Abstract
Proteasomes are multicatalytic proteinase complexes within cells that selectively degrade ubiquitinated proteins. We have recently demonstrated that fatty acids, major components of cell membranes, are able to regulate the proteasomal degradation of tyrosinase, a critical enzyme required for melanin biosynthesis, in contrasting manners by relative increases or decreases in the ubiquitinated tyrosinase. In the present study, we show that altering the intracellular composition of fatty acids affects the post-Golgi degradation of tyrosinase. Incubation with linoleic acid (C18:2) dramatically changed the fatty acid composition of cultured B16 melanoma cells, i.e. the remarkable increase in polyunsaturated fatty acids such as linoleic acid and arachidonic acid (C20:4) was compensated by the decrease in monounsaturated fatty acids such as oleic acid (C18:1) and palmitoleic acid (C16:1), with little effect on the proportion of saturated to unsaturated fatty acid. When the composition of intracellular fatty acids was altered, tyrosinase was rapidly processed to the Golgi apparatus from the ER (endoplasmic reticulum) and the degradation of tyrosinase was increased after its maturation in the Golgi. Retention of tyrosinase in the ER was observed when cells were treated with linoleic acid in the presence of proteasome inhibitors, explaining why melanin synthesis was decreased in cells treated with linoleic acid and a proteasome inhibitor despite the abrogation of tyrosinase degradation. These results suggest that the intracellular composition of fatty acid affects the processing and function of tyrosinase in connection with the ubiquitin–proteasome pathway and suggest that this might be a common physiological approach to regulate protein degradation.
Keywords: fatty acid composition, linoleic acid, melanin, palmitic acid, tyrosinase, ubiquitin–proteasome pathway
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; endo H, endoglycosidase H; ER, endoplasmic reticulum; ERAD, ER-associated degradation; FBS, fetal bovine serum; OCA, oculocutaneous albinism; PNGase F, peptide N-glycosidase F
INTRODUCTION
Fatty acids are major components of biological cell membranes that play important roles in intracellular signalling and as precursors for ligands that bind to nuclear receptors [1–3]. Several studies have revealed that the composition of intracellular fatty acids can be changed by the dietary intake of various kinds of lipids [4–8]; however, little is known about the relationship between intracellular fatty acids and the protein turnover that is controlled by a balance between synthesis and degradation. In general, abnormal proteins misfolded in the ER (endoplasmic reticulum) and short-lived proteins are selectively degraded by the ubiquitin–proteasome pathway [9–11]. Proteasomes are multicatalytic proteinase complexes located within cells that selectively degrade ubiquitinated proteins.
Tyrosinase, the membrane glycoprotein critically associated with melanin biosynthesis, is actively degraded by proteasomes [12]. The degradation of tyrosinase is involved in several pigmentary disorders such as OCA (oculocutaneous albinism) that are caused by the dysfunction of tyrosinase. One of the causes of OCA is the aberrant retention of tyrosinase in the ER, which results in its hyperdegradation by proteasomes [12–14]. We recently demonstrated that fatty acids regulate the degradation of tyrosinase by modulating the ubiquitination of tyrosinase, which leads to an increase or a decrease in its degradation by proteasomes [15]. These findings imply that intracellular fatty acids regulate the degradation of glycoproteins by modulating their ubiquitination. It is worth noting that the ubiquitin–proteasome pathway, which is intimately connected to various diseases, might be regulated by intracellular fatty acids. In the present study, we explored the possibility of whether modulating the intracellular composition of fatty acids affects the function of membrane glycoproteins, using the model of the fatty acid-induced regulation of tyrosinase degradation by the ubiquitin–proteasome pathway.
EXPERIMENTAL
Materials
αPEP7, a rabbit polyclonal antibody generated against a synthetic peptide corresponding to the C-terminus of mouse tyrosinase, was produced in our laboratory [16]. Mouse monoclonal antibodies used were Bip/GRP78 (BD Transduction Laboratories, Lexington, KY, U.S.A.) and Vti1b (BD Transduction Laboratories), which are reactive with the ER [17] and Golgi apparatus [18] respectively. MG132 (benzyloxycarbonyl-Leu-Leu-leucinal, also known as Cbz-LLL), a proteasome inhibitor, was obtained from Calbiochem (San Diego, CA, U.S.A.). Saturated palmitic acid (hexadecanoic acid) and unsaturated linoleic acid (cis-9,12-octadecadienoic acid) were from Sigma (St. Louis, MO, U.S.A.).
Cell cultures and conditions
B16F10 mouse melanoma cells were cultured in Eagle's minimal essential medium (Sigma) containing 10% (v/v) heat-inactivated (56 °C, 30 min) FBS (fetal bovine serum; HyClone, Logan, UT, U.S.A.) at 37 °C in a humidified atmosphere with 5% CO2. Non-esterified fatty acids and MG132 were dissolved in DMSO. Final DMSO concentrations were 0.1% or less, levels that did not influence cell viability or tyrosinase function. We showed previously that 25 μM of each fatty acid had no effect on the proliferation of B16F10 cells [19]. For confocal microscopy, cells were treated with linoleic acid (25 μM) or palmitic acid (25 μM) for 72 h in the presence or absence of MG132 (120 nM) for the last 24 h before fixation.
Fatty acid analysis
After incubation with linoleic acid or palmitic acid for 72 h, cells were washed three times with Eagle's minimal essential medium containing 10% FBS to remove residual fatty acids from the cell surface. The cells were then further washed three times with 0.1 M Hepes (pH 7.5). Fatty acids were determined by GLC analysis after transmethylation with the addition of tricosanoic acid (C23:0) as an internal standard [20]. Briefly, aliquots of lipid extracts were transmethylated by reaction with BF3/methanol (14%, w/v) at 100 °C for 2 h under a nitrogen atmosphere. Fatty acid methyl esters were extracted with hexane and were then analysed using a Hewlett–Packard 5890 gas chromatograph equipped with a flame ionization detector (Hewlett–Packard, Palo Alto, CA, U.S.A.) and a fused-silica DB-FFAP capillary column (30 m×0.25 mm internal diameter, 0.25 μm; J&W Scientific, Folsom, CA, U.S.A.) as described previously [21]. The oven temperature was programmed from 130 to 180 °C at 4 °C/min, from 180 to 215 °C at 1 °C/min and then raised to 245 °C at 30 °C/min, and then maintained at 245 °C for 15 min. The injector and detector temperatures were set at 250 °C. Hydrogen was used as a carrier gas with a linear velocity of 54 cm/s. Individual fatty acids were identified by comparing the retention times with known fatty acid standards GLC-411 (Nu-Chek Prep, Elysian, MN, U.S.A.).
Carbohydrate cleavage
After treatment with fatty acids, 1 μg of protein from the cell extracts was digested with 1000 units of endo H (endoglycosidase H) or PNGase F (peptide N-glycosidase F; New England Biolabs, Beverly, MA, U.S.A.) for 3 h at 37 °C according to the manufacturer's instructions. After the digestion, cell extracts were mixed with 2×Tris/glycine SDS sample buffer (Invitrogen, Carlsbad, CA, U.S.A.) supplemented with 10% (v/v) 2-mercaptoethanol and were boiled at 95 °C for 5 min. Samples were subjected to SDS/PAGE, and immunoreactive bands were detected by Western blotting using the αPEP7 antibody.
Western-blot analysis
Cells were solubilized in lysis buffer consisting of 0.1 M Tris/HCl (pH 7.4) containing 1% Nonidet P40, 0.01% SDS, 2.4 μM MG132 and complete protease inhibitor cocktail (Roche, Mannheim, Germany) for 30 min at 4 °C. After centrifugation (15000 g for 10 min at 4 °C), the cell extracts were mixed with 2×Tris/glycine SDS sample buffer (Invitrogen) supplemented with 10% 2-mercaptoethanol and were boiled at 95 °C for 5 min. Total protein (5 μg) from each cell extract was separated by SDS/PAGE on 8–16% gradient Tris/glycine gels (Invitrogen). The Immobilon-P membranes (Millipore, Bedford, MA, U.S.A.) were blocked in 10% (w/v) non-fat dry milk in TBS-T (20 mM Tris/HCl, pH 7.4, 137 mM NaCl and 0.1% Tween 20) overnight at 4 °C and were then incubated with primary antibodies diluted (as noted in the Figure legends) in 2% non-fat dry milk in TBS-T for 2 h. After five washes with TBS-T, the blotting membranes were incubated in HRP (horseradish peroxidase)-linked anti-rabbit or anti-mouse whole antibodies (1:10000; Amersham Biosciences, Piscataway, NJ, U.S.A.) in TBS-T for 2 h. After five washes with TBS-T, the immunoreactivities of the blots were detected using an ECL®-plus Western Blotting Detection system (Amersham Biosciences).
Immunofluorescence microscopy
Confocal microscopy was performed using double indirect immunofluorescence as described previously [22]. Cells were seeded in 2-well Lab-Tek chamber slides (Nalge Nunc International, Naperville, IL, U.S.A.) and incubated with linoleic acid or palmitic acid for 72 h at 37 °C in Eagle's minimum essential medium containing 10% heat-inactivated FBS. At 24 h before fixation (48 h after cell seeding), 120 nM MG132 was added to some plates to stabilize the ubiquitinated proteins. After three washes in PBS at 37 °C, the cells were fixed in 4% (w/v) paraformaldehyde for 15 min at 4 °C. After three further washes in PBS at room temperature (23 °C), the cells were permeabilized with 100% (v/v) methanol for 15 min at 4 °C and were then blocked with 5% (v/v) normal goat serum and 5% (v/v) normal horse serum in PBS for 1 h at room temperature. The cells were then incubated with a mixture of polyclonal and monoclonal antibodies diluted (as noted in the Figure legends) in PBS containing 2% normal goat serum and 2% normal horse serum overnight at 4 °C. After five washes in PBS at room temperature, polyclonal antibodies were detected using goat anti-rabbit IgG labelled with Texas red (1:100) and monoclonal antibodies were detected using horse anti-mouse IgG labelled with fluorescein (1:100; Vector Laboratories, Burlingame, CA, U.S.A.), followed by nuclear counterstaining with 4′,6-diamidino-2-phenylindole (Vector). All preparations were examined with a confocal microscope (LSM 510; Zeiss), equipped with He-Ne, argon and krypton laser sources.
Metabolic labelling and immunoprecipitation
Metabolic labelling and immunoprecipitation experiments were performed as described previously [23]. Cells grown in 25 cm2 flasks, pretreated with linoleic acid or palmitic acid for 72 h, were incubated in methionine/cysteine-free DMEM (Dulbecco's modified Eagle's medium) containing 10% dialysed FBS (Gibco, Grand Island, NY, U.S.A.) for 30 min at 37 °C. Cells were then metabolically labelled for 30 min at 37 °C with 0.3 mCi 35S-methionine/cysteine (Amersham Biosciences) in methionine/cysteine-free DMEM containing 10% dialysed FBS. For chase treatments, cells were incubated for specific periods (0.5–4 h) at 37 °C in Eagle's minimal essential medium containing 10% FBS (HyClone) supplemented with 1 mM unlabelled methionine, with 25 μM linoleic acid or palmitic acid. Cells were harvested and solubilized for 30 min at 4 °C in 1 ml of lysis buffer as detailed in the Western-blot analysis subsection. After centrifugation (15000 g for 10 min at 4 °C), the supernatants were used as cell extracts. Cell extracts were incubated with 20 μl of normal rabbit serum (Sigma) with continuous mixing for 2 h at 4 °C. Protein G–Sepharose 4 Fast Flow (150 μl; Amersham Biosciences) pre-equilibrated with lysis buffer (1:1, v/v) was added and the cell extracts were subjected to further mixing for 2 h at 4 °C. After centrifugation (2000 g for 1 min at 4 °C), the supernatants were used as precleared cell extracts. The radioactivity of each precleared cell extract was measured and 5×106 c.p.m. of each extract was transferred and adjusted in volume to 400 μl. Precleared cell extracts were then mixed with 10 μl of αPEP7. After continuous mixing for 2 h at 4 °C, 20 μl of Protein G–Sepharose suspended in lysis buffer was added and the extracts were further mixed for 2 h at 4 °C. After the antigen–antibody complexes were precipitated by brief centrifugation (2000 g for 1 min at 4 °C), the pellets were washed six times with 800 μl of lysis buffer. Finally, absorbed proteins were eluted with 50 μl of Tris/glycine SDS sample buffer with 2-mercaptoethanol at 95 °C for 5 min. Each supernatant (20 μl) was separated on SDS/8–16% polyacrylamide gels (Invitrogen) and the separated protein bands were visualized by fluorography using Enlightning (NEN Life Science Products, Boston, MA, U.S.A.), dried and exposed to a Bio-Max MR film (Eastman Kodak, Rochester, NY, U.S.A.) for 1–3 days at −80 °C. The expression level of tyrosinase was quantified by measuring the absorbance of specific bands using an image analysis system with NIH Image software (v 1.62).
Melanin measurement
Melanin content was measured spectrophotometrically as follows: after washing twice with PBS, cells were suspended in ethanol/diethyl ether (1:1, v/v) at room temperature for 15 min to remove opaque substances other than melanin, which remain insoluble [24]. After further centrifugation, the precipitates were solubilized with 1 M NaOH containing 10% (v/v) DMSO at 80 °C for 30 min. Absorbance was then measured at 470 nm, and melanin content per cell was calculated and expressed as a percentage of control cells [25].
RESULTS
Alteration of the intracellular composition of fatty acids after treatment with linoleic acid or palmitic acid
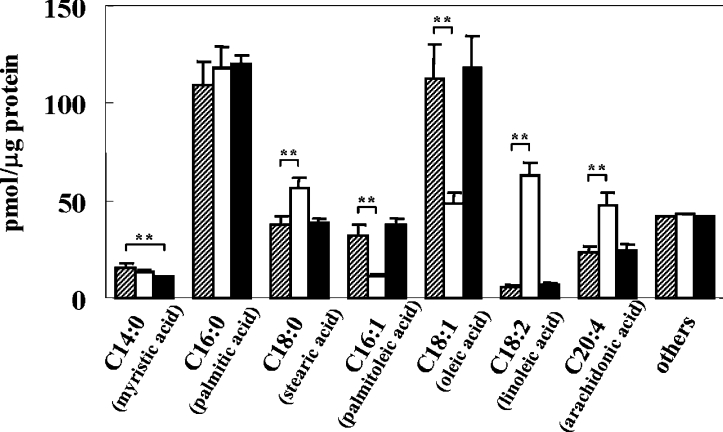
To determine the incorporation and metabolism of fatty acids in B16 mouse melanoma cells, the intracellular composition of fatty acids was analysed after incubation of the cells with 25 μM linoleic acid or palmitic acid for 72 h. In the control cells, the most abundant fatty acids were palmitic acid (C16:0) and oleic acid (C18:1), i.e. the amount and percentage of those fatty acids were 109.0 pmol/μg of protein (28.9%) and 112.6 pmol/μg of protein (29.9%) respectively (Figure 1). After incubation with linoleic acid for 72 h, the amount and percentage of intracellular linoleic acid were increased significantly to 62.7 pmol/μg of protein and 15.7% compared with the control (6.1 pmol/μg of protein and 1.6%). Arachidonic acid (C20:4), a metabolite of linoleic acid, was also increased (from 22.7 pmol/μg of protein in the control to 47.9 pmol/μg of protein), while levels of monounsaturated fatty acids such as oleic acid and palmitoleic acid (C16:1) were decreased significantly (from 112.6 and 31.7 pmol/μg of protein respectively in the controls to 48.3 and 10.7 pmol/μg of protein) following incubation with linoleic acid. In contrast, incubation with palmitic acid for 72 h showed a slight increase in its pre-existing content; however, there was little appreciable difference in other species of fatty acids in cells treated with palmitic acid.
Figure 1. Incubation of linoleic acid dramatically alters the intracellular composition of fatty acids.
Bar graphs of fatty acid composition of B16 melanoma cells after incubation with 25 μM linoleic acid (open bar) or palmitic acid (closed bar) for 72 h, as compared with the control (shaded bar). Results shown are mean±S.D. for triplicate determinations. Student's t test was used for statistical analysis of the data (**P<0.01 versus the control).
Interestingly, the proportion of saturated to unsaturated fatty acids changed very little, even after incubation with linoleic acid, i.e. the dramatic increase in polyunsaturated fatty acids such as linoleic acid and arachidonic acid was compensated by the decrease in monounsaturated fatty acids such as oleic acid and palmitoleic acid (Figure 1). The percentage of unsaturated fatty acids in the total fatty acid content after incubation with linoleic acid or palmitic acid for 72 h was changed only slightly (to 52.3 and 55.4% respectively) compared with the control (55.9%). The total amount of fatty acids after incubation with linoleic acid or palmitic acid was slightly increased to 399.8 and 397.7 pmol/μg of protein respectively as compared with the control (376.6 pmol/μg of protein), indicating that linoleic acid and palmitic acid are incorporated into the cells.
Fatty acids regulate the post-Golgi degradation of tyrosinase after rapid trafficking from the ER
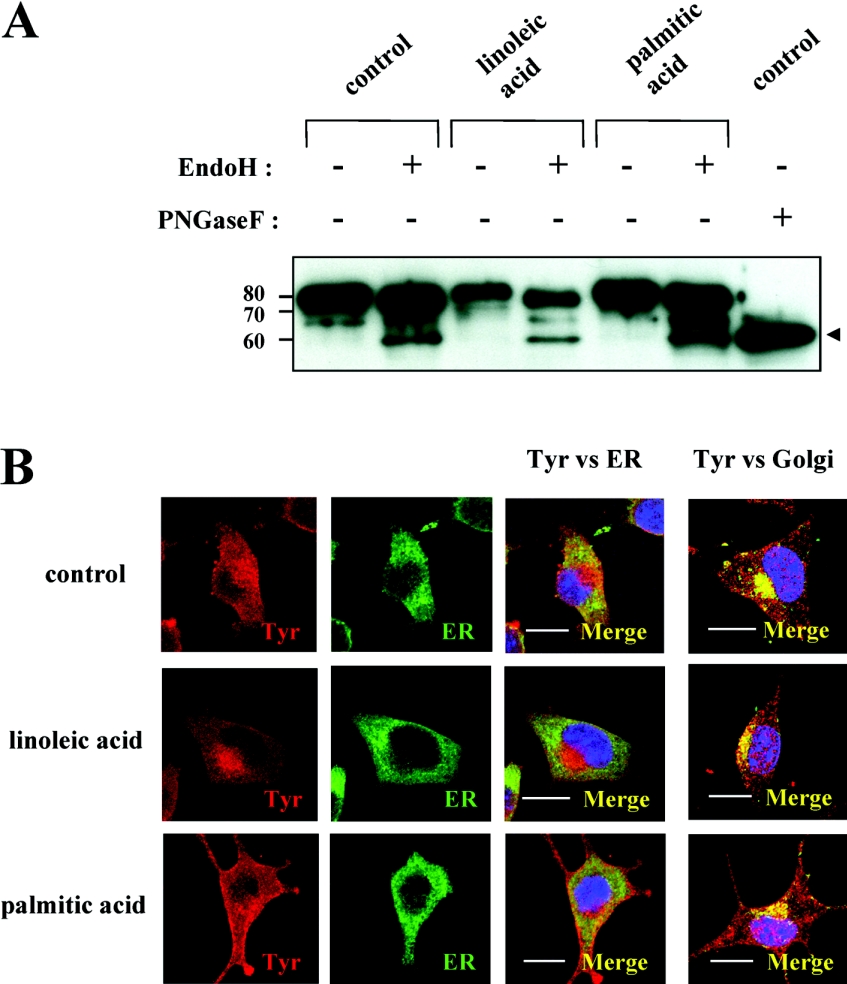
Glycosidase digestion was then used to evaluate whether aberrant trafficking of tyrosinase occurs in fatty acid-treated melanoma cells, as has been observed in amelanotic OCA melanocytes [12–14,17]. Endo H cleaves tyrosinase with early high mannose when it is immature in the ER, whereas PNGase F cleaves immature and mature forms containing complex carbohydrates once tyrosinase has been processed through the Golgi apparatus [26]. Digestion with endo H generated a 60 kDa band, termed the endo H-sensitive band (Figure 2A, arrowhead). Because endo H cleaves immature tyrosinase, the appearance of the endo H-sensitive band represents tyrosinase that has not been processed beyond the ER. Digestion with PNGase F also generated a 60 kDa band representing the fully deglycosylated core polypeptide of tyrosinase. Although the total amount of tyrosinase was decreased by treatment with linoleic acid, those cells contained comparable proportions of endo H-resistant and -sensitive bands. Treatment with palmitic acid increased the total amount of tyrosinase, but also did not affect the endo H-resistant/endo H-sensitive proportion. Comparing the endo H-sensitive bands of control and fatty acid-treated cells, the ratios of those bands were not changed appreciably (20.5–26.8% compared with the endo H-resistant band), indicating that tyrosinase trafficking from the ER to the Golgi is little affected by fatty acids.
Figure 2. Tyrosinase is rapidly processed to the Golgi with little change in sensitivity to glycosidase digestion.
(A) Western blotting of tyrosinase with (+) or without (−) glycosidase digestion with endo H or PNGase F in whole cell lysates (1 μg of protein) of B16 melanoma cells pretreated with linoleic acid (25 μM) or palmitic acid (25 μM) for 72 h. Arrowhead indicates the position of endo H-sensitive and PNGase F-digested band. Numbers on the left indicate masses of protein in kDa. (B) Immunofluorescence confocal microscopy showing the intracellular distribution of tyrosinase, ER and Golgi after incubation with fatty acids. B16 mouse melanoma cells were treated with linoleic acid (25 μM; middle row panels), palmitic acid (25 μM; lower row panels) or DMSO only (control; upper row panels) for 72 h. Cells were stained with antibodies for tyrosinase (αPEP7 at 1:20), ER (Bip/GRP78 at 1:10) or Golgi (Vti1b at 1:10). Reactivity was classified into three categories, according to whether they showed red (tyrosinase), green (ER) or yellow fluorescence. The left two column panels show tyrosinase (Tyr) and ER respectively, while the right two column panels represent the merged images indicating co-localization of tyrosinase with the ER and Golgi respectively as yellow fluorescence. Three independent experiments were performed with similar results and panels showing only single representative cells are shown. Scale bar, 10 μm.
To evaluate the intracellular localization of tyrosinase in the ER and Golgi after treatment with fatty acids, we used confocal immunohistochemistry with αPEP7 (tyrosinase), Bip/GRP78 (a marker of the ER) and Vti1b (a marker of the Golgi). Tyrosinase staining is detected by red fluorescence and ER/Golgi staining is detected by green fluorescence, while in the merged images, yellow fluorescence indicates co-localization of the two signals. Little co-localization of tyrosinase and the ER was observed in the presence or absence of fatty acids (Figure 2B, the second right cascade panels). In contrast, the co-localization of tyrosinase in the Golgi was similarly apparent in the control and fatty acid-treated cells (Figure 2B, the right cascade panels). Similar results were obtained in three independent experiments. These results indicate that tyrosinase is processed rapidly from the ER to the Golgi regardless of the treatment with fatty acids, which is consistent with the glycosidase digestion analyses above.
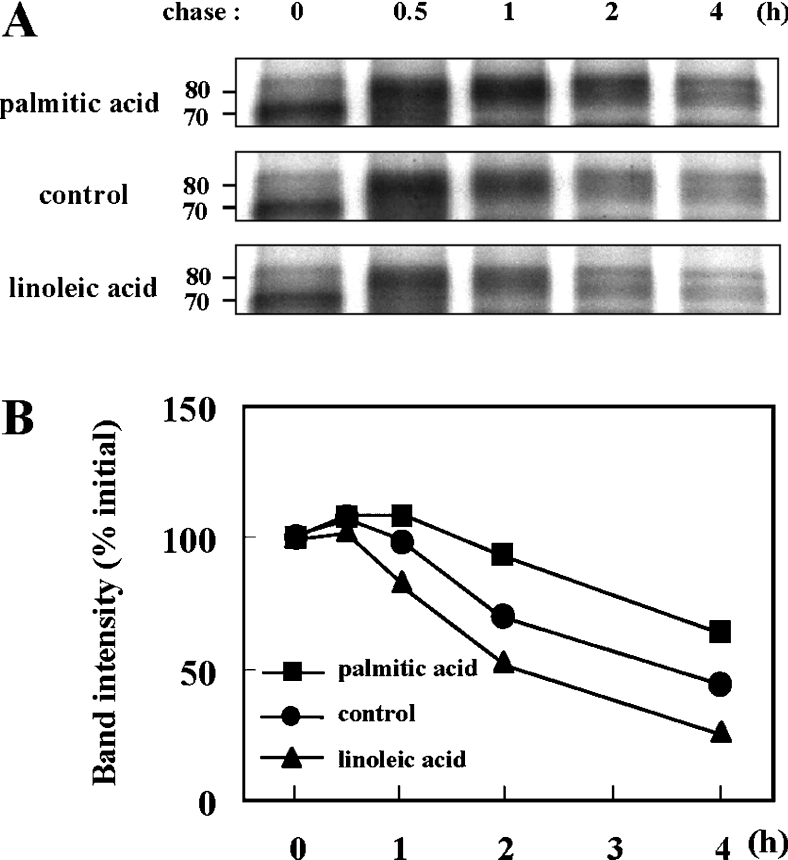
To further characterize the mechanism(s) involved in the regulation of tyrosinase degradation induced by fatty acids, metabolic labelling and immunoprecipitation analyses were performed. The molecular mass of the immature form of tyrosinase is 70 kDa, which results from processing in the ER by the co-translational addition of multiple N-linked glycans to the 60 kDa core polypeptide of tyrosinase [13]. Complex sugar modifications that occur subsequently in the Golgi further increase the molecular mass of tyrosinase to the mature form of 80 kDa [27]. In cells pretreated with linoleic acid or palmitic acid for 72 h, most of the 35S-labelled tyrosinase was of the 70 kDa immature form after a 30 min pulse labelling (no chase). The fully glycosylated mature form of tyrosinase at 80 kDa appeared after a 30 min chase and then the bands decreased in a time-dependent manner (Figure 3A). The band intensities of immature (0 h chase at 70 kDa) and mature (0.5–4 h chase at 80 kDa) tyrosinase were measured and their intensities compared with the initial intensity of each treatment (0 h chase) were normalized to 100% to evaluate the relative stability of tyrosinase (Figure 3B). Our recent experiments using cycloheximide have shown that linoleic acid accelerated, while palmitic acid decelerated, the proteolysis of tyrosinase compared with the control [15]; however, the initial location where the tyrosinase degradation occurs has not been determined. The results obtained in the present study have revealed that the degraded tyrosinase is the fully glycosylated mature form of tyrosinase and that the rate of tyrosinase degradation after incubation with linoleic acid or palmitic acid varied during the first hour of chase at a post-Golgi stage.
Figure 3. Fatty acids regulate the degradation of the glycosylated form of mature tyrosinase.
(A) Metabolic labelling and immunoprecipitation analysis of tyrosinase after pretreatment with linoleic acid (25 μM) or palmitic acid (25 μM) for 72 h, showing the processing and degradation of tyrosinase in B16 melanoma cells. Cells were pulsed for 30 min and then chased for the indicated times. Numbers on the left indicate masses of protein in kDa. Three independent experiments were performed and representative blots are shown. (B) Line graphs of band intensities of immature (0 h, 70 kDa) and glycosylated (0.5–4 h, 80 kDa) tyrosinase pretreated with linoleic acid (▲) or palmitic acid (■) or untreated controls (●). The intensities of the 70 and 80 kDa bands were measured individually. The initial intensity of each band (0 h) is adjusted to 100% and the relative intensities are recalculated to determine tyrosinase degradation. Shown are representative results from three independent experiments.
A proteasome inhibitor elicits retention of tyrosinase in the ER of linoleic acid-treated cells
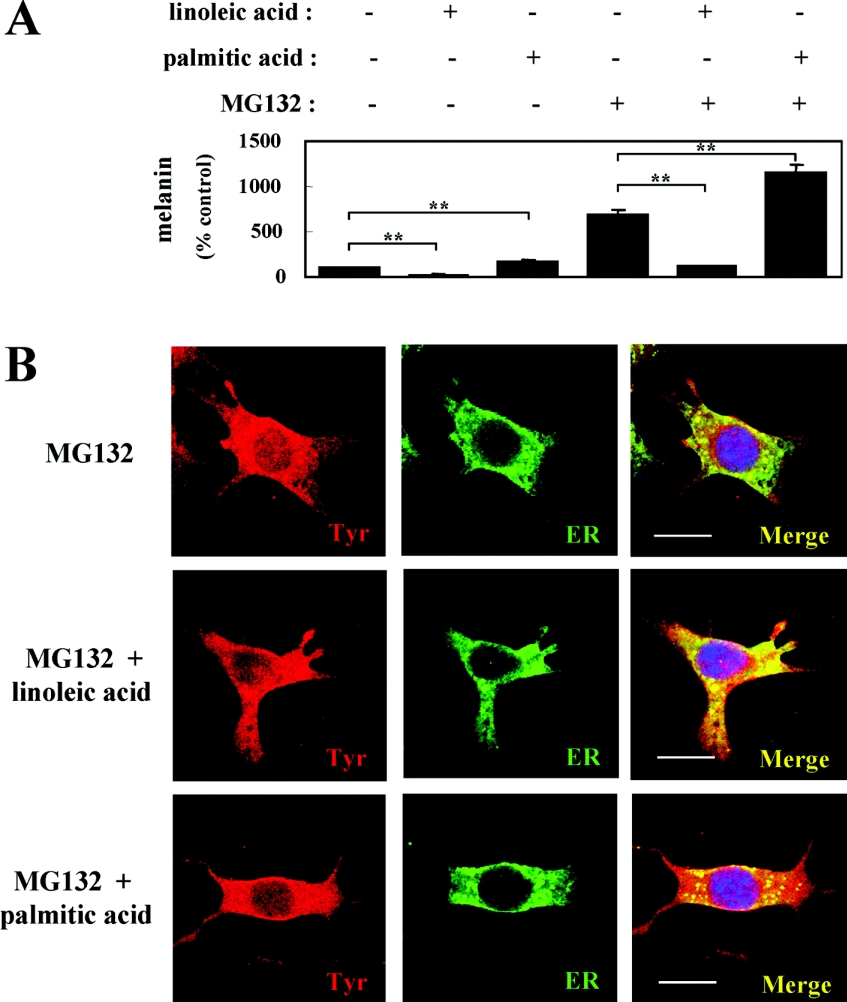
We previously determined that MG132 at a concentration of 120 nM completely blocks the degradation of tyrosinase in B16 melanoma cells [15]. Although tyrosinase degradation could be abrogated by treatment with MG132, the melanin content of cells treated with linoleic acid and MG132 was decreased compared with the control treated with MG132 only, similar to what occurred in the absence of MG132 (Figure 4A).
Figure 4. ER retention of tyrosinase is elicited by linoleic acid incubation when proteasome activity is inhibited.
(A) Melanin measurements of B16 melanoma cells after incubation with (+) or without (−) fatty acids, in the presence (+) or absence (−) of MG132, for 72 h. Results shown are mean±S.D. for triplicate determinations. Student's t test was used for statistical analysis of the data (**P<0.01 versus untreated control or MG132-treated control). (B) Immunofluorescence confocal microscopy showing the intracellular distribution of tyrosinase and the ER after incubation with fatty acids in the presence of MG132. B16 melanoma cells were treated with linoleic acid (25 μM; middle row panels), palmitic acid (25 μM; lower row panels) or DMSO only (control; upper row panels) for 72 h in the presence of MG132 (120 nM) for the last 24 h of treatment before fixation. The merged images indicate co-localization of tyrosinase and the ER as yellow fluorescence. Three independent experiments were performed with similar results and panels showing only single representative cells are shown. Scale bar, 10 μm.
The linoleic acid-induced decrease in melanin content despite the presence of a stable amount of tyrosinase led us to examine whether tyrosinase was retained in the ER during proteasome inhibition. Therefore, to evaluate the intracellular localization of tyrosinase in the ER after incubation with fatty acids in the presence of MG132, we used confocal immunohistochemistry with αPEP7 (tyrosinase) and Bip/GRP78 (an ER marker). In the control cells treated with MG132 only (Figure 4B, upper row panels) and in the cells treated with palmitic acid and MG132 (Figure 4B, lower row panels), the co-localization of the two signals was limited. In contrast, in cells treated with linoleic acid and MG132 (Figure 4B, middle row panels), the co-localization of tyrosinase in the ER was dramatically increased, showing that the retention of tyrosinase in the ER occurred in the presence of linoleic acid and a proteasome inhibitor. Similar results were obtained in three independent experiments. These results indicate that the stabilized tyrosinase induced by MG132 in the linoleic acid-treated cells could be accumulated in the ER and that leads to the decrease in melanin synthesis.
DISCUSSION
Among factors that regulate the ubiquitin–proteasome pathway, fatty acids and amino acids taken from dietary foods are known to be relevant physiological factors [15,28–30]. Our study has revealed that treatment of melanocytic cells with representative fatty acids, e.g. linoleic acid (unsaturated) and palmitic acid (saturated), not only altered the ubiquitination of tyrosinase but also altered the ubiquitination of many kinds of cellular proteins [15]. This prompted us to clarify whether altering the intracellular composition of fatty acids might affect protein turnover mediated by the ubiquitin–proteasome pathway, since fatty acids are known to play an important role in regulating cellular functions. The present study shows that altering the intracellular composition of fatty acids elicits regulatory effects on the proteasomal degradation of tyrosinase at a post-Golgi stage. The dramatic increase in polyunsaturated fatty acids in the intracellular component is coupled with the accelerated degradation of tyrosinase, while the abundance of saturated and monounsaturated fatty acids decelerates its degradation. The contrasting opposite effects of saturated and unsaturated fatty acids on cellular functions have been reported by others [31,32]. For example, saturated fatty acids were found to elevate the serum cholesterol concentration, whereas unsaturated fatty acids were found to decrease it [31]. Further, another recent study revealed that saturated palmitic acid induced the expression of interleukin-6 in human myotubes, whereas unsaturated linoleic acid inhibited the induction of interleukin-6 production by palmitic acid [32]. However, to our knowledge, there have been no reports to date on the reverse modulation of ubiquitin–proteasome function in connection with the intracellular composition of fatty acids.
One interesting point of the present study is that the ratio of saturated to unsaturated fatty acids of total cellular components after incubation with linoleic acid does not change, although remarkable changes of intracellular fatty acid composition occurred in those cells. This indicates that the degradation of tyrosinase is controlled by the individual fatty acid composition, not by the proportion of saturated to unsaturated fatty acids. Our results are consistent with previous studies showing that homoeostatic mechanisms may act to buffer the proportion of saturated to unsaturated fatty acids [33,34]. In those studies, when the experimental oral diets given to rats or monkeys for 5 months were varied in lipid composition (saturated to unsaturated fatty acid ratios), there was little effect on the proportion of total saturated to total unsaturated fatty acids in the cell membranes, although the composition of fatty acids was changed. It is likely that an appropriate proportion of saturated to unsaturated intracellular fatty acids must be regulated to maintain cellular functions. These results may, at least in part, explain why a significant change of fatty acid composition was not observed by treatment with palmitic acid in the present study, i.e. the abundant pre-existing palmitic acid prevents the alteration, possibly via the homoeostatic mechanism.
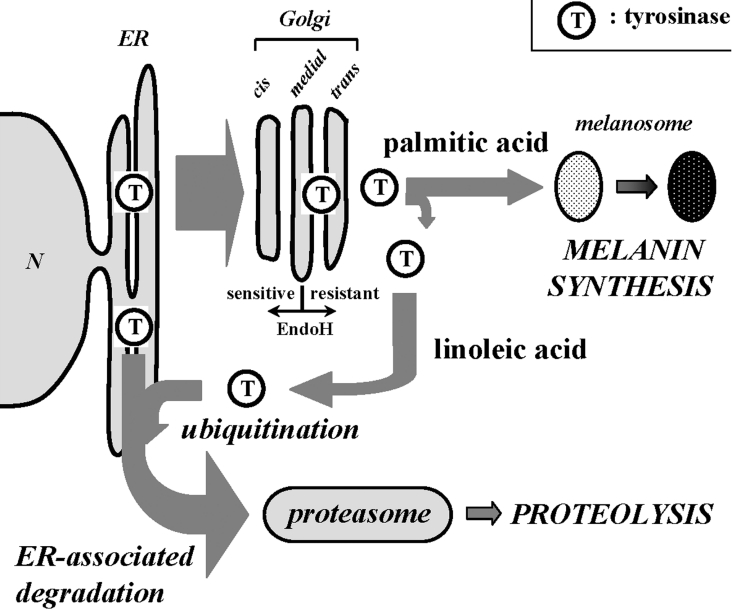
To date, several physiologically relevant factors and synthetic reagents, such as transforming growth factor-β1, unsaturated fatty acid, 2,2′-dihydroxy-5,5′-dipropyl-biphenyl, PMA, 25-hydroxycholesterol and phenylthiourea have been reported to induce the degradation of tyrosinase [35–40]; however, the subcellular location where the degradation takes place was not fully understood. Studies on carbohydrate modifications have revealed that a portion of tyrosinase in the ER is retro-translocated to the cytosol and degraded by the proteasome via ERAD (ER-associated degradation) [41,42]. Recently, it was also found that tyrosinase degradation occurs following complete maturation in the Golgi, suggesting that tyrosinase is also subject to post-Golgi quality control [39,40], but the mechanism of tyrosinase degradation near the Golgi complex has not been clarified. Our study clearly shows that tyrosinase is rapidly processed to the Golgi from the ER during the incubation with fatty acids and that linoleic acid-treated tyrosinase destined to be degraded via the ubiquitin–proteasome pathway appeared in the ER when the proteasome activity was inhibited. Although the precise mechanism(s) by which linoleic acid elicits ER retention of tyrosinase when the degradation is abrogated still remains to be determined, our results suggest that ERAD may play a role, at least in part, in the post-Golgi degradation of tyrosinase (presented schematically in Figure 5).
Figure 5. Scheme depicting the trafficking of tyrosinase after incubation with linoleic acid or palmitic acid.
After rapid processing of tyrosinase from the ER to the Golgi, linoleic acid enhances the ubiquitination of mature tyrosinase and the ubiquitinated tyrosinase could be integrated into the ERAD. In contrast, palmitic acid diminishes the proteasomal degradation of tyrosinase, leading to melanin formation in melanosomes.
The retention of tyrosinase in the ER, as observed in OCA, was elicited only when the cells were cultured with linoleic acid and MG132. The decrease in melanin synthesis despite comparable amounts of tyrosinase due to the abrogation of tyrosinase degradation could be explained by the accumulation of tyrosinase in the ER, and its lack of sorting to melanosomes where melanin is synthesized. The recent demonstration that linoleic acid enhances the ubiquitination of tyrosinase [15] suggests that the ubiquitinated tyrosinase could be retained in the ER. The notion that ubiquitinated tyrosinase may be retained in the ER is also supported by studies showing that ubiquitination is required for the retro-translocation of a glycoprotein to the cytosol for degradation by proteasomes [43] and that the full-length tyrosinase in the ER could be retro-translocated to the cytosol [44]. In contrast, in the cells treated with MG132 alone or in combination with palmitic acid, large amounts of melanin were produced, and the quick movement of tyrosinase from the ER to the cytosol was observed. Similar results have been reported in previous studies, i.e. tyrosinase could be detected in the cytosol after inhibition of proteasome activity [44,45] or of vacuolar proton ATPase [46]. Taking these findings into consideration, the retention of tyrosinase in the ER would probably not be elicited only by inhibiting proteasome activity. Although we do not completely understand the interaction between the retention of tyrosinase in the ER and ubiquitination, we believe that the movement of tyrosinase out of the ER can be interrupted by the dysfunction of proteasomes when a substrate is ubiquitinated. Further study will be needed to clarify the precise mechanism(s) involved in this process and to explain why ubiquitinated tyrosinase could be retained in the ER when the proteasome activity is inhibited.
In summary, the present study investigated the relationship between intracellular fatty acid composition and the degradation of proteins via the ubiquitin–proteasome pathway, focusing on tyrosinase as a model protein. Our results suggest that altering the intracellular fatty acid composition, but not the proportion of saturated to unsaturated fatty acid, may affect the degradation of membrane glycoproteins such as tyrosinase. In addition, the dysfunction of proteasomes elicits the retention of tyrosinase in the ER when polyunsaturated fatty acids are increased and monounsaturated fatty acids are decreased in the intracellular component. In the present study, we only analysed the total cellular fatty acid composition. Further studies of the fatty acid composition of subcellular organelles will be required for a deeper understanding of their roles in regulating the trafficking of tyrosinase and other proteins. Since the composition of fatty acids in living cells in vivo could be altered by dietary intake, our in vitro system to modulate cellular components of fatty acids will be a useful model to study the roles of fatty acids in cellular functions.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Tsujishita Y., Asaoka Y., Nishizuka Y. Regulation of phospholipase A2 in human leukemia cell lines: its implication for intracellular signaling. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6274–6278. doi: 10.1073/pnas.91.14.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 3.Clarke S. D. The multi-dimensional regulation of gene expression by fatty acids: polyunsaturated fats as nutrient sensors. Curr. Opin. Lipidol. 2004;15:13–18. doi: 10.1097/00041433-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Tahin Q. S., Blum M., Carafoli E. The fatty acid composition of subcellular membranes of rat liver, heart, and brain: diet-induced modifications. Eur. J. Biochem. 1981;121:5–13. doi: 10.1111/j.1432-1033.1981.tb06421.x. [DOI] [PubMed] [Google Scholar]
- 5.Awad A. B., Chattopadhyay J. P. Effect of dietary saturated fatty acids on intracellular free fatty acids and kinetic properties of hormone-sensitive lipase of rat adipocytes. J. Nutr. 1986;116:1095–1100. doi: 10.1093/jn/116.6.1095. [DOI] [PubMed] [Google Scholar]
- 6.Benediktsdottir V. E., Gudbjarnason S. Modification of the fatty acid composition of rat heart sarcolemma with dietary cod liver oil, corn oil or butter. J. Mol. Cell. Cardiol. 1988;20:141–147. doi: 10.1016/s0022-2828(88)80027-0. [DOI] [PubMed] [Google Scholar]
- 7.Piche L. A., Mahadevappa V. G. Modification of rat platelet fatty acid composition by dietary lipids of animal and vegetable origin. J. Nutr. 1990;120:444–449. doi: 10.1093/jn/120.5.444. [DOI] [PubMed] [Google Scholar]
- 8.Marteinsdottir I., Horrobin D. F., Stenfors C., Theodorsson E., Mathe A. A. Changes in dietary fatty acids alter phospholipid fatty acid composition in selected regions of rat brain. Prog. Neuropsychopharmacol. Biol. Psychiatry. 1998;22:1007–1021. doi: 10.1016/s0278-5846(98)00052-9. [DOI] [PubMed] [Google Scholar]
- 9.Hiller M. M., Finger A., Schweiger M., Wolf D. H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 12.Halaban R., Cheng E., Zhang Y., Moellmann G., Hanlon D., Michalak M., Setaluri V., Hebert D. N. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halaban R., Svedine S., Cheng E., Smicun Y., Aron R., Hebert D. N. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5889–5894. doi: 10.1073/pnas.97.11.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyofuku K., Wada I., Valencia J. C., Kushimoto T., Ferrans V. J., Hearing V. J. Oculocutaneous albinism types 1 and 3 are ER retention diseases: mutation of tyrosinase or Tyrp1 can affect the processing of both mutant and wild-type proteins. FASEB J. 2001;15:2149–2161. doi: 10.1096/fj.01-0216com. [DOI] [PubMed] [Google Scholar]
- 15.Ando H., Watabe H., Valencia J. C., Yasumoto K., Furumura M., Funasaka Y., Oka M., Ichihashi M., Hearing V. J. Fatty acids regulate pigmentation via proteasomal degradation of tyrosinase. J. Biol. Chem. 2004;279:15427–15433. doi: 10.1074/jbc.M313701200. [DOI] [PubMed] [Google Scholar]
- 16.Jiménez M., Tsukamoto K., Hearing V. J. Tyrosinases from two different loci are expressed by normal and by transformed melanocytes. J. Biol. Chem. 1991;266:1147–1156. [PubMed] [Google Scholar]
- 17.Watabe H., Valencia J. C., Yasumoto K., Kushimoto T., Ando H., Vieira W. D., Mizogichi M., Appella E., Hearing V. J. Regulation of tyrosinase processing and trafficking by organellar pH and by proteasome activity. J. Biol. Chem. 2004;279:7971–7981. doi: 10.1074/jbc.M309714200. [DOI] [PubMed] [Google Scholar]
- 18.Chen K., Manga P., Orlow S. J. Pink-eyed dilution protein controls the processing of tyrosinase. Mol. Biol. Cell. 2002;13:1953–1964. doi: 10.1091/mbc.02-02-0022.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ando H., Itoh A., Mishima Y., Ichihashi M. Correlation between the number of melanosomes, tyrosinase mRNA levels, and tyrosinase activity in cultured murine melanoma cells in response to various melanogenesis regulatory agents. J. Cell. Physiol. 1995;163:608–614. doi: 10.1002/jcp.1041630322. [DOI] [PubMed] [Google Scholar]
- 20.Wen Z., Kim H. Y. Alterations in hippocampal phospholipids profile by prenatal exposure to ethanol. J. Neurochem. 2004;89:1368–1377. doi: 10.1111/j.1471-4159.2004.02433.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim H. Y., Salem N. Separation of lipid classes by solid phase extraction. J. Lipid Res. 1990;31:2285–2289. [PubMed] [Google Scholar]
- 22.Kushimoto T., Basrur V., Matsunaga J., Vieira W. D., Muller J., Appella E., Hearing V. J. A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10698–10703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasumoto K., Watabe H., Valencia J. C., Kushimoto T., Kobayashi T., Appella E., Hearing V. J. Epitope mapping of the melanosomal matrix protein gp100 (PMEL17): rapid processing in the endoplasmic reticulum and glycosylation in the early Golgi compartment. J. Biol. Chem. 2004;279:28330–28338. doi: 10.1074/jbc.M401269200. [DOI] [PubMed] [Google Scholar]
- 24.Oikawa A., Nakayasu M. Quantitative measurement of melanin as tyrosine equivalents and as weight of purified melanin. Yale J. Biol. Med. 1973;46:500–507. [PMC free article] [PubMed] [Google Scholar]
- 25.Ando H., Ryu A., Hashimoto A., Oka M., Ichihashi M. Linoleic acid and α-linolenic acid lightens ultraviolet-induced hyperpigmentation of the skin. Arch. Dermatol. Res. 1998;290:375–381. doi: 10.1007/s004030050320. [DOI] [PubMed] [Google Scholar]
- 26.Costin G. E., Valencia J. C., Vieira W. D., Lamoreux M. L., Hearing V. J. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J. Cell Sci. 2003;116:3203–3212. doi: 10.1242/jcs.00598. [DOI] [PubMed] [Google Scholar]
- 27.Halaban R., Pomerantz S. H., Marshall S., Lambert D. T., Lerner A. B. Regulation of tyrosinase in human melanocytes grown in culture. J. Cell Biol. 1983;97:480–488. doi: 10.1083/jcb.97.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlmann B., Rutschmann M., Kuehn L., Reinauer H. Activation of the multicatalytic proteinase from rat skeletal muscle by fatty acids or sodium dodecyl sulphate. Biochem. J. 1985;228:171–177. doi: 10.1042/bj2280171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe N., Yamada S. Activation of 20S proteasomes from spinach leaves by fatty acids. Plant Cell Physiol. 1996;37:147–151. doi: 10.1093/oxfordjournals.pcp.a028925. [DOI] [PubMed] [Google Scholar]
- 30.Hamel F. G., Upward J. L., Siford G. L., Duckworth W. C. Inhibition of proteasome activity by selected amino acids. Metabolism. 2003;52:810–814. doi: 10.1016/s0026-0495(03)00094-5. [DOI] [PubMed] [Google Scholar]
- 31.Dietschy J. M. Dietary fatty acids and the regulation of plasma low density lipoprotein cholesterol concentrations. J. Nutr. 1998;128:444S–448S. doi: 10.1093/jn/128.2.444S. [DOI] [PubMed] [Google Scholar]
- 32.Weigert C., Brodbeck K., Staiger H., Kausch C., Machicao F., Haring H. U., Schleicher E. D. Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappaB. J. Biol. Chem. 2004;279:23942–23952. doi: 10.1074/jbc.M312692200. [DOI] [PubMed] [Google Scholar]
- 33.Gibson R. A., McMurchie E. J., Charnock J. S., Kneebone G. M. Homeostatic control of membrane fatty acid composition in the rat after dietary lipid treatment. Lipids. 1984;19:942–951. doi: 10.1007/BF02534730. [DOI] [PubMed] [Google Scholar]
- 34.McMurchie E. J., Gibson R. A., Charnock J. S., McIntosh G. H. Mitochondrial membrane fatty acid composition in the marmoset monkey following dietary lipid supplementation. Lipids. 1986;21:315–323. doi: 10.1007/BF02535693. [DOI] [PubMed] [Google Scholar]
- 35.Martínez-Esparza M., Jiménez-Cervantes C., Beermann F., Aparicio P., Lozano J. A., García-Borrón J. C. Transforming growth factor-β1 inhibits basal melanogenesis in B16/F10 mouse melanoma cells by increasing the rate of degradation of tyrosinase and tyrosinase-related protein-1. J. Biol. Chem. 1997;272:3967–3972. doi: 10.1074/jbc.272.7.3967. [DOI] [PubMed] [Google Scholar]
- 36.Ando H., Funasaka Y., Oka M., Ohashi A., Furumura M., Matsunaga J., Matsunaga N., Hearing V. J., Ichihashi M. Possible involvement of proteolytic degradation of tyrosinase in the regulatory effect of fatty acids on melanogenesis. J. Lipid Res. 1999;40:1312–1316. [PubMed] [Google Scholar]
- 37.Nakamura K., Yoshida M., Uchiwa H., Kawa Y., Mizoguchi M. Down-regulation of melanin synthesis by a biphenyl derivative and its mechanism. Pigment Cell Res. 2003;16:494–500. doi: 10.1034/j.1600-0749.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 38.Kageyama A., Oka M., Okada T., Nakamura S., Ueyama T., Saito N., Hearing V. J., Ichihashi M., Nishigori C. Down-regulation of melanogenesis by phospholipase D2 through ubiquitin proteasome-mediated degradation of tyrosinase. J. Biol. Chem. 2004;279:27774–27780. doi: 10.1074/jbc.M401786200. [DOI] [PubMed] [Google Scholar]
- 39.Hall A. M., Krishnamoorthy L., Orlow S. J. 25-Hydroxycholesterol acts in the Golgi compartment to induce degradation of tyrosinase. Pigment Cell Res. 2004;17:396–406. doi: 10.1111/j.1600-0749.2004.00161.x. [DOI] [PubMed] [Google Scholar]
- 40.Hall A. M., Orlow S. J. Degradation of tyrosinase induced by phenylthiourea occurs following Golgi maturation. Pigment Cell Res. 2005;18:122–129. doi: 10.1111/j.1600-0749.2005.00213.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Androlewicz M. J. Oligosaccharide trimming plays a role in the endoplasmic reticulum-associated degradation of tyrosinase. Biochem. Biophys. Res. Commun. 2000;271:22–27. doi: 10.1006/bbrc.2000.2577. [DOI] [PubMed] [Google Scholar]
- 42.Svedine S., Wang T., Halaban R., Hebert D. N. Carbohydrates act as sorting determinants in ER-associated degradation of tyrosinase. J. Cell Sci. 2004;117:2937–2949. doi: 10.1242/jcs.01154. [DOI] [PubMed] [Google Scholar]
- 43.de Virgilio M., Weninger H., Ivessa N. E. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J. Biol. Chem. 1998;273:9734–9743. doi: 10.1074/jbc.273.16.9734. [DOI] [PubMed] [Google Scholar]
- 44.Mosse C. A., Meadows L., Luckey C. J., Kittlesen D. J., Huczko E. L., Slingluff C. L., Shabanowitz J., Hunt D. F., Engelhard V. H. The class I antigen-processing pathway for the membrane protein tyrosinase involves translation in the endoplasmic reticulum and processing in the cytosol. J. Exp. Med. 1998;187:37–48. doi: 10.1084/jem.187.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosse C. A., Hsu W., Engelhard V. H. Tyrosinase degradation via two pathways during reverse translocation to the cytosol. Biochem. Biophys. Res. Commun. 2001;285:313–319. doi: 10.1006/bbrc.2001.5153. [DOI] [PubMed] [Google Scholar]
- 46.Halaban R., Patton R. S., Cheng E., Svedine S., Trombetta E. S., Wahl M. L., Ariyan S., Hebert D. N. Abnormal acidification of melanoma cells induces tyrosinase retention in the early secretory pathway. J. Biol. Chem. 2002;277:14821–14828. doi: 10.1074/jbc.M111497200. [DOI] [PubMed] [Google Scholar]