Abstract
The bacterium Proteus mirabilis expresses a cytosolic class beta glutathione S-transferase (PmGST B1-1) that is part of a family of multifunctional detoxication enzymes. Like other cytosolic GSTs, PmGST B1-1 possesses two local structural motifs, an N-capping box and a hydrophobic staple motif, both of which are located between amino acids 151 and 156. The N-capping box consists of a reciprocal hydrogen bonding interaction of Thr152 with Asp155, whereas the hydrophobic staple motif consists of a hydrophobic interaction between Phe151 and Ala156. By contrast with other GSTs, PmGST B1-1 displays distinct hydrogen bond interactions in the N-capping box. In mammalian GSTs these structural elements are critical for protein folding and stability. To investigate the role played by these two motifs in a distantly related organism on the evolutionary scale, site-directed mutagenesis was used to generate several mutants of both motifs in PmGST B1-1. All mutants were efficiently overexpressed and purified, but they were quite unstable, although at different levels, indicating that protein folding was significantly destabilized. The analysis of the T152A and D155G variants indicated that the N-capping box motif plays an important role in the stability and correct folding of the enzyme. The analysis of F151A and A156G mutants revealed that the hydrophobic staple motif influences the structural maintenance of the protein and is implicated in the folding process of PmGST B1-1. Finally, the replacement of Thr152 and Asp155, as well as Phe151 and Ala156 residues influences the catalytic efficiency of the enzyme.
Keywords: N-capping box, hydrophobic staple motif, Proteus mirabilis GST B1-1 (PmGST B1-1), site-directed mutagenesis, bacterial glutathione S-transferase (GST)
Abbreviations: CDNB, 1-chloro-2,4-dinitrobenzene; DTT, dithiothreitol; GST, glutathione S-transferase; NCBI, National Center for Biotechnology Information; PmGST B1-1, Proteus mirabilis GST B1-1; H-bond, hydrogen bond
INTRODUCTION
The superfamily of GSTs (glutathione transferases; EC 2.5.1.18) is composed of a large group of enzymes, the common feature of which is to catalyse the nucleophilic attack of GSH on the electrophilic groups of a wide range of hydrophobic toxic compounds [1–5]. GSTs are also involved in other cell functions, such as peroxidase and isomerase activities, and are capable of binding a large number of endogenous and exogenous compounds noncatalytically [1–4]. GSTs are divided into at least three major families of proteins, namely cytosolic, mitochondrial and microsomal GSTs [5]. The cytosolic GSTs have been sub-grouped into several divergent classes on the basis of their physical, chemical, immunological and structural properties [5–6]. Despite their low inter-class sequence identity (often less than 20%), crystallographic studies have shown that the overall GST fold is conserved among the different classes [3,6]. The cytosolic GSTs are dimeric proteins with each subunit divided in to two domains. The N-terminal domain adopts an α/β topology similar to the fold of thioredoxins, whereas the C-terminal domain is completely α helical. The active site of GSTs consists of two binding sites: the G-site where GSH binds and the H-site where hydrophobic electrophiles bind [3,6].
It has previously been shown that at the N-terminal region of the α6-helix, two local structural motifs, an N-capping box (S/TXXD) and a hydrophobic staple motif, are strictly conserved in the cytosolic GSTs [7–10]. The N-capping box is characterized by a reciprocal backbone–side-chain hydrogen bond interaction between the N-cap (Ser/Thr) and N3 (Glu/Asp) regions. Previous studies indicated that this motif is involved in the formation and stability of α-helices [11,12]. The hydrophobic staple motif consists of a specific hydrophobic interaction between a residue located within the α6-helix (N4) and a residue adjacent to the N-cap (N′) [13]. It has been shown that when this motif is combined with a capping box it produces a co-operative effect in defining the α-helix starting point [13]. The nomenclature used for helices and their flanking residues has been proposed by Richardson and Richardson [14] and it is as follows: ….-N″-N′-Ncap-N1-N2-N3-N4-….. Recent mutagenesis studies on two members of mammalian cytosolic GSTs, human hGSTP1-1 and hGSTA1-1, demonstrated that these two local structural motifs have a critical role in protein folding and stability [8–10]. In fact, the removal of the capping residues greatly destabilizes the final structure of the two cytosolic GSTs, as well as their folding pathway [8,10]. It has also been observed that in mammalian GSTs the hydrophobic staple motif represents an evolutionarily conserved determinant for rapid folding of the enzyme [9,10].
In the present work, in order to verify the common role played by the residues forming these two structural motifs a distant organism in the evolutionary scale has been used. Our model is the bacterial GST from Proteus mirabilis (PmGST B1-1) a member of the cytosolic beta class GSTs [15–23]. PmGST B1-1 is characterized by the presence of a GSH molecule covalently bound to Cys10 per subunit, although the enzyme has GSH-conjugating activity [19,24]. Furthermore, PmGST B1-1 is also able to perform redox activity with comparable efficiency [25]. Recent results suggest that this bacterial enzyme has an active role in protection against oxidative stress generated by hydrogen peroxide [26]. A possible role for this enzyme in the detoxification of antimicrobial agents has been suggested previously [17,26,27].
In PmGST B1-1 the N-capping box consists of a Thr-Xaa-Xaa-Asp motif, whereas a phenylalanine and an alanine residue constitute the hydrophobic staple motif (Figure 1). To examine the role of these conserved residues in bacterial GSTs they have been replaced by site-directed mutagenesis, and the effect of these variations has been examined. The results obtained support the idea that these residues were conserved during evolution because of their involvement in the folding and stability of cytosolic GSTs.
Figure 1. Comparison of the amino acid sequence of PmGST B1-1 with representative eukaryotic and bacterial members of the cytosolic class GST.
Sequences were aligned manually. The position of the first residue shown relative to the full-length protein is indicated to the left of each sequence. Sequences were retrieved from the NCBI data bases. The N-capping box residues are shown in grey boxes. The hydrophobic staple motif residues are shown in black boxes. The glycine residue located four residues before the N-cap is shown in grey. The nomenclature is N″-N′-Ncap-N1-N2-N3-N4 as proposed by Richardson and Richardson [14].
EXPERIMENTAL
Site-directed mutagenesis, expression and purification of wild-type and mutant PmGST B1-1 enzymes
The DNA encoding PmGST B1-1 in pBtac1 (pGPT1) [17] was used as a template in the mutagenesis procedure. The following forward oligonucleotides were used for the mutations: T152A, 5′-GTGGTGATCACTTTGCTGTGGCGGATGC-3′; D155A, 5′-CTTTACTGTGGCGGCTGCGTATCTGTTT-3′; D155G, 5′-CTTTACTGTGGCGGGTGCGTATCTGTTT-3′; F151A, 5′-GTTTGTGGTGATCACGCTACTGTGGCGGATG-3′; A156G, 5′-ACTGTGGCGGATGGGTATCTGTTTACG-3′; T152A/D155A, 5′-G-TGGTGATCACTTTGCTGTGGCGGCTGCGTATC-3′; F151A/A156G, 5′-GATCACGCTACTGTGGCGGATGGGTATCTGTTT-3′. The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used according to the manufacturer's instructions. Clones with the required mutation were confirmed by DNA sequencing [28]. To induce gene transcription, IPTG (isopropyl-β-D-thiogalactopyranoside) (Sigma-Aldrich, Milano, Italy) was added to a final concentration of 1 mM when Escherichia coli XL1-Blue strains, grown at 37 °C in Luria–Bertani medium [29] and supplemented with tetracycline (Sigma-Aldrich) and ampicillin (Sigma-Aldrich), reached an approximate A550 of 0.4. The incubation time was prolonged for a further 16 h after switching the temperature to 25 °C. The purification of enzymes was performed as reported previously [15]. Briefly, the bacterial cells were collected by centrifugation, washed twice and resuspended in 10 mM potassium phosphate buffer (pH 7.0) containing 1 mM EDTA (buffer A) and 2 mM dithiothreitol and disrupted by cold sonication. The particulate material was removed by centrifugation and the resulting supernatant was loaded on to a column (10×1 cm internal diameter) packed with glutathione-Sepharose 6B (Sigma–Aldrich) that was pre-equilibrated with buffer A [30]. The column was exhaustively washed with buffer A supplemented with 50 mM KCl. The enzyme was eluted with 50 mM Tris/HCl buffer (pH 9.6), containing 5 mM GSH. The fractions containing GST activity were pooled, concentrated, dialysed against buffer A by ultrafiltration and subjected to further analyses. SDS/PAGE in a discontinuous slab gel was performed by the method of Laemmli [31]. Protein concentration was determined by the method of Bradford [32] with γ-globulin as standard.
Enzyme assays
GST activity with CDNB (1-chloro-2,4-dinitrobenzene) was assayed at 30 °C according to the method of Habig and Jakoby [33]. For the enzyme kinetic determinations either CDNB or GSH was held constant at 1 and 5 mM respectively, whilst the concentration of the other substrate was varied (0.1–5 mM for GSH and 0.1–1.6 mM for CDNB). Each initial velocity was measured at least in triplicate. The KaleidaGraph Software package (Synergy Software, Reading, PA, U.S.A.) was used to estimate the Michaelis constant (Km) and Vmax values.
To study refolding, all mutants (7 μM), as well as wild-type enzyme were first incubated for 30 min at 25, 37 and 40 °C in 0.1 M potassium phosphate buffer (pH 6.5) containing 1 mM EDTA and 5 mM DTT (dithiothreitol) with 4 M GdmCl. At the end of the incubation period, denatured proteins were rapidly diluted 40-fold in 0.1 M potassium phosphate buffer (pH 6.5) containing 1 mM EDTA and 5 mM DTT at the same temperature of denaturation. The residual concentration of GdnHCl was 0.1 M. Aliquots of the refolding reaction mixture were taken at different times and assayed for enzymatic activity. Thermal stability measurements of mutant enzymes (0.7 μM) were carried out by incubating the samples at each temperature for 15 min. GST activity was determined at the end of the incubation.
Spectroscopic studies
Fluorescence measurements were performed on a Spex spectrofluorimeter (model Fluoromax) equipped with a thermostatically controlled sample holder at 25 °C. The enzyme samples were in buffer A. The intrinsic fluorescence spectra of the proteins (excitation at 280 nm) were recorded in 1 nm wavelength increments and the signal for 1 s was acquired at each wavelength. Spectra were corrected by subtraction of the corresponding spectra for blank samples.
CD spectra in the far-UV region from 190–250 nm were acquired with a Jasco-810 spectropolarimeter (Jasco Europe, Cremella, Lecco, Italy). Samples containing wild-type and mutant enzymes at a concentration of 17 μM were scanned at least five times at a rate of 20 nm/min and then averaged. The temperature of the sample compartment was maintained at 20 °C using a circulating-water bath. The cuvette used had a light path of 0.1 cm. Each spectrum was corrected by subtracting the corresponding spectrum for a blank sample.
Database searches and molecular modelling
GST sequences were retrieved from the NCBI (National Center for Biotechnology Information) data bases (www.ncbi.nlm.nih.gov/Entrez/index). Local similarity analysis of sequences was performed using the Pattern Hit Initiated BLAST (PHI-BLAST) method [34] at Institut Pasteur, Paris (http://bioweb.pasteur.fr/seqanal/interfaces/phiblast.html). The structure of PmGST B1-1 was rendered using PyMOL [35]. Co-ordinates of PmGST B1-1 are available from the Protein Data Bank (PDB code 1PMT).
RESULTS AND DISCUSSION
Analysis of PmGST B1-1 structure
Figure 1 shows the alignment of representative cytosolic GSTs highlighting the two conserved structural motifs h-S/T-X-X-D-h (where h is a hydrophobic residue and X is a non-conserved residue) in all cytosolic classes from mammals to bacteria. A glycine residue located four amino acids before the N-cap residue is also conserved. This residue is part of a buried local sequence: G-X-X-h-S/T-X-X-D-h that is maintained in all GSTs and adopts a high energy stereochemistry allowed in glycines only [19,36]. Previous studies showed the presence of this conserved module in 115 GST sequences [36]. Our PHI-BLAST analysis [34] using the sequence of PmGST B1-1 and the pattern G-X-X-h-[ST]-X-X-D-h revealed that the conserved module could be extended to more than two hundred sequences of cytosolic GSTs with stringent E-values of 0.001.
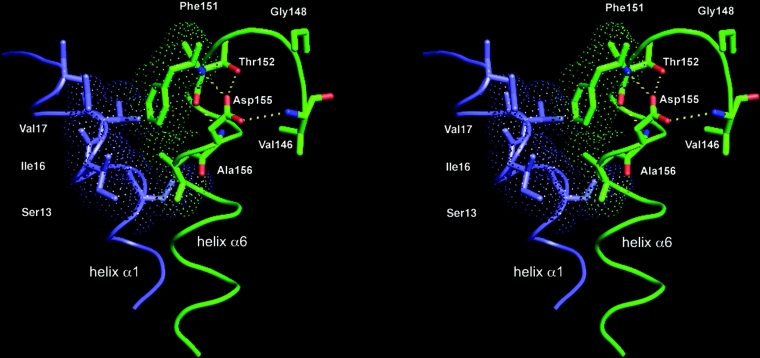
In contrast with other cytosolic GSTs, Tau and Lambda classes show different patterns. In Tau class GSTs, at the N-cap position the Ser/Thr residue is replaced by a glycine residue [37]. This is in agreement with earlier studies where it was observed that glycine residues occur frequently at the N-cap [12,38]. Furthermore, in lambda class GSTs the conserved glycine is located three residues before the N-cap [39]. The analysis of the structure of PmGST B1-1 revealed different H-bond interactions for the N-capping box (Figure 2). Commonly, in an N-capping box, the side chain of N3 forms an H-bond with the backbone of N-cap and, reciprocally, the side chain of N-cap forms an H-bond with the backbone of N3 [11]. Conversely, in PmGST B1-1 whereas the side chain of N3 (Asp) forms an H-bond with the backbone of N-cap (Thr), the backbone of N-cap forms an H-bond with the backbone of N3. Doig and co-workers observed that this backbone-to-backbone H-bond network is very common and it constitutes an alternative hydrogen bonding pattern present in 30% of the proteins analysed with Asp/Ser/Thr as N-cap residues [38]. An additional H-bond between the side-chain oxydril group of Thr152 and the side chain carboxyl group of Asp155 was also seen. It probably participates in stabilizing the α6-helix. Furthermore, the structural analysis of PmGST B1-1 showed that the side chain of Asp155 forms a H-bond with the backbone amide nitrogen of Val146, positioned in the loop preceding the α6-helix (Figure 2). Interestingly this interaction has also been observed in several other GST classes [8,10,40]. This residue and its equivalents in the other classes appear to have an important role in the structural stability of the protein [8,10,40]. The second motif present in PmGST B1-1 corresponds to a specific interaction between the residues Phe151 (N′) and Ala156 (N4) and possesses all the structural characteristics of a hydrophobic staple [13]. As shown in Figure 2, the Ala156 side-chain makes a hydrophobic interaction with the aromatic ring of Phe151. The two hydrophobic residues are located within a distance of 4 Å (1 Å=0.1 nm) from each other and adopt dihedral angles in the β-region of the Ramachandran plot. Additionally, Ala156 forms favourable hydrophobic contacts with Ser13, Ile16 and Val17, three residues of the α1-helix that are an important structural element supporting the active site [19].
Figure 2. Stereoscopic view of the PmGST B1-1 region with residues that form the N-capping box and the hydrophobic staple motif.
α1-helix (in blue) and α6-helix (in green) are shown in loop representation and key residues are shown as sticks. Thr152 and Asp155 correspond to the N-cap and N3 residues, respectively. Broken lines indicate hydrogen bonds (in yellow). Oxygen and nitrogen atoms are indicated in red and blue respectively. Hydrophobic interactions formed by the Phe151 (N′) and Ala156 (N4) residues, as well as between Ala156 and α1-helix residues are emphasized by the Connolly surface representation (dot frame).
Enzyme expression and purification
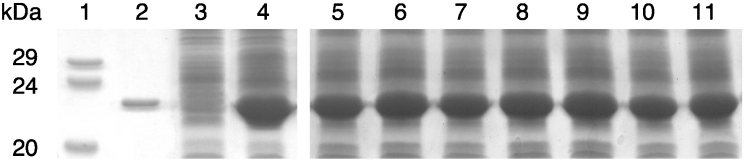
The residues of the N-capping box and the hydrophobic staple motif were replaced with alanine. The Asp155 residue was also converted into glycine. The alanine residue in position 156 was substituted with a glycine residue that does not contribute to the formation of hydrophobic interactions. Two double mutants, T152A/D155A and F151A/A156G, were also obtained. Previous studies showed that in mammalian GSTs the N-cap and hydrophobic staple mutations resulted in temperature-sensitive species, consequently these variants were expressed at the permissive temperature of 25 °C [8–10]. Similar results were obtained with PmGST B1-1 mutants. When growing the bacteria at 30 and 37 °C the production of all single and double mutant enzymes was lower than at 25 °C, showing the influence of temperature on the protein yield for all variants (results not shown). Therefore protein expression of all mutants, as well as wild-type enzymes was carried out at 25 °C. In Figure 3 the total cellular extracts from induced E. coli XL1-Blue cells examined by SDS/PAGE are shown. All mutants were efficiently overexpressed and they co-migrated with the wild-type protein with comparable levels of expression. In the cytosol, very low levels of enzymatic activity were detected with CDNB as the substrate for all mutants (Table 1). In fact, the A156G variant, as well as double mutants showed only 1% specific activity compared with wild-type. Moreover, D155A and D155G showed 3 and 6% of the activity of the wild-type GST respectively. Higher levels of enzymatic activity were obtained using T152A (26%) and F151A (20%). The enzymes were purified to homogeneity by glutathione-affinity chromatography. In each case, neither the flow-through nor the washed material showed any GST activity, indicating that mutation of these residues did not influence the binding of the variant enzymes to the affinity column. The electrophoretic mobility and apparent molecular mass, as well as the immunological properties [15] of the purified variants, were indistinguishable from those of the wild-type enzyme (results not shown). The D155A, D155A/T152A and F151A/A156G mutants were quite unstable and occurred in very low abundance. In fact, the purified enzymes showed low activity which was lost upon overnight storage at −20 °C. Chemical cellular disruption, as well as the addition of 10% glycerol did not lead to an increase and/or preservation of enzymatic activity. Thus the purified enzymes could not be extensively studied. The weak stability of these mutants is clearly a consequence of the structural instability of the enzyme. Although the yields of T152A, D155G, F151A and A156G mutants were very low, these variants could be used for further analysis.
Figure 3. SDS/PAGE of total-cellular extracts from wild-type and mutant enzymes.
Proteins were detected by Coomassie Brilliant Blue R-250 staining. Lane 1, molecular mass markers; lane 2, purified wild-type enzyme; lane 3, total cellular extract of non-transformed E. coli XL1Blue; lane 4, wild-type enzyme; lane 5, D155A; lane 6, D155G; lane7, T152A; lane 8, F151A; lane 9, A156G; lane 10, D155A/T152A; lane 11, F151A/A156G.
Table 1. Specific activity and kinetic constants for wild-type and mutant enzymes with CDNB as the second substrate.
Protein yield represents the fraction of GST recovered after affinity chromatography given as a percentage of total cytosolic proteins. ND, not detectable as reported in the text. The data are the means for at least three independent determinations; S.D. values do not exceed 10% of the means.
| Specific activity (μmol·min−1·mg−1) | GSH | CDNB | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Cytosol | Affinity chromatography | Protein yield (%) | Km (mM) | kcat (s−1) | kcat/Km (mM−1·s−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1·s−1) |
| Wild-type | 0.3 | 1.09 | 15.50 | 0.68 | 0.93 | 1.36 | 0.73 | 1.18 | 1.62 |
| T152A | 0.08 | 0.79 | 0.89 | 0.72 | 0.88 | 1.22 | 3.55 | 5.15 | 1.45 |
| D155A | 0.02 | ND | ND | ||||||
| D155G | 0.01 | 0.60 | 0.61 | 4.63 | 0.37 | 0.08 | 1.70 | 0.33 | 0.19 |
| F151A | 0.06 | 0.71 | 3.72 | 0.63 | 1.03 | 1.63 | 2.16 | 3.41 | 1.57 |
| A156G | 0.002 | 0.33 | 0.09 | 2.53 | 0.51 | 0.20 | 1.14 | 0.81 | 0.71 |
| D155A/T152A | 0.002 | ND | ND | ||||||
| F151A/A156G | 0.003 | ND | ND | ||||||
Enzymatic studies
In Table 1, the specific activities and kinetic constants of the N-capping and hydrophobic staple mutant enzymes in comparison with the wild-type enzyme using GSH and CDNB as substrates are shown. The replacement of Thr152, Asp155 or Phe151 with alanine resulted in a 30–50% decrease in activity. Even lower activity was observed for A156G, with a decrease of approx. 70%. The substitution of Phe151 and Thr152 with alanine does not affect the kinetic parameters for GSH when compared with the wild-type enzyme. Instead, D155G exhibited an increased KmGSH value of approx. 7-fold coupled with a decreased catalytic efficiency of 17-fold relative to the wild-type enzyme. A similar trend was observed for A156G, with an increased KmGSH value of approx. 4-fold and a decreased catalytic efficiency of 7-fold. Thus the effect of mutations on the kinetics for CDNB was also reported. The F151A and T152A mutants showed comparable kinetic constants for CDNB. In fact, both mutants had KmCDNB and kcat values approx. 3- and 4-fold higher, whereas no difference in the kcat/Km values was observed when compared with the wild-type enzyme. However, unlike the other two mutants, D155G and A156G showed similarly higher KmCDNB values of approx. 2-fold accompanied by a drastic reduction in the catalytic activities of approx. 2- and 8-fold respectively, as compared with the wild-type enzyme. Taken together, these data indicate that the mutation of the Thr152 and Phe151 residues resulted in a moderate change in kinetic parameters. On the other hand, the replacement of Asp155 and Ala156 has a greater influence on the catalytic properties of the enzyme. The effects on the catalytic efficiency for GSH and CDNB may be explained by local structural perturbations of the residues involved in the active site, as a consequence of the mutations.
Structural studies
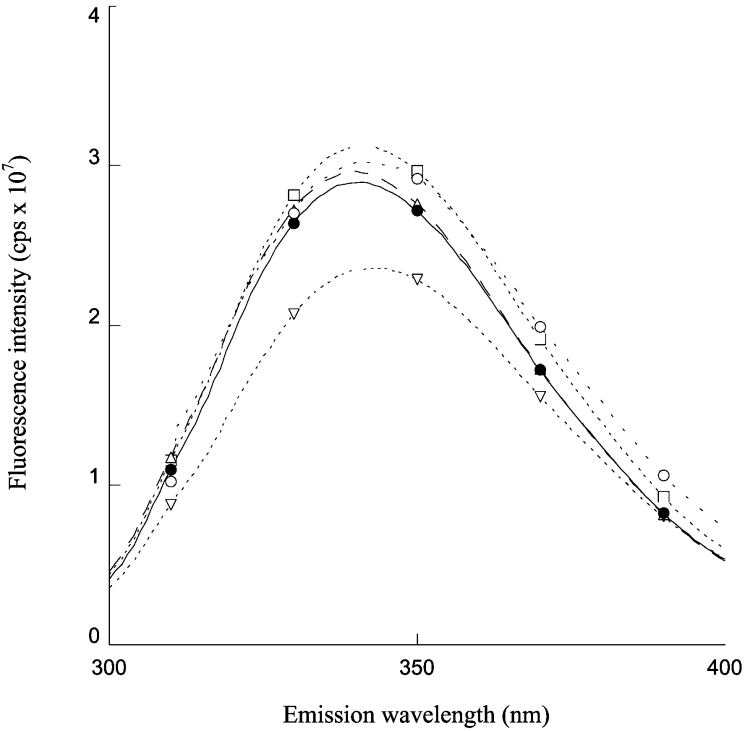
Spectroscopic properties of the mutant enzymes were analysed. The structure of the four mutants in comparison with wild-type enzyme was monitored by far-UV CD spectroscopy. All mutants possessed very similar spectra to that of the wild-type enzyme, indicating that the amino acid substitutions had no effect on the secondary structure and the dimeric arrangement of the GST variants (results not shown). In order to obtain information on the tertiary structure of the variants, fluorescence emission spectra were also measured. The spectra were obtained after excitation at 280 nm. The fluorescence spectra of F151A, T152A and A156G mutants had analogous fluorescence intensity to the wild-type enzyme, with a maximum at 340 nm (Figure 4). A different result was obtained for the D155G mutant. In fact, a decrease of approx. 20% in the fluorescence intensity for this mutant was observed. In addition, the emission maximum of D155G was red-shifted by approx. 4 nm, suggesting that slight conformational changes occur in the mutant enzyme.
Figure 4. Fluorescence emission spectra of wild-type and mutant enzymes.
Wild-type (●), D155G (▽), T152A (△), A156G (○) and F151A (□). Protein concentration was 3 μM in 10 mM phosphate buffer (pH 7.0) containing 1 mM EDTA. Excitation was at 280 nm.
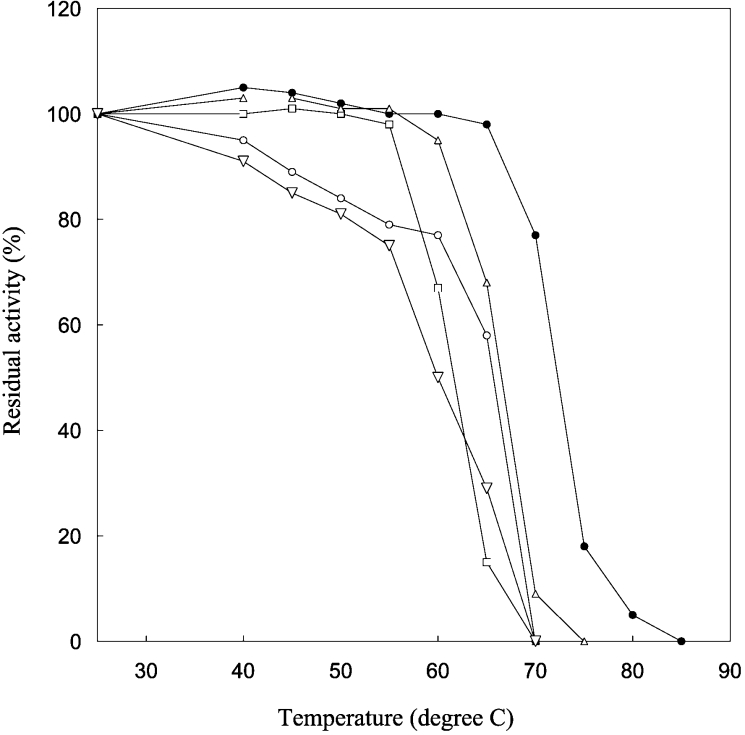
The thermal stability of each of the four mutant enzymes was then examined. PmGST B1-1 is a very thermostable enzyme, retaining more than 80% of its activity after 15 min of incubation in 10 mM potassium phosphate buffer (pH 7.0) at 70 °C [23]. All mutant enzymes were less stable than wild-type enzyme upon incubation at various temperatures between 25 and 85 °C, indicating that both N-cap and hydrophobic staple residues have a crucial role in the overall stability of the enzyme (Figure 5). F151A and T152A mutants lost 50% of their activity at 60 and 65 °C respectively and were completely inactivated at 70 °C. Furthermore, D155G and A156G mutants were more unstable than the wild-type enzyme. In fact, Figure 5 shows that after incubation at 50 °C, both mutants were already inactivated by approx. 20% and were completely inactivated at 70 °C. These results are consistent with previous kinetic data and suggest that each single residue substitution produces significant effects on the active site although it is located far from these residues.
Figure 5. Effects of temperature on the stability of wild-type and mutants.
The enzyme activity at 25 °C was taken as 100%. Wild-type (●), D155G (▽), T152A (△), A156G (○) and F151A (□).
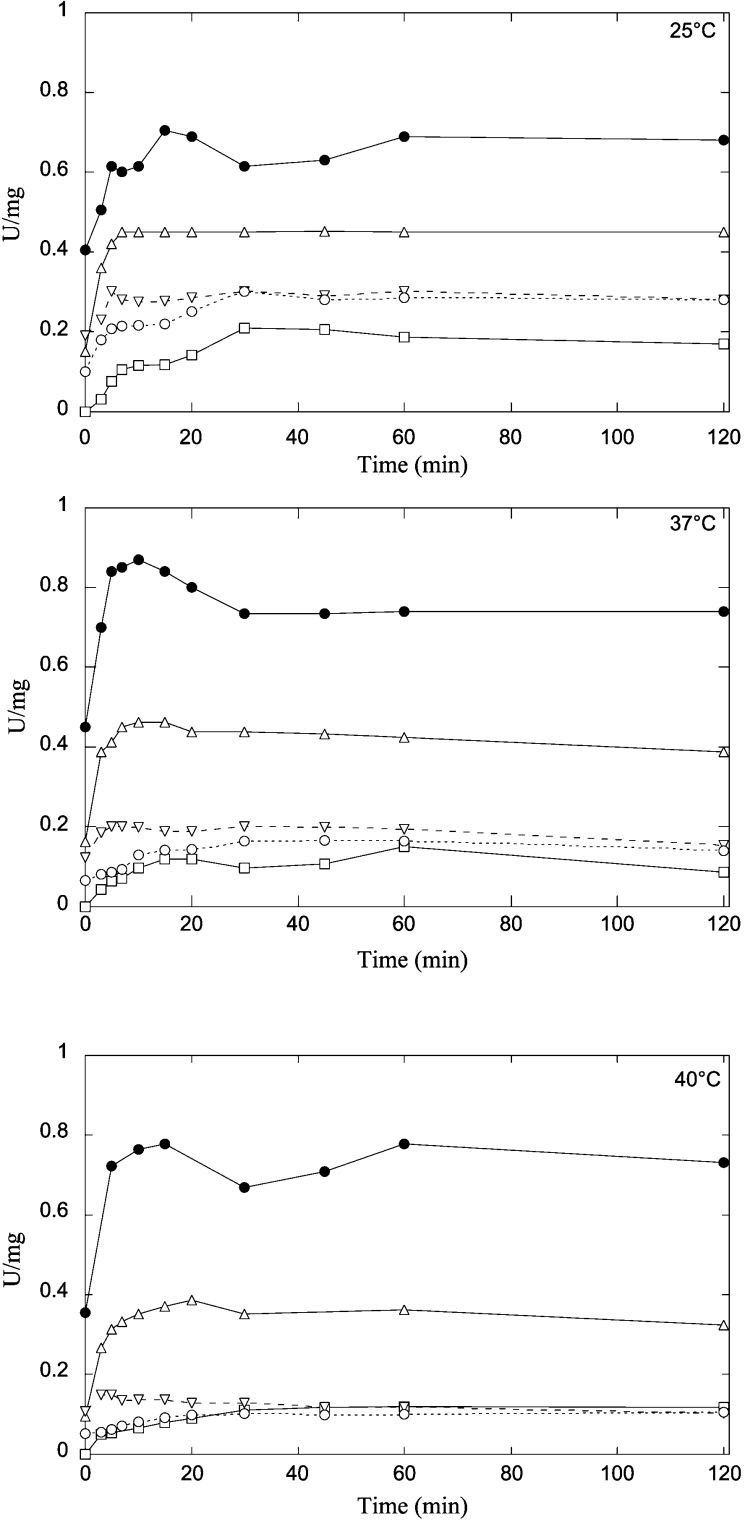
In order to further investigate the role of these residues in maintaining the structure of the protein fold, the reactivation of wild-type and mutants has been studied after unfolding in 4 M GdnHCl, and refolding by dilution. Considering that mutation of N-cap and hydrophobic staple residues in mammalian GSTs produced temperature-sensitive fold-mutants that showed low or no efficiency in refolding [8–10], our experiments were performed at three different temperatures. In 4 M GdnHCl all mutants, as well as the wild-type protein were completely unfolded, as monitored by far-UV CD spectroscopy (results not shown). The refolding rates of N-cap mutants were very similar to wild-type enzyme. As can be seen in Figure 6, wild-type and mutant enzymes reached a maximum of reactivation in a few minutes. Different trends were observed for the reactivation yields of the N-cap mutants when compared with the wild-type enzyme (Figure 6). The reactivation yields of the wild-type enzyme were similar at 25, 37 and 40 °C, indicating that the enzyme was unaltered by temperature. On the contrary, the reactivation yield of D155G and T152A mutants was decreased progressively by increasing the temperature towards physiological values. In particular, the D155G mutant displayed lower refolding yields than the T152A mutant. Thus this motif influences the refolding kinetics of PmGST B1-1 suggesting that N-cap residues are critical for proper folding of the enzyme. In Figure 6 the refolding kinetics of hydrophobic staple motif mutants are also shown. Unlike N-cap mutants, the refolding rates of F151A and A156G mutants reached the maximum of reactivation more slowly than the wild-type enzyme revealing a loss of stability in the protein. This suggests that a single replacement of these two residues affects the refolding process, accelerating the rate of enzyme reactivation. Furthermore, the reactivation yield of both hydrophobic staple mutants was significantly lower than that of wild-type enzyme and it decreased gradually with increasing temperatures from 25 to 40 °C.
Figure 6. Refolding kinetics of wild-type (●) and D155G (▽), T152A (△), A156G (○) and F151A (□) mutants monitored by changes in enzymatic activity at different temperatures.
All enzymes were unfolded in 4 M GdnHCl in 0.1 M potassium phosphate buffer, 1 mM EDTA and 5 mM DTT (pH 6.5) for 30 min. Refolding was initiated by rapid dilution in the same buffer. The residual concentration of GdnHCl was 0.1 M. Aliquots of the refolding reaction mixture were taken at different times and assayed for GST activity.
Conclusions
The present results extend previous data regarding the role of two conserved motifs in mammalian GSTs. Although significant structural and kinetic differences have been found between mammalian and bacterial GSTs [15–19,24,25], the N-capping box and the hydrophobic staple motifs are conserved in PmGST B1-1 and play a role in protein stability and refolding similar to that found in mammalian GSTs.
Analysis of the N-cap mutants revealed that both variant enzymes were more thermolabile than the wild-type enzyme. The loss of the H-bond interactions that characterize the N-capping box is the most obvious reason for the decreased stability of the mutant enzymes. The kinetic analysis of the mutant enzymes also revealed that the active site is directly influenced by this structural perturbation. As previously observed, these two residues destabilize the α6-helix, which makes several inter-domain contacts with the α1-helix, a structural element of the active site [19]. Thus although the N-cap residues are located in the interior of the molecule their replacement affects the catalytic efficiency of PmGST B1-1. Our studies also demonstrate that the N-capping box contributes to the folding process. In fact, both Thr152 and, to a greater extent, Asp155 exhibited a very low refolding yield at physiological temperatures.
The replacement of residues Phe151 and Ala156 that form the hydrophobic staple motif, also affected the stability of the protein. In particular, the A156G mutation resulted in a greater destabilization of the protein, reflecting the loss of hydrophobic interactions. The hydrophobic staple variants also displayed different kinetic properties when compared with the wild-type enzyme. In particular, the replacement of Ala156 with glycine considerably decreased the enzymatic activity. This drastic reduction can be explained by the loss of the hydrophobic contacts between Ala156 and the α1-helix suggesting that these interactions play an important role in maintaining the infra-structure of PmGST B1-1 and consequently for the overall stability of the enzyme. The analysis of refolding kinetics showed that both hydrophobic staple residues contribute to stabilizing the PmGST B1-1 structure, as highlighted by the effect that their replacement has on the refolding process.
The GST C-terminal domain, which constitutes many of the residues of the H-site, varies widely between classes, explaining the different substrate specificities. Therefore, the evolutionary conservation in the C-terminal domain of the N-capping box and the hydrophobic staple motif is remarkable and appears to be the result of a selective pressure for preserving both the structure and the function of cytosolic GSTs.
Acknowledgments
This work was partially supported by grants from Ministero dell' Istruzione dell' Universita' e della Ricerca.
References
- 1.Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 2.Hayes J. D., McLellan L. I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Rad. Res. 1999;31:273–300. doi: 10.1080/10715769900300851. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong R. N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 4.Mannervik B., Danielson U. H. Glutathione transferases: structure and catalytic activity. Crit. Rev. Biochem. Mol. Biol. 1988;23:283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- 5.Hayes J. D., Flanagan J. U., Jowsey I. R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 6.Sheehan D., Meade G., Foley V. M., Dowd C. A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aceto A., Dragani B., Melino S., Allocati N., Masulli M., Di Ilio C., Petruzzelli R. Identification of an N-capping box that affects the α6-helix propensity in glutathione S-transferase superfamily proteins: a role for an invariant aspartic residue. Biochem. J. 1997;322:229–234. doi: 10.1042/bj3220229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragani B., Stenberg G., Melino S., Petruzzelli R., Mannervik B., Aceto A. The conserved N-capping box in the hydrophobic core of glutathione S-transferase P1-1 is essential for refolding. Identification of a buried and conserved hydrogen bond important for protein stability. J. Biol. Chem. 1997;272:25518–25523. doi: 10.1074/jbc.272.41.25518. [DOI] [PubMed] [Google Scholar]
- 9.Stenberg G., Dragani B., Cocco R., Mannervik B., Aceto A. A conserved ‘hydrophobic staple motif’ plays a crucial role in the refolding of human glutathione transferase P1-1. J. Biol. Chem. 2000;275:10421–10428. doi: 10.1074/jbc.275.14.10421. [DOI] [PubMed] [Google Scholar]
- 10.Cocco R., Stenberg G., Dragani B., Rossi Principe D., Paludi D., Mannervik B., Aceto A. The folding and stability of human alpha class glutathione transferase A1-1 depend on distinct roles of a conserved N-capping box and hydrophobic staple motif. J. Biol. Chem. 2001;276:32177–32183. doi: 10.1074/jbc.M104057200. [DOI] [PubMed] [Google Scholar]
- 11.Harper E. T., Rose G. D. Helix stop signals in proteins and peptides: the capping box. Biochemistry. 1993;32:7605–7609. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- 12.Aurora R., Rose G. D. Helix capping. Protein Science. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz V., Blanco F. J., Serrano L. The hydrophobic-staple motif and a role for loop-residues in alpha-helix stability and protein folding. Nat. Struct. Biol. 1995;2:380–385. doi: 10.1038/nsb0595-380. [DOI] [PubMed] [Google Scholar]
- 14.Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of a helices. Science (Washington, D.C.) 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 15.Di Ilio C., Aceto A., Piccolomini R., Allocati N., Faraone A., Cellini L., Ravagnan G., Federici G. Purification and characterization of three forms of glutathione transferase from Proteus mirabilis. Biochem. J. 1988;255:971–975. doi: 10.1042/bj2550971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mignogna G., Allocati N., Aceto A., Piccolomini R., Di Ilio C., Barra D., Martini F. The amino acid sequence of glutathione transferase from Proteus mirabilis, a prototype of a new class of enzymes. Eur. J. Biochem. 1993;211:421–425. doi: 10.1111/j.1432-1033.1993.tb17566.x. [DOI] [PubMed] [Google Scholar]
- 17.Perito B., Allocati N., Casalone E., Masulli M., Dragani B., Polsinelli M., Aceto A., Di Ilio C. Molecular cloning and overexpression of a glutathione transferase gene from Proteus mirabilis. Biochem. J. 1996;318:157–162. doi: 10.1042/bj3180157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casalone E., Allocati N., Ceccarelli I., Masulli M., Rossjohn J., Parker M. W., Di Ilio C. Site-directed mutagenesis of the Proteus mirabilis glutathione transferase B1–1 G-site. FEBS Lett. 1998;423:122–124. doi: 10.1016/s0014-5793(98)00080-5. [DOI] [PubMed] [Google Scholar]
- 19.Rossjohn J., Polekhina G., Feil S. C., Allocati N., Masulli M., Di Ilio C., Parker M. W. A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications. Structure. 1998;6:721–734. doi: 10.1016/s0969-2126(98)00074-4. [DOI] [PubMed] [Google Scholar]
- 20.Allocati N., Casalone E., Masulli M., Ceccarelli I., Carletti E., Parker M. W., Di Ilio C. Functional of the evolutionarily conserved proline 53 residue in Proteus mirabilis glutathione transferase B1–1. FEBS Lett. 1999;445:347–350. doi: 10.1016/s0014-5793(99)00147-7. [DOI] [PubMed] [Google Scholar]
- 21.Allocati N., Casalone E., Masulli M., Polekhina G., Rossjohn J., Parker M. W., Di Ilio C. Evaluation of the role of two conserved active-site residues in beta class glutathione S-transferases. Biochem J. 2000;351:341–346. [PMC free article] [PubMed] [Google Scholar]
- 22.Allocati N., Masulli M., Casalone E., Santucci S., Favaloro B., Parker M. W., Di Ilio C. Glutamic acid 65 is an essential residue for catalysis in Proteus mirabilis glutathione S-transferase B1–1. Biochem J. 2002;363:189–193. doi: 10.1042/0264-6021:3630189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allocati N., Masulli M., Pietracupa M., Favaloro B., Federici L., Di Ilio C. Contribution of the two conserved tryptophan residues to the catalytic and structural properties of Proteus mirabilis glutathione S-transferase B1–1. Biochem J. 2005;385:37–43. doi: 10.1042/BJ20040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caccuri A. M., Antonini G., Allocati N., Di Ilio C., Innocenti F., De Maria F., Parker M. W., Masulli M., Polizio F., Federici G., Ricci G. Properties and utility of the peculiar mixed disulfide in the bacterial glutathione transferase B1-1. Biochemistry. 2002;41:4686–4693. doi: 10.1021/bi0158425. [DOI] [PubMed] [Google Scholar]
- 25.Caccuri A. M., Antonini G., Allocati N., Di Ilio C., De Maria F., Innocenti F., Parker M. W., Masulli M., Lo Bello M., Turella P., Federici G., Ricci G. GSTB1-1 from Proteus mirabilis: a snapshot of an enzyme in the evolutionary pathway from a redox enzyme to a conjugating enzyme. J. Biol. Chem. 2002;277:18777–18784. doi: 10.1074/jbc.M201137200. [DOI] [PubMed] [Google Scholar]
- 26.Allocati N., Favaloro B., Masulli M., Alexeyev M. F., Di Ilio C. Proteus mirabilis glutathione S-transferase B1-1 is involved in protective mechanisms against oxidative and chemical stresses. Biochem. J. 2003;373:305–311. doi: 10.1042/BJ20030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccolomini R., Di Ilio C., Aceto A., Allocati N., Faraone A., Cellini L., Ravagnan G., Federici G. Glutathione transferase in bacteria: subunit composition and antigenic characterization. J. Gen. Microbiol. 1989;135:3119–3125. doi: 10.1099/00221287-135-11-3119. [DOI] [PubMed] [Google Scholar]
- 28.Sanger F., Nicklen S., Coulsen A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J., Russell D. W. 3rd edn. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 2001. Molecular cloning: A Laboratory Manual. [Google Scholar]
- 30.Simons P. C., Vander Jagt D. L. Purification of glutathione S-transferases from human liver by glutathione-affinity chromatography. Anal Biochem. 1977;82:334–341. doi: 10.1016/0003-2697(77)90169-5. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Habig W. H., Jakoby W. B. Assay for differentiation of glutathione S-transferases. Methods Enzymol. 1981;77:398–405. doi: 10.1016/s0076-6879(81)77053-8. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search Programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano W. L. San Carlos, CA, U.S.A.: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 36.Kong G. K., Polekhina G., McKinstry W. J., Parker M. W., Dragani B., Aceto A., Paludi D., Principe D. R., Mannervik B., Stenberg G. Contribution of glycine 146 to a conserved folding module affecting stability and refolding of human glutathione transferase P1–1. J. Biol. Chem. 2003;278:1291–1302. doi: 10.1074/jbc.M209581200. [DOI] [PubMed] [Google Scholar]
- 37.Thom R., Cummins I., Dixon D. P., Edwards R., Cole D. J., Lapthorn A. J. Structure of a Tau class glutathione S-transferase from wheat active in herbicide detoxification. Biochemistry. 2002;41:7008–7020. doi: 10.1021/bi015964x. [DOI] [PubMed] [Google Scholar]
- 38.Doig A. J., MacArthur M. W., Stapley B. J., Thornton J. M. Structures of N-termini of helices in proteins. Protein Sci. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon D. P., Davis B. G., Edwards R. Functional divergence in the glutathione transferase superfamily in plants. J. Biol. Chem. 2002;277:30859–30869. doi: 10.1074/jbc.M202919200. [DOI] [PubMed] [Google Scholar]
- 40.Wongtrakul J., Sramala I., Prapanthadara L., Ketterman A. J. Intra-subunit residue interactions from the protein surface to the active site of glutathione S-transferase AdGSTD3-3 impact on structure and enzyme properties. Insect Biochem. Mol. Biol. 2005;35:197–205. doi: 10.1016/j.ibmb.2004.11.003. [DOI] [PubMed] [Google Scholar]