Abstract
Patched1 (PTCH1) is a human tumour suppressor that acts as an HH (Hedgehog) receptor protein and is important for embryonic patterning. PTCH1 mediates its effects through SMO (Smoothened) and represses the expression of HH target genes such as the transcription factor GLI1 (glioma 1) as well as PTCH1. Up-regulation of these genes has been observed in several cancer forms, including basal cell carcinoma, digestive track tumours and small cell lung cancer. The fact that PTCH1 down-regulates its own expression via ‘negative feedback’ is an important feature in HH signalling, as it keeps the balance between HH and PTCH1 activities that are essential for normal development. In the present study, we provide evidence that a novel mechanism allowing PTCH1 to maintain this balance may also exist. We show that gene activation by GLI1, the transcriptional effector of the pathway, can be down-regulated by PTCH1 without involvement of the canonical cascade of HH signalling events. Specifically, the SMO antagonist cyclopamine has no appreciable effects in blocking this PTCH1-mediated inhibition. Moreover, the negative GLI1 regulator SUFU (Suppressor of Fused) was also found to be dispensable. Additionally, deletion mapping of PTCH1 has revealed that the domains encompassed by amino acids 180–786 and 1058–1210 are of highest significance in inhibiting GLI1 gene activation. This contrasts with the importance of the PTCH1 C-terminal domain for HH signalling.
Keywords: gene activation, GLI1, Patched1 (PTCH1) tumour suppressor gene, Smoothened, Sonic Hedgehog (SHH), Suppressor of Fused
Abbreviations: AP, alkaline phosphatase; Ci, Cubitus interruptus; Cos2, Costal-2; GLI1, glioma 1; HA, haemagglutinin; HEK-293 cells, human embryonic kidney 293 cells; HH, Hedgehog; PTCH1, Patched1; RT, reverse transcriptase; SHH, Sonic HH; SMO, Smoothened; SUFU, Suppressor of Fused; TK, thymidine kinase; TM, transmembrane domain; Z-VAD-FMK, benzyloxycarbonyl-valylalanyl-DL-aspartylfluoromethane
INTRODUCTION
The HH (Hedgehog) signalling pathway has diverse roles in human development and cancer biology [1], and is evolutionarily conserved from insects to vertebrates. The endpoint of the HH signal-transduction cascade, representing the nuclear component of the pathway, is the zinc-finger transcription factor GLI [Ci (Cubitus interruptus) in Drosophila]. Early work in the fly has shown that Ptch (Patched), a 12 transmembrane protein, inhibits the seven transmembrane signalling protein Smo (Smoothened) [1a]. Upon binding of the Hh ligand to its Ptch receptor, Smo is derepressed and a large complex containing Cos2 (Costal-2), a protein with kinesin-like motif, the serine/threonine kinase, Fu (Fused), Sufu (Suppressor of Fused) and Ci is released from the microtubules allowing active Ci to translocate to the nucleus. Mammals have three different types of HH proteins, SHH (Sonic HH), IHH (Indian HH) and DHH (Desert HH), two patched receptors, PTCH1 and PTCH2, and three Ci-like proteins, GLI1 (glioma 1), GLI2 and GLI3 [2]. All GLIs bind to DNA through five Zn-finger domains that recognize the consensus sequence 5′-TGGGTGGTC [3]. GLI1 is a transcriptional activator, whereas GLI2 and GLI3 can act as both activators and repressors [4]. The GLI proteins can be found in the cytoplasm as well as in the nucleus depending on context [5]. The mechanisms of these translocations may involve the regulation of interactions of the GLIs with anchoring protein factors, or the modification of the GLI proteins themselves, including the possibility of proteolytic processing. According to recent models, GLI1 activity is mainly regulated by nuclear–cytoplasmic shuttling that is mediated by interaction with SUFU, a conserved negative regulator of GLI1 [6].
PTCH1 itself is a target gene of HH signalling and acts in a negative feedback loop within the pathway [7,8]. Increased levels of PTCH1 prevent transcription of both GLI1 and PTCH1, two genes that are normally induced by SHH [9]. SMO is apparently involved in this inhibition; however, the mechanistic details are not clearly known. A recent study has shown that free PTCH1 (unbound by HH) acts sub-stoichiometrically to suppress SMO activity, possibly through changes in distribution or concentration of a small molecule, and thus is critical in specifying the level of pathway activity [10]. PTCH1 also functions in the receptor-mediated endocytosis of SHH protein [11], which might limit the range of signalling by degradation of the secreted ligand.
In the present study, we show that PTCH1 can inhibit GLI1 activation of the promoter of PTCH2, an additional target gene. We could also show that this inhibition is directly mediated through GLI binding sites and is not due to apoptotic effects. Moreover, by using mutated GLI1 expression constructs, which are not inhibited by SUFU, as well as Sufu−/− cells, we could demonstrate that the PTCH1 down-regulation of GLI1 activity does not involve SUFU and thus must occur through a novel mechanism. Additionally, no role for the signalling molecule SMO was observed, as blocking SMO activity did not alter the PTCH1 effects. Finally, by performing deletion analysis of PTCH1, the domains responsible for this inhibition were mapped.
MATERIALS AND METHODS
Expression constructs
To generate the PTCH1 deletion constructs, plasmids containing the full-length PTCH1 cDNA with exon 1B as the first exon, flanked by restriction enzyme cleavage sites NheI and NotI, were digested with NotI and subsequently partially with XhoI. The mixture was incubated at 37 °C and 20 μl samples were taken at different time points after the addition of XhoI. The cleavage was stopped by 0.5 M EDTA. The reactions were loaded on to a 0.8% soft agarose gel and the XhoI–NotI partially digested fragment containing the vector backbone and the 5′-end of PTCH1 was extracted and cleaned, using the Qiagen PCR Preps DNA Purification System.
The 3′-deletion fragments from PTCH1 were amplified using the Advantage HF™ PCR kit (ClonTech Laboratories). The PCR products were cleaved with XhoI and NotI and ligated to the XhoI–NotI partially digested fragment from above. The primer sequences used for PCR are listed in Table 1.
Table 1. Sequences of the PCR primers used for generating deletion constructs.
| Primer name | Primer sequence |
|---|---|
| PTCH1,XhoΔ2F | 5′-AAAGCAGCGAACCTCGAG |
| PTCH1,NotΔ2R | 5′-GCGCGGCCGCCTAGTCCAGGTGTTGTAGGAG |
| PTCH1,XhoΔ8,Δ9,ΔCF | 5′-AGCCTCCACTGCCTCGA |
| PTCH1,NotΔCR | 5′-GCGCGGCCGCCTACGGCATGGCGAAGCGGAC |
| PTCH1,NotΔ8R | 5′-GCGCGGCCGCCTATCTGGTTTCCCGAGGTAC |
| PTCH1,NotΔ9R | 5′-GCGCGGCCGCCTACATCACAATGATCCCGGC |
Other expression constructs used were the human full-length HA (haemagglutinin)-tagged GLI1 and Myc-tagged SUFU [6] as well as the cytochrome CYP3A4 [12].
Immunofluorescence
COS7 or HEK-293 (human embryonic kidney 293) cells transfected with Myc- and FLAG-tagged PTCH1 [13] or HA-tagged GLI1 expression constructs were permeabilized and blocked in PBS+0.5% Triton X-100+10% (v/v) donkey or rabbit serum for 1 h at room temperature (20 °C). Subsequently, cells were incubated with primary followed by secondary antibodies diluted in blocking solution [10% of appropriate serum in PBS (depending on secondary antibody)] for 1 h at room temperature. Washes with PBS were performed twice between the incubations. The following antibodies were used: HA antibody (Santa Cruz Biotechnology) at 1:400 dilution; Anti-FLAG® M2 Antibody (Sigma-Aldrich; Stratagene) at 1:400; Patched (G-19) (Santa Cruz Biotechnology) antibody at 1:200; Rhodamine Red™-X-conjugated donkey anti-rabbit IgG (Jackson Immunoresearch Laboratories) at 1:400; fluorescein-conjugated donkey anti-mouse IgG (Jackson Immunoresearch Laboratories) at 1:400; and fluorescein-conjugated donkey anti-goat IgG (Jackson Immuno-research Laboratories) at 1:400. For nuclear staining, cells were incubated with 5 μM DRAQ™ (Alexis Biochemical) in PBS for 10 min, after incubation with fluorescein antibody.
Transfection assays
Confluent HEK-293, NIH-3T3 or Sufu−/− cells were plated at 1:6 dilution on to 24-well plates. On the next day, cells were transfected with 0.2 μg of PTCH2-luc reporter [14], the 12GLI RETKO luciferase construct [6], or the 8×GLIBS-luc reporter construct [15], 0.2 μg of GLI1 cDNA, additional expression constructs as indicated and 50 ng of Renilla luciferase (pRL-SV40) as transfection control. FuGENE™ (3 μl; Roche) was used, with equal amounts of DNA per well (1.45, 1.25 or 1.20 μg) using pcDNA3.1/His B (Invitrogen), CMV5 or pCI vector as necessary. One day after transfection, the medium was changed to Dulbecco's modified Eagle's medium containing 0.5% fetal bovine serum with or without 10 μM cyclopamine or 1 μM of the apoptotic inhibitor Z-VAD-FMK (benzyloxycarbonyl-valylalanyl-DL-aspartylfluoromethane). After incubation for 24 h, cells were washed with PBS to remove apoptotic and necrotic cells and normalized luciferase activity was determined with the dual-luciferase reporter assay system (Promega) using the Microplate Luminometer (Berthold Detection System). The experiments and the individual measurements were performed at least twice.
Apoptosis assays
HEK-293 cells were seeded in 96-well plates and treated for 24 h with 5 μg/ml mitomycin C as well as with 1 μM of the caspase inhibitor Z-VAD-FMK (Alexis Biochemicals). Caspase induction was measured luminometrically using the Caspase-Glo 3/7 Assay from Promega.
Repression of endogenous Gli1 expression
One day after transfection, total RNA was isolated by the RNeasy kit (Qiagen, Hilden, Germany) and reverse transcription was performed as described previously [16]. The amount of single-stranded cDNA used for subsequent PCR amplification was equalized by monitoring the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene expression as a quantitative control. RT (reverse transcriptase)–PCR products were generated with primers obtained from Cybergene AB (Huddinge, Sweden). Each reaction mixture consisted of 1× buffer B (Takara, Japan), 2 mM MgCl2, 0.2 mM of each dNTP, 1.0 μM of each primer (Gli1: 5′-AGTTTCCAGCCTGGACCACG and 5′-GAGGTCCGGATTACGGTTT), 1.0 μl of Taq DNA polymerase (5 units/μl, MBI Fermentas) and 1 ng of cDNA in a total volume of 25 μl. Thirty cycles with 30 s at 95 °C, 30 s at 54 °C and 1 min at 72 °C were performed on a PerkinElmer thermocycler. For nested PCR, 0.5 μl of the initial amplification products was used with a nested primer set (Gli1: 5′-CACCGCGCCCGACGGAG and 5′-ATCAGAAAGGGGCGAGATGG). The nested products were analysed on a 4% NuSieve 3:1 agarose gel (FMC BioProducts, Rockland, ME, U.S.A.). All PCR products were sequence-verified.
Differentiation assays
The differentiation assays were performed 4 days after transfection of GLI1, PTCH1, CYP3A4 or SUFU expression constructs into C3H10T1/2 cells. AP (alkaline phosphatase) staining was performed using a commercial detection kit (Chemicon International) according to the manufacturer's instructions. Images from differentiated colonies expressing AP (red cells), versus non-differentiated colonies (colourless cells), were taken from selected areas (see Figure 10). For the quantitative analysis, cells were fixed and stained for AP activity with BCIP (5-bromo-4-chloroindol-3-yl phosphate) and Nitro Blue Tetrazolium (Sigma) according to the manufacturer's instructions. The amount of reaction products was calculated by taking readings at 595 nm. At least three independent experiments were compiled for the quantitative analysis.
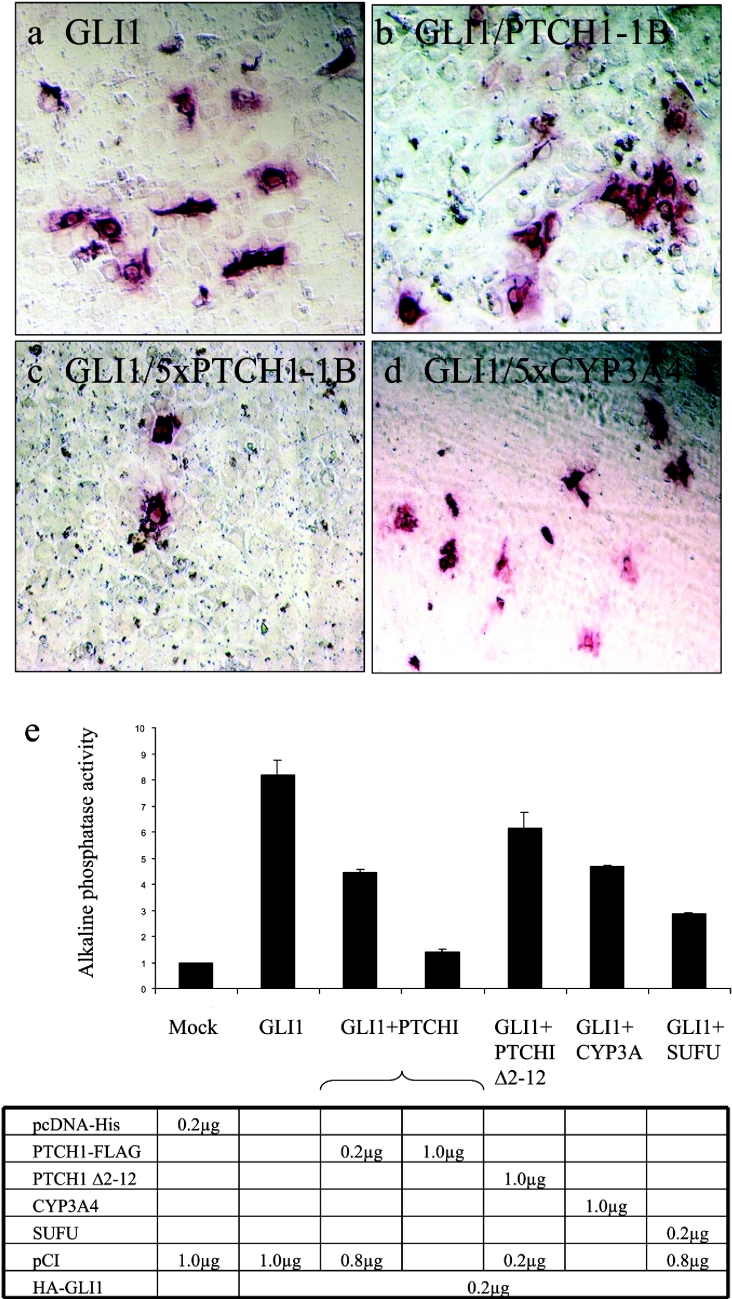
Figure 10. Inhibition of GLI1-induced osteogenic differentiation by PTCH1.
Osteogenic differentiation of C3H10T1/2 cells, as detected by AP staining. Cells were transfected with (a) GLI-1, (b) GLI1 and PTCH1, (c) GLI1 and 5×PTCH1, and (d) GLI1 and 5×CYP3A4. Note that only 5×PTCH1-1B could significantly inhibit the differentiation in these cells. (e) Quantification of differentiation assays, based on three independent experiments. The y-axis represents the AP activity in arbitrary units that reflect absorbance values. Note that GLI1 strongly induces osteogenic differentiation. This could be inhibited by five times higher amounts of full-length but not Δ2–12-truncated PTCH1. The GLI1-dependent differentiation was also reduced by co-expression of SUFU but not five times higher amounts of CYP3A4.
RESULTS
Inhibition of GLI1 activity by PTCH1
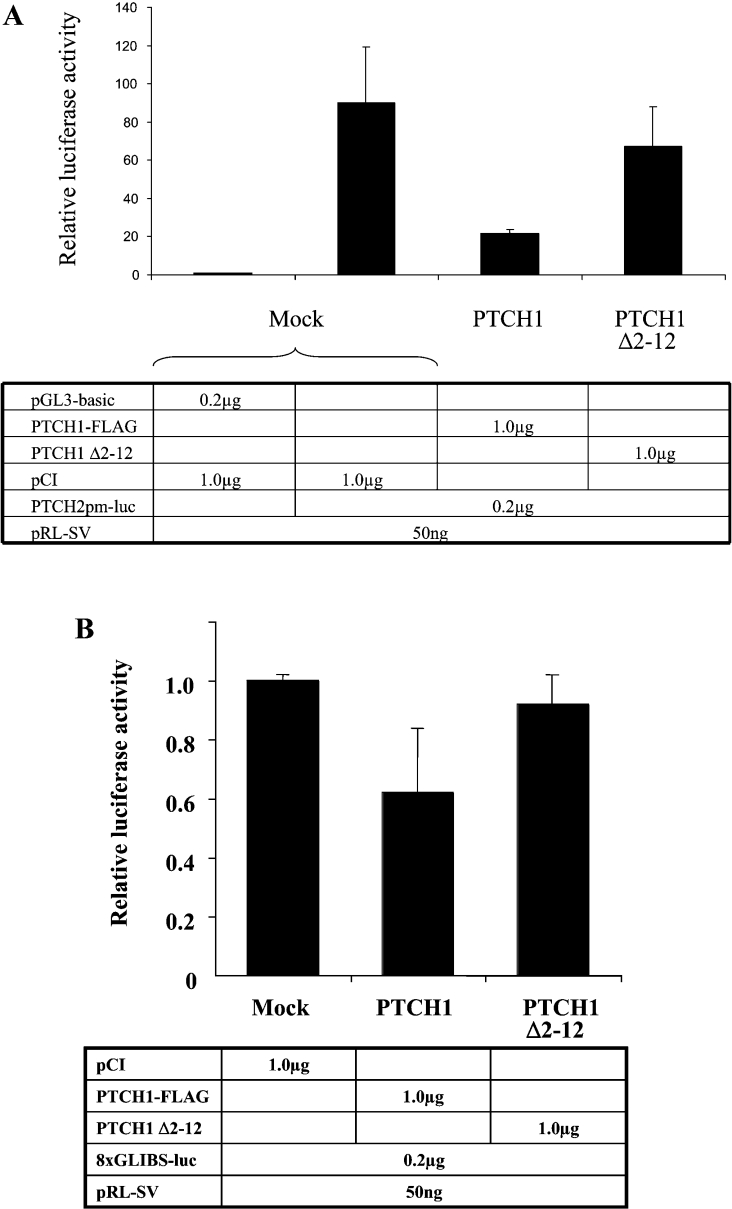
During the analysis of the GLI1 up-regulation of the PTCH2 promoter [14], a surprising finding regarding the role of PTCH1 was made. GLI1 induction was apparently inhibited by PTCH1 after co-transfection in HEK-293 cells. This inhibition could not be observed when equal amounts of GLI1 and PTCH1 cDNAs were co-transfected. However, it was obvious with amounts of PTCH1 at least three times higher than that of GLI1. To verify that this inhibition is PTCH1-specific, we used as a control a CYP3A4 expression construct that is unrelated to the SHH signalling pathway [12]. In contrast with PTCH1, CYP3A4 could not inhibit the GLI1 activity even at the highest amounts used (Figure 1A).
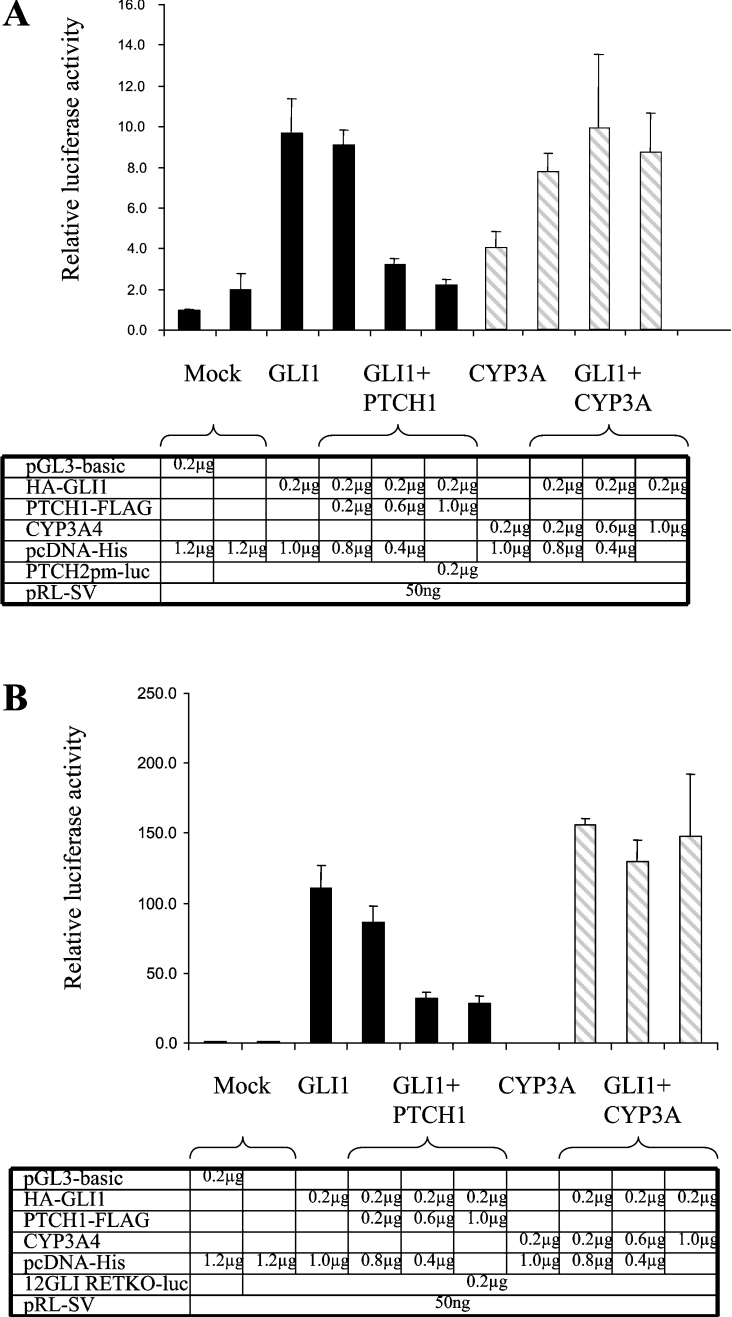
Figure 1. Inhibition of GLI1 activation of reporter constructs by PTCH1.
(A) HEK-293 cells were transfected with PTCH2 reporter, Renilla luciferase transfection control and expression constructs as indicated. PTCH1 could strongly inhibit the PTCH2 promoter up-regulation by GLI1. The inhibition could be observed only when the amount of the PTCH1 construct was at least three times higher than that of GLI1. No inhibition could be observed using a CYP3A4 expression construct instead of PTCH1. (B) As in (A), except that the reporter construct was a synthetic enhancer consisting of 12 copies of the GLI-binding consensus site linked to the TK (thymidine kinase) basal promoter and the luciferase reporter gene. The results were similar to those obtained with the PTCH2 promoter except that GLI1 activated this construct to a higher extent (100–150-fold). The observed activation could be suppressed by PTCH1 but not by CYP3A4.
Inhibition of GLI1 activity by PTCH1 is via GLI-binding sites
To examine whether the observed inhibition is directly mediated through a GLI-binding site such as the one present in the PTCH2 promoter [14], we performed the same experiment using the 12GLI RETKO luciferase construct [6]. This construct contains 12 GLI-binding sites as a tandem repeat. Similarly to the results obtained above, the same inhibition pattern was observed. The GLI1 activity was inhibited with amounts of PTCH1 at least three times higher than GLI1. Again CYP3A4 was used as a control and no inhibition could be observed with any amount of construct used (Figure 1B).
To verify that these observations are not cell- or vector-specific, the NIH-3T3 cell line and PTCH1 constructs with different vector backbones were also used. These experiments confirmed the results regardless of cell type or vector construct (results not shown).
Inhibition of GLI1 activity by PTCH1 is apoptosis-independent
PTCH1 has recently been shown to also have the capacity to induce an apoptotic response [17]. To examine whether apoptosis may be involved in the observed down-regulation of GLI1 activity, HEK-293 cells were transfected with the same constructs as above but also in the presence of the caspase inhibitor Z-VAD-FMK. No difference in the PTCH inhibition of GLI1 activity was observed with this treatment, implying that apoptosis has no role in mediating the PTCH effects on GLI1 (Figure 2).
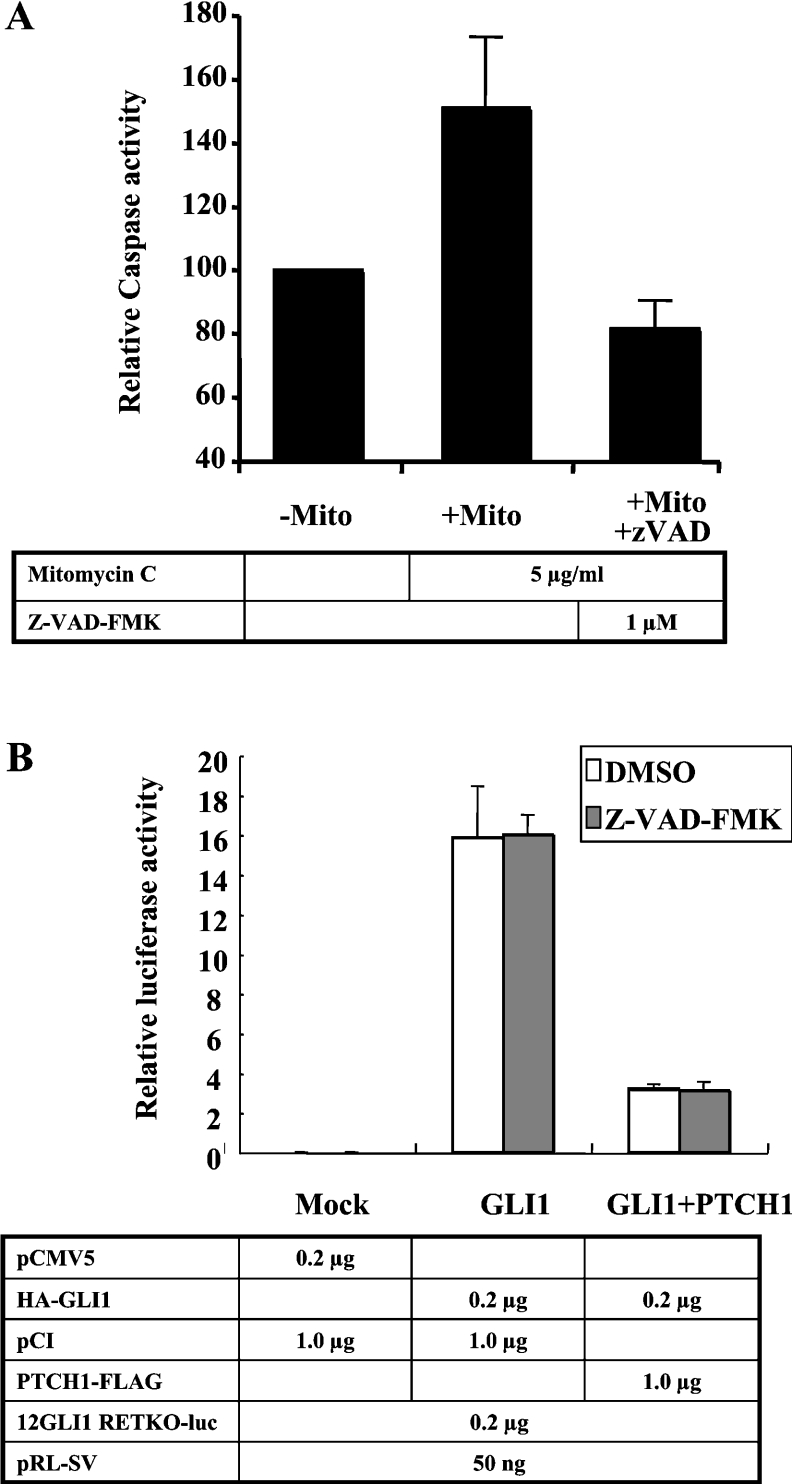
Figure 2. Apoptosis is not involved in the PTCH1 inhibition of GLI1 activity.
(A) Mitomycin C (‘Mito’) induced apoptosis in HEK-293 cells as measured by a caspase 3/7 luminometric assay. This was inhibited in the presence of 1 μM of the caspase inhibitor Z-VAD-FMK. (B) PTCH1 inhibition of GLI1 activity is not blocked by the presence of 1 μM of the caspase inhibitor Z-VAD-FMK. HEK-293 cells were transfected as in Figure 1(B) but with or without the presence of the caspase inhibitor.
Cellular localization of GLI1 and PTCH1
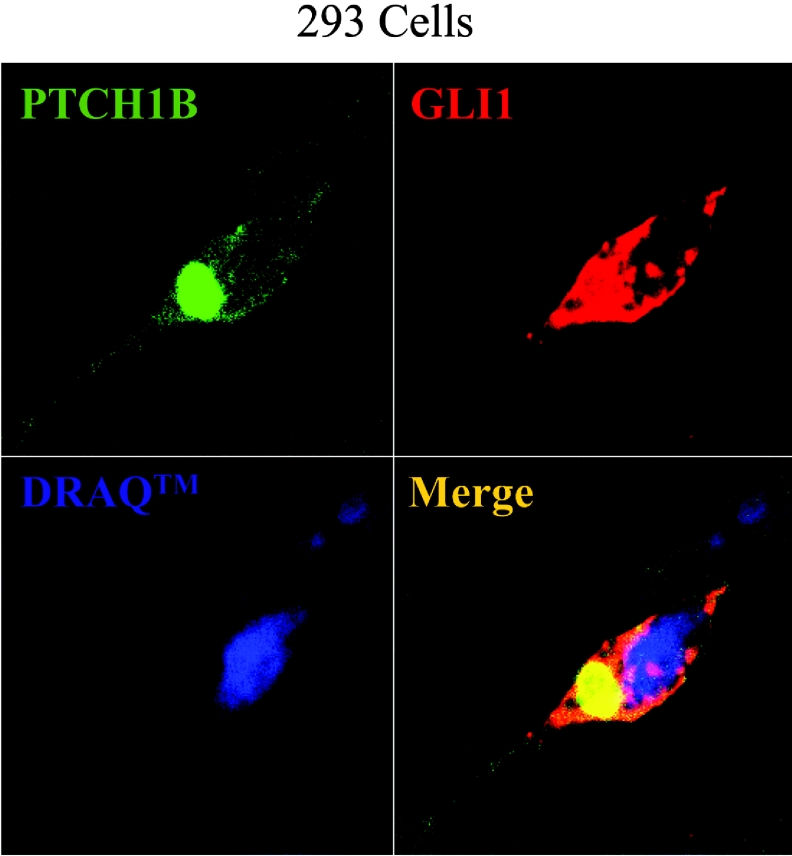
It has been shown that the GLI1 protein activity can be inhibited by a change in subcellular localization from the nucleus to the cytoplasm [6]. Therefore HEK-293 cells were co-transfected with FLAG-tagged PTCH1 and HA-tagged GLI1 followed by immunofluorescence analysis, using antibodies against the epitope tags. GLI1 and PTCH1 were both mainly localized in the cytoplasm; however, GLI1 had a more widespread pattern while PTCH1, as reported previously, was found to be more concentrated near the nucleus [10]. Thus a co-localization of these two proteins was only observed in the regions where PTCH1 was detected (Figure 3) and no change of GLI1 localization could be observed in the presence or absence of PTCH1, even when the nuclear export inhibitor leptomycin B was used (results not shown). Additionally, we were unable to detect strong interactions between PTCH1 and GLI1 using immunoprecipitation assays (results not shown).
Figure 3. Subcellular localization of co-transfected PTCH1 and GLI1.
HEK-293 cells were transfected with FLAG-tagged PTCH1 with HA-tagged GLI1 and stained with antibodies to the respective tag. The proteins were visualized by immunofluorescence confocal microscopy. Dual-channel imaging indicates co-localized region, with GLI1 shown in red and PTCH1 in green. Nuclear staining with DRAQ™ is shown in blue.
Inhibition of GLI1 activity by PTCH1 is SUFU-independent
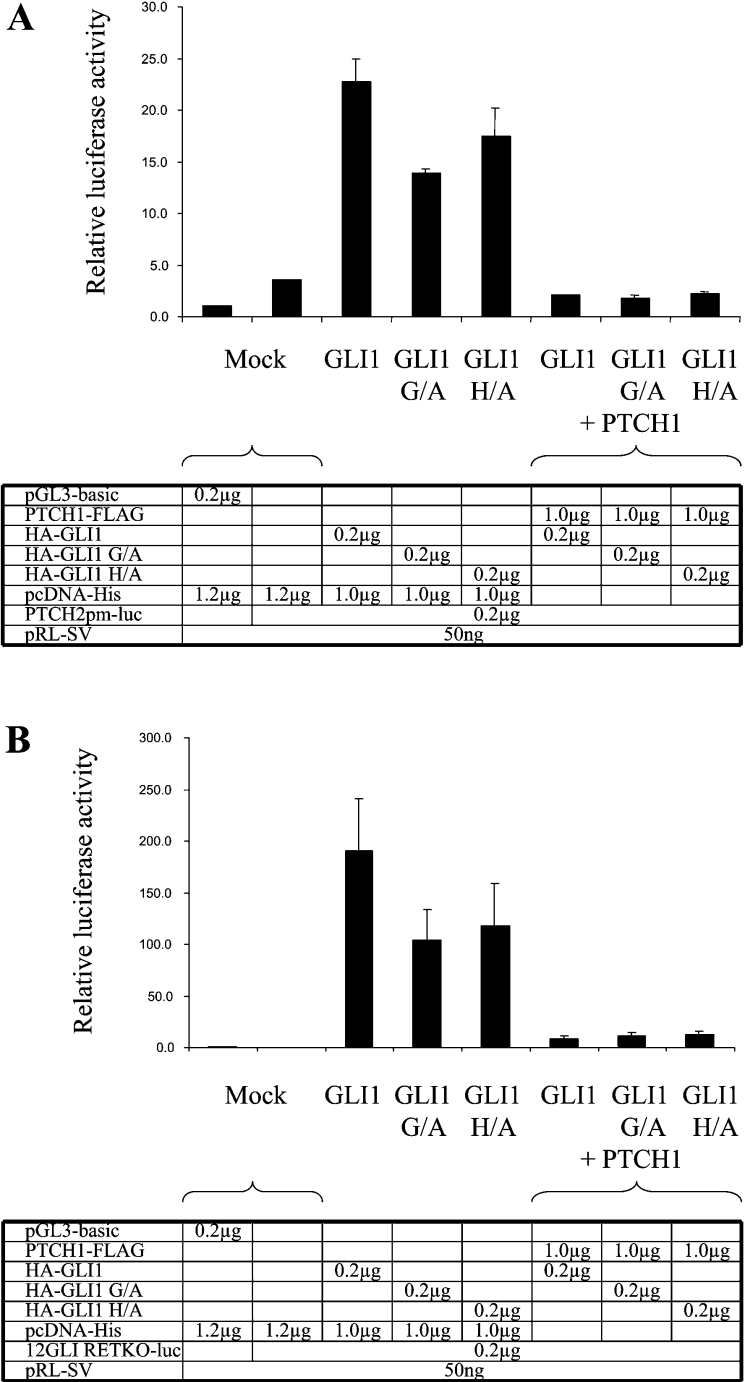
Mutational studies have revealed that Gly122 and His123 in GLI1 are crucial for binding to SUFU. Functional analysis has also shown that the HH target gene activation by GLI1 with alanine substituting for either Gly122 or His123 is no longer suppressed by co-transfection of SUFU [18]. To examine whether the PTCH1 inhibition of the GLI1 activity occurs via SUFU, we used expression vectors encoding these mutated GLI1 proteins in the transfection assays. It was found that the GLI1 activation of the PTCH2 promoter could be inhibited by PTCH1 regardless of the mutation in the SUFU-binding site. Similar results were observed when these experiments were performed using the 12GLI RETKO luciferase construct (Figures 4A and 4B).
Figure 4. Analysis of the inhibition of mutated GLI1 activity by PTCH1.
(A) Effects of PTCH1 on the transcriptional activity of wild-type and mutated forms of GLI1 in HEK-293 cells. Both the Gly122 to alanine and His123 to alanine mutated GLI1 constructs, which are resistant to SUFU inhibition, can be inhibited by PTCH1. (B) As in (A), except that the reporter construct was a synthetic enhancer consisting of 12 copies of the GLI-binding consensus site linked to the TK basal promoter and the luciferase reporter gene. The results were similar to those obtained with the PTCH2 promoter.
Inhibition of GLI1 activity by PTCH1 is SMO-independent
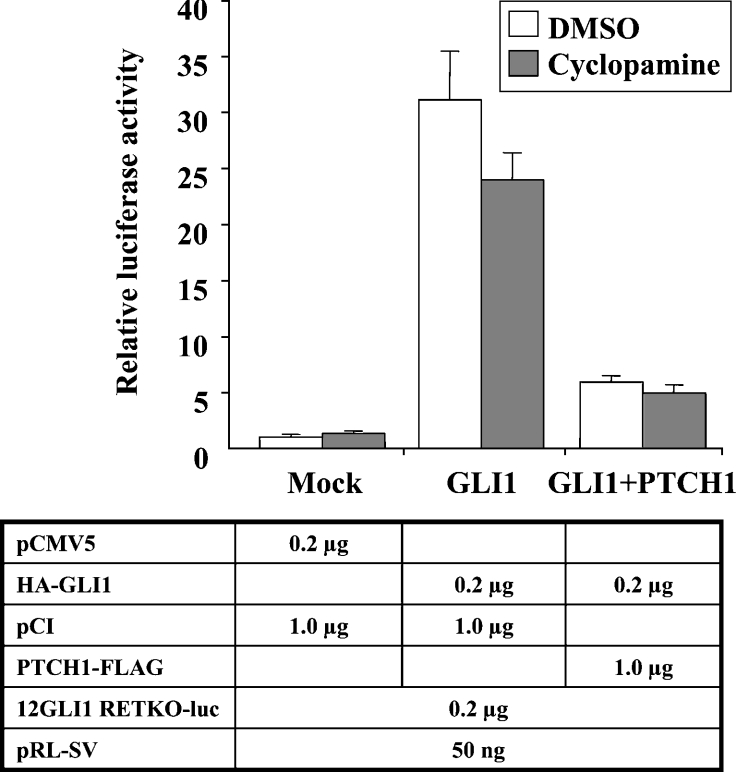
Cyclopamine is a plant-derived molecule that acts as a SMO antagonist, blocking signalling activation [19,20]. To examine whether the PTCH1 inhibition of GLI1 activity is dependent on SMO, NIH-3T3 cells were treated with cyclopamine prior to the analysis of the 12GLI RETKO construct. In these cells, cyclopamine at the range of concentrations tested, from 2.5 to 40 μM, completely blocks SHH signalling activation, as determined by GLI1 reporter constructs (results not shown). However, the PTCH1 inhibition of GLI1 activity was found to be independent of cyclopamine treatment (Figure 5). Similar results were also obtained with HEK-293 cells and the PTCH2 promoter construct (results not shown). Thus these observations provide strong evidence that SMO signalling blockage has no role in the inhibitory effects of PTCH1 on GLI1 activity.
Figure 5. Cyclopamine does not influence the PTCH1 inhibition of GLI1 activity.
NIH-3T3 cells were transfected with the 12GLI RETKO luciferase construct, Renilla luciferase transfection control and expression constructs as indicated, and treated with or without the SMO antagonist cyclopamine at 10 μM concentration. Note that this treatment has no appreciable effects on the PTCH1 inhibition of GLI1 activity.
Mapping the PTCH1 domains responsible for GLI1 inhibition
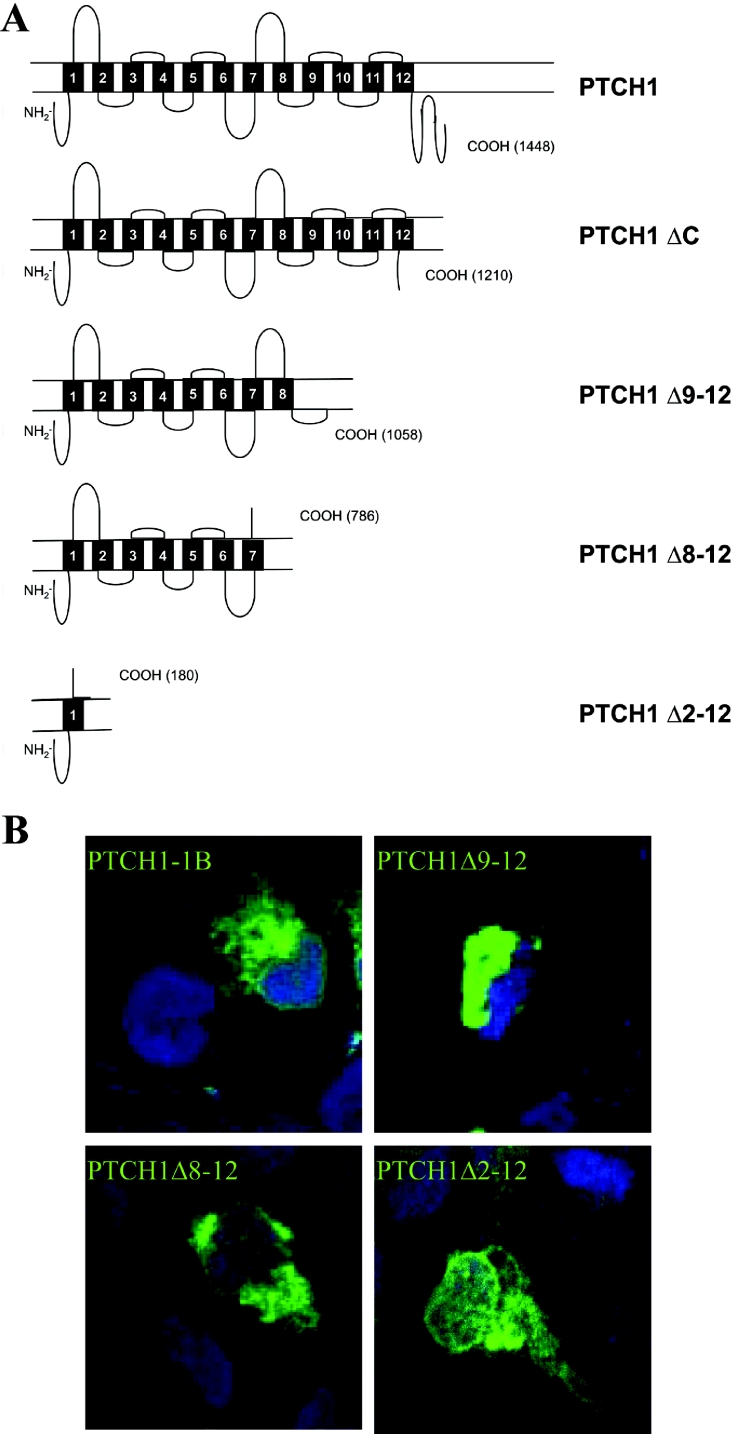
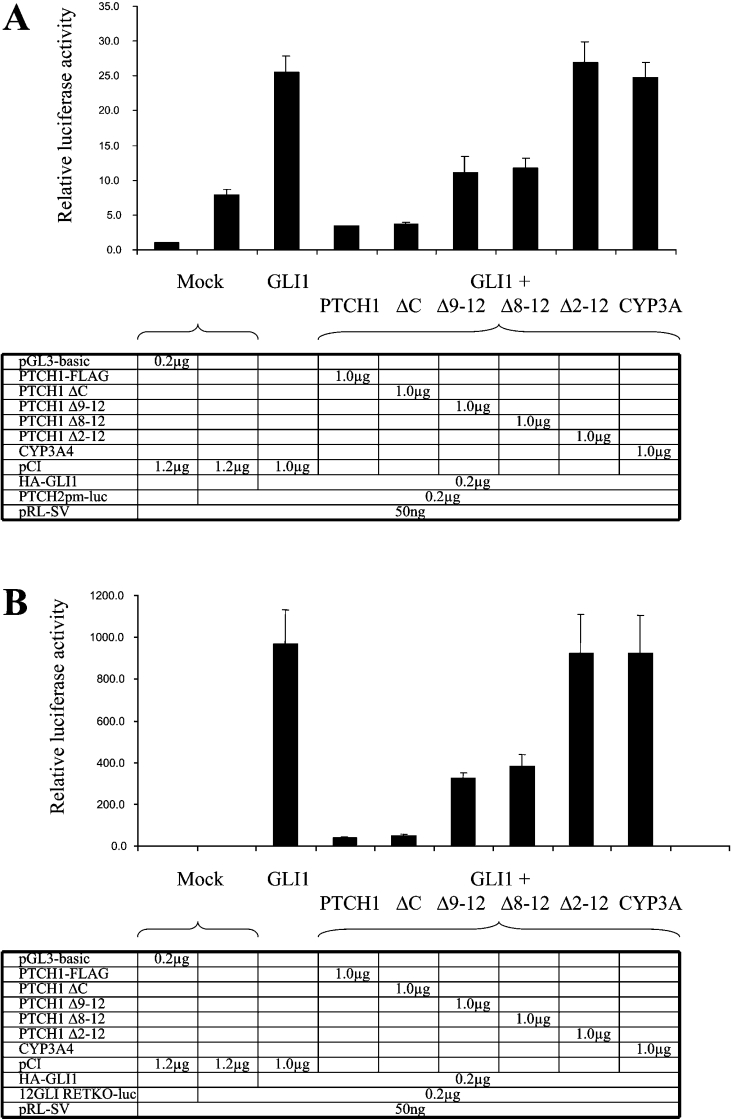
To map the region of PTCH1 required for GLI1 inhibition, PTCH1 C-terminal deletion constructs were generated (Figure 6A), and by using an immunofluorescence assay, their expression was found to be comparable (Figure 6B). PTCH1 and the deletion constructs were then co-transfected into HEK-293 cells along with GLI1 and using as a reporter the PTCH2 promoter construct. PTCH1 Δ2–12, which lacks TMs (transmembrane domains) 2–12 (amino acids 180–1448), failed to inhibit GLI1 activity. Similar results were also obtained using the 12GLI RETKO luciferase construct instead of the PTCH2 promoter. The other deletion constructs, PTCH1 Δ8–12 lacking TM 8–12 (amino acid 786–1448), PTCH1 Δ9–12 lacking TM 9–12 (amino acids 1058–1448) and PTCH1 ΔC lacking only the last 238 amino acids, were still able to inhibit the GLI1 activation, albeit to different extents (Figures 7A and 7B). The fact that PTCH1 ΔC inhibited GLI1 to the same extent as the full-length PTCH1 provides additional evidence that apoptosis is not involved in mediating these effects, as the C-terminal region contains the so-called ‘pro-apoptotic domain’ of PTCH1 [17]. Thus, based on these experiments, we conclude that the regions between amino acids 180–768 and 1058–1210 are of critical importance for the observed inhibition by PTCH1.
Figure 6. Cellular expression of PTCH1 deletion constructs.
(A) Diagram representing the C-terminal deletion constructs of PTCH1 that were used in the present study. The end of each construct is indicated by the corresponding amino acid based on the numbering of GenBank® entry NM_000264. (B) COS7 cells transfected with PTCH1, PTCH1 Δ9–12, PTCH1 Δ8–12 or PTCH1 Δ2–12 constructs. The cells were stained with antibodies recognizing epitopes at the PTCH1 N-terminal domain. The proteins were visualized by immunofluorescence microscopy. The number of transfected cells and the amount of protein expressed in individual cells were comparable with each of the four constructs used. The cells were also stained with the nuclear marker DRAQ™.
Figure 7. Mapping of the domains of PTCH1 responsible for GLI1 inhibition.
(A) HEK-293 cells were transfected with PTCH2 reporter, Renilla luciferase transfection control and expression constructs as indicated. Note the increased loss of PTCH1 inhibition when specific domains of the protein were deleted. (B) As in (A), except that the reporter construct was a synthetic enhancer consisting of 12 copies of the GLI-binding consensus site linked to the TK basal promoter and the luciferase reporter gene. The results were similar to those obtained with the PTCH2 promoter.
Endogenous Gli1 activity is inhibited by PTCH1
We also used mouse embryonic fibroblasts with the Sufu gene inactivated for analysis of the PTCH1 expression constructs. These Sufu−/− cells are characterized by a high expression of Gli1, in line with the lack of the Gli1 negative regulator, Sufu (J. Svärd, K. H. Henricson, M. Persson-Lek, B. Rozell, M. Lauth, J. Ericson, R. Toftgård and S. Teglund, unpublished work). As anticipated, PTCH1 could inhibit the endogenous Gli1 activation of the PTCH2 promoter construct while PTCH1 Δ2–12 lacked this capacity (Figure 8A). We could also observe an inhibition by PTCH1 when an 8×GLI-reporter construct [15] instead of the PTCH2 promoter was used (Figure 8B), although this did not reach statistical significance. The observation of only a weak inhibition may be rationalized by the recently obtained evidence that these cells also express increased levels of endogenous PTCH1, raising the possibility that these are masking the effects of exogenously added PTCH1. Thus these experiments provide additional evidence that PTCH1 can inhibit GLI1 activity independently of SUFU.
Figure 8. Analysis of endogenous Gli1 inhibition by PTCH1 in Sufu−/− cells.
(A) Sufu−/− mouse embryonic fibroblast cells were transfected with PTCH2 reporter, Renilla luciferase transfection control and expression constructs as indicated. Note that PTCH1 inhibited the endogenous Gli1 activation of the reporter construct, while PTCH1 Δ2–12 did not. (B) As in (A), except that the reporter construct (8×GLIBS) consisted of eight copies of the GLI-binding consensus site linked to the luciferase gene.
Endogenous Gli1 expression, activated by GLI1, is inhibited by PTCH1
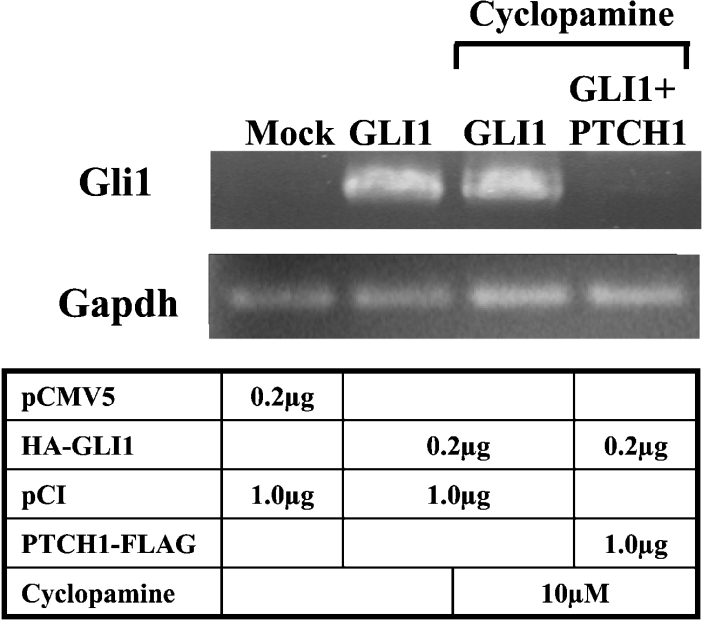
To investigate whether endogenous Gli1 expression is subjected to the PTCH1 inhibition of GLI1 activity, cyclopamine-treated NIH-3T3 cells were used. Transfection with GLI1 induced endogenous expression of the Gli1 mRNA, as revealed by RT–PCR analysis, in line with the fact that Gli1 is a target gene. Moreover, co-transfection with PTCH1 inhibited the observed increase in Gli1 mRNA levels. Thus the PTCH1 effects on GLI1 may influence not only activities of reporter constructs but also endogenous gene expression as well (Figure 9).
Figure 9. GLI1 activation of endogenous Gli1 gene expression is inhibited by PTCH1.
NIH-3T3 cells were transfected with GLI1 and PTCH1 expression constructs as indicated and the endogenous Gli1 expression was monitored by RT–PCR. Note that GLI1 induced endogenous Gli1 expression in cyclopamine-treated cells, while co-transfection of PTCH1 inhibited this capacity of GLI1.
PTCH1 inhibits GLI1-dependent differentiation
To determine whether PTCH1 has the capacity to also inhibit a GLI1-dependent cellular response, we used the C3H10T1/2 cells osteogenic differentiation assay [6]. Transfection of GLI1 induced cellular differentiation, as measured by AP staining. Co-transfection of five times higher amounts of PTCH1 Δ2–12 or CYP3A4 expression constructs did not result in major changes of this response. However, the use of five times higher amounts of PTCH1 or equal amounts of the GLI1 negative regulator SUFU did result in significant inhibition of the observed differentiation (Figure 10). Thus these findings indicate that the influence of PTCH1 on GLI1 extends beyond the activities of reporter constructs and endogenous gene expression to encompass distinct biological responses.
DISCUSSION
Our results demonstrate that the capacity of GLI1 for transcriptional activation can be inhibited by PTCH1 using mechanisms that apparently do not involve the known sequence of HH signalling events. This inhibition was observed when the amount of transfected PTCH1 cDNA was in excess relative to GLI1, in contrast with the effectiveness of lower PTCH1 cDNA amounts in inhibiting HH/SMO activation [14]. It has been shown that overexpression of PTCH1 is sufficient to inhibit expression of SHH target genes such as GLI1 and PTCH1 in the neural tube [9]. Several mechanisms for the regulation of HH signalling by PTCH1 have been proposed. One suggests that the increased amount of PTCH1 may bind SHH and alter ligand accessibility for different target cells by, for example, endocytosis [11]. PTCH1 might also regulate the level of signal activity by forcing SMO to a subcellular compartment either containing small molecule antagonists or lacking small molecule agonists [10]. It has also been shown that PTCH1 induces apoptotic cell death unless its ligand SHH is present [17]. However, apoptosis is apparently not involved in mediating these novel PTCH1 effects on GLI1, as the use of the caspase inhibitor Z-VAD-FMK does not alleviate the observed PTCH1 inhibition. An additional negative regulator of SHH signalling that has been reported is SUFU [6]. This acts either by retaining the active GLI1 protein in the cytoplasm or by direct binding to GLI1 in the nucleus. To check if SUFU may be involved in the observed PTCH1 inhibition of GLI1 activity, we tested two mutated GLI1 constructs that have been shown not to be sensitive for SUFU inhibition but still retaining full GLI1 activity [18]. To our surprise, both the Gly122 to alanine and the His123 to alanine constructs could be inhibited by PTCH1. These results as well as the finding that endogenous Gli1 in Sufu−/− cells is inhibited by PTCH1 indicate that the mechanism of this inhibition does not include the SHH signalling component SUFU [21]. Additionally, the use of the SMO antagonist cyclopamine did not alter the inhibitory effects of PTCH1 on GLI1 activity, supporting the notion that the capacity for active signalling is dispensable for this inhibition. Thus at relatively high PTCH1 to GLI1 levels, novel mechanisms that influence gene activation by the transcription factor GLI1 may be involved. Conceivably, these may represent an additional means for PTCH1 to down-regulate signalling when its levels exceed a certain threshold.
To test whether the PTCH1 effects are due to localization changes of GLI1 protein, we used immunofluorescence in HEK-293 cells. These cells were transfected with GLI1 in the presence or absence of PTCH1. No changes could be observed in the subcellular staining of GLI1, indicating that major localization changes are not involved in the mechanism of this inhibition. Recent studies have also shown that the components of HH signalling are interacting in patterns that are beyond a simple linear order, as Ci was found to bind Smo via Cos2 [22]. Thus some unidentified component(s) may be involved in mediating the PTCH1 effects on GLI1, as no major direct protein–protein interactions could be revealed in an immunoprecipitation assay. Moreover, by generating deletion constructs of PTCH1, the domains responsible for inhibition of GLI1 activation were mapped. Deletion of the regions encompassed by amino acids 180–786 and 1058–1210 were found to negatively influence the inhibition of GLI1 by PTCH1. It should be mentioned that the former segment encodes the SSD (sterol-sensing domain) that is important for signal transduction [23,24]. However, the second extracellular loop between TM7 and TM8, which is thought to interact with HH ligands [25,26], is not critical for the observed inhibition. Additionally, the C-terminal region that has a role in blocking HH/SMO signalling [27,28] and, moreover, was recently shown to also be involved in an apoptotic response [17] is dispensable for this PTCH1-mediated inhibition. Therefore these findings highlight the distinct downstream effects that unique PTCH1 domains may elicit.
In summary, the present study reveals an additional function of PTCH1. This protein acts not only as an HH receptor, but also as a factor that has the capacity to limit GLI1 activity not only through the known, canonical signalling cascade but also by novel mechanisms that are SUFU/SMO-independent. Additionally, two distinct domains in PTCH1 responsible for the observed GLI1 inhibition were delineated.
Acknowledgments
This study was supported by the Swedish Cancer Fund. P. K. acknowledges support from the Estonian Science Foundation grant no. 5552.
References
- 1.McMahon A. P., Ingham P. W., Tabin C. J. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 1a.Chen Y., Struhl G. In vivo evidence that Patched and Smoothened constitute distinct binding and transducing components of a Hedgehog receptor complex. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- 2.Bai C. B., Joyner A. L. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler K. W., Ruppert J. M., Bigner S. H., Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature (London) 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki H., Nishizaki Y., Hui C., Nakafuku M., Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 6.Kogerman P., Grimm T., Kogerman L., Krause D., Unden A. B., Sandstedt B., Toftgård R., Zaphiropoulos P. G. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 7.Ingham P. W., Taylor A. M., Nakano Y. Role of the Drosophila patched gene in positional signalling. Nature (London) 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R. L., Grenier J. K., Scott M. P. Patched overexpression alters wing disc size and pattern: transcriptional and post-transcriptional effects on hedgehog targets. Development. 1995;121:4161–4170. doi: 10.1242/dev.121.12.4161. [DOI] [PubMed] [Google Scholar]
- 9.Goodrich L. V., Jung D., Higgins K. M., Scott M. P. Overexpression of ptc1 inhibits induction of Shh target genes and prevents normal patterning in the neural tube. Dev. Biol. 1999;211:323–334. doi: 10.1006/dbio.1999.9311. [DOI] [PubMed] [Google Scholar]
- 10.Taipale J., Cooper M. K., Maiti T., Beachy P. A. Patched acts catalytically to suppress the activity of Smoothened. Nature (London) 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 11.Incardona J. P., Lee J. H., Robertson C. P., Enga K., Kapur R. P., Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered Sonic hedgehog by Patched-1. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finta C., Zaphiropoulos P. G. Intergenic mRNA molecules resulting from trans-splicing. J. Biol. Chem. 2002;277:5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 13.Kogerman P., Krause D., Rahnama F., Kogerman L., Unden A. B., Zaphiropoulos P. G., Toftgård R. Alternative first exons of PTCH1 are differentially regulated in vivo and may confer different functions to the PTCH1 protein. Oncogene. 2002;21:6007–6016. doi: 10.1038/sj.onc.1205865. [DOI] [PubMed] [Google Scholar]
- 14.Rahnama F., Toftgård R., Zaphiropoulos P. G. Distinct roles of PTCH2 splice variants in Hedgehog signalling. Biochem. J. 2004;378:325–334. doi: 10.1042/BJ20031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki H., Hui C., Nakafuku M., Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 16.Shimokawa T., Rahnama F., Zaphiropoulos P. G. A novel first exon of the Patched1 gene is upregulated by Hedgehog signaling resulting in a protein with pathway inhibitory functions. FEBS Lett. 2004;578:157–162. doi: 10.1016/j.febslet.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Thibert C., Teillet M. A., Lapointe F., Mazelin L., Le Douarin N. M., Mehlen P. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science. 2003;301:843–846. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 18.Dunaeva M., Michelson P., Kogerman P., Toftgård R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 2003;278:5116–5122. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 19.Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P., Beachy P. A. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature (London) 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 20.Chen J. K., Taipale J., Cooper M. K., Beachy P. A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalderon D. Hedgehog signaling: costal-2 bridges the transduction gap. Curr. Biol. 2004;14:R67–R69. doi: 10.1016/j.cub.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 22.Ruel L., Rodriguez R., Gallet A., Lavenant-Staccini L., Therond P. P. Stability and association of Smoothened, Costal2 and fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- 23.Martin V., Carrillo G., Torroja C., Guerrero I. The sterol-sensing domain of patched protein seems to control smoothened activity through patched vesicular trafficking. Curr. Biol. 2001;11:601–607. doi: 10.1016/s0960-9822(01)00178-6. [DOI] [PubMed] [Google Scholar]
- 24.Strutt H., Thomas C., Nakano Y., Stark D., Neave B., Taylor A. M., Ingham P. W. Mutations in the sterol-sensing domain of patched suggest a role for vesicular trafficking in smoothened regulation. Curr. Biol. 2001;11:608–613. doi: 10.1016/s0960-9822(01)00179-8. [DOI] [PubMed] [Google Scholar]
- 25.Marigo V., Davey R. A., Zuo Y., Cunningham J. M., Tabin C. J. Biochemical evidence that patched is the Hedgehog receptor. Nature (London) 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 26.Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H., et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature (London) 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 27.Johnson R. L., Milenkovic L., Scott M. P. In vivo functions of the patched protein: requirement of the C terminus for target gene inactivation but not Hedgehog sequestration. Mol. Cell. 2000;6:467–478. doi: 10.1016/s1097-2765(00)00045-9. [DOI] [PubMed] [Google Scholar]
- 28.Makino S., Masuya H., Ishijima J., Yada Y., Shiroishi T. A spontaneous mouse mutation, mesenchymal dysplasia (mes), is caused by a deletion of the most C-terminal cytoplasmic domain of patched (ptc) Dev. Biol. 2001;239:95–106. doi: 10.1006/dbio.2001.0419. [DOI] [PubMed] [Google Scholar]