sites of persistently unrepaired breaks, but requires the assistance of early steps of the DNA repair cascade for targeting to breaks or resolution of integration. In contrast, in the present study, the cells had a largely intact DNA repair apparatus, so breaks were presumably repaired swiftly.
Endonuclease-dependent integration also predominates in published surveys of L1 insertions in the human genome and in tissue culture assays. An in silico survey of 399 human-specific L1 insertions identified only two with possible endonuclease-independent features (40). Because large deletions often accompany endonuclease-independent L1 insertions, it is possible that such insertions were selected against during evolution. However, the analysis of new L1 insertions in cultured cells also reveals a predominance of what appear to be endonuclease-dependent integration events (6,7,14). Thus, when faced with multiple DNA breaks, a cell appears to use conserved mechanism of DNA repair, if intact. L1 mediated DNA break repair appears to be infrequent compared to conventional, endonuclease-dependent retrotransposition.
Genotoxic agents and L1 mobility
That gamma radiation increases L1 retrotransposition has potential implications for individuals who have been exposed to radiation (41,42). Previous studies have documented L1's ability to insert into pre-existing DNA breaks (24), whereas the present study implies that most L1 insertions in cells subjected to gamma radiation occur via an endonuclease-dependent pathway. The L1 endonuclease is potentially highly damaging to genomic integrity, and, like gamma irradiation, produces a cellular response in the form of γH2AX foci (43).
Evidence from the literature suggests that gamma radiation is not unique in mobilizing L1. An increase in L1 retrotransposition has been associated with chronic exposure to the carcinogens mercury, cadmium and nickel, but not other heavy metals (44). In Drosophila, activation of L1-like retrotransposons (I-factors) can be enhanced by treating reactive females with gamma radiation or with inhibitors of nucleotide synthesis (45). In mouse cells exposed to etoposide, a human Alu SINE (thought to utilize L1 retrotransposition machinery) exhibited increased endonuclease-dependent retrotransposition (46).
It is possible that some forms of genotoxic stress cannot mobilize L1s, either because they cause different types of DNA damage or induce a different cellular response to the damage. This could make some forms of genotoxic therapy safer than others in terms of their risk of producing secondary genetic changes due to L1 mobilization. Alternatively or in addition, some genotoxic agents may cause toxicity or cell death before exerting a measurable influence on L1 retrotransposition. Given the potential toxicity of L1 mobilization, it seems reasonable to speculate that cells with a greater resistance to cell cycle check point arrest and apoptosis would be more likely to survive genotoxic stress and L1 mobilization. Unfortunately, the cells most likely to achieve this pernicious state of L1 and DNA damage induced genomic instability are probably the tumor cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Yang-Zhu Du, Andrew Fesnak and Peffin Lee for help with the inverse PCR and Janet Sallit and Sarah Fox for skilled technical assistance. We thank John Moran for the neomycin tagged L1.3 D205A endonuclease mutant and for many helpful discussions. We also thank the Flow Cytometry Core Facility and the Molecular Diagnosis and Genotyping Facility at the University of Pennsylvania. Calicheamicin γ1 was a gift from Wyeth Pharmaceuticals. Finally, we would like to thank the reviewers of this manuscript for their helpful suggestions and improvements to the manuscript. These studies were supported by NIH grants T32H10791, CA83977 and CA108812. S.H. was supported by T32 CA09140. Additional funds for this work were provided by the American Cancer Society (ACS-IRG-78-002-25). Funding to pay the Open Access publication charges for this article was provided by Dr Luning Prak.
Conflict of interest statement. None declared.
REFERENCES
- 1.Smit A.F., Toth G., Riggs A.D., Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 4.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazazian H.H., Jr Mobile elements and disease. Curr. Opin. Genet. Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert N., Lutz-Prigge S., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 7.Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 8.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 9.Cost G.J., Golding A., Schlissel M.S., Boeke J.D. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res. 2001;29:573–577. doi: 10.1093/nar/29.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen S.M., Eickbush T.H. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol. Cell Biol. 2005;25:6617–6628. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szak S.T., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052.1–research0052.18. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of l1 retrotransposition intermediates in cultured human cells. Mol. Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Q., Schumann G., Boeke J.D. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl Acad. Sci. USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin S.L., Bushman F.D. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostertag E.M., Kazazian H.H., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin S.L., Li W.L., Furano A.V., Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet. Genome Res. 2005;110:223–228. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 19.Zingler N., Willhoeft U., Brose H.P., Schoder V., Jahns T., Hanschmann K.M., Morrish T.A., Lower J., Schumann G.G. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 21.Yu X., Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 22.Teng S.C., Kim B., Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y., Waldman A.S. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 25.Eickbush T.H. Repair by retrotransposition. Nature Genet. 2002;31:126–127. doi: 10.1038/ng897. [DOI] [PubMed] [Google Scholar]
- 26.Ostertag E.M., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prak E.T., Dodson A.W., Farkash E.A., Kazazian H.H., Jr Tracking an embryonic L1 retrotransposition event. Proc. Natl Acad. Sci. USA. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badge R.M., Alisch R.S., Moran J.V. ATLAS: a system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W.M., Chu W.M., Choudary P.V., Schmid C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elmroth K., Nygren J., Martensson S., Ismail I.H., Hammarsten O. Cleavage of cellular DNA by calicheamicin gamma1. DNA Repair (Amst) 2003;2:363–374. doi: 10.1016/s1568-7864(02)00235-5. [DOI] [PubMed] [Google Scholar]
- 31.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 32.Rogakou E.P., Boon C., Redon C., Bonner W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farley A.H., Luning Prak E.T., Kazazian H.H., Jr More active human L1 retrotransposons produce longer insertions. Nucleic Acids Res. 2004;32:502–510. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dombroski B.A., Scott A.F., Kazazian H.H., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl Acad. Sci. USA. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimberland M.L., Divoky V., Prchal J., Schwahn U., Berger W., Kazazian H.H., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 36.Prak E.T., Kazazian H.H., Jr Mobile elements and the human genome. Nature Rev. Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- 37.Han J.S., Szak S.T., Boeke J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 38.Han J.S., Boeke J.D. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 39.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers J.S., Vincent B.J., Udall H., Watkins W.S., Morrish T.A., Kilroy G.E., Swergold G.D., Henke J., Henke L., Moran J.V., et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sankaranarayanan K. Transposable genetic elements, spontaneous mutations and the doubling-dose method of radiation genetic risk evaluation in man. Mutat. Res. 1986;160:73–86. doi: 10.1016/0027-5107(86)90031-x. [DOI] [PubMed] [Google Scholar]
- 42.Hagan C.R., Rudin C.M. Mobile genetic element activation and genotoxic cancer therapy: potential clinical implications. Am. J. Pharmacogenomics. 2002;2:25–35. doi: 10.2165/00129785-200202010-00003. [DOI] [PubMed] [Google Scholar]
- 43.Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.01.089. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kale S.P., Moore L., Deininger P.L., Roy-Engel A.M. Heavy metals stimulate human L1 retrotransposition. Int. J. Environ. Res. Public Health. 2005;2:14–23. doi: 10.3390/ijerph2005010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bregliano J.C., Laurencon A., Degroote F. Evidence for an inducible repair-recombination system in the female germ line of Drosophila melanogaster. I. Induction by inhibitors of nucleotide synthesis and by gamma rays. Genetics. 1995;141:571–578. doi: 10.1093/genetics/141.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagan C.R., Sheffield R.F., Rudin C.M. Human Alu element retrotransposition induced by genotoxic stress. Nature Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.