Abstract
A 65 kDa GPI (glycosylphosphatidyl-inositol)-anchored ALP (alkaline phosphatase) was characterized as a functional receptor of the Bacillus thuringiensis subsp. israelensis Cry11Aa toxin in Aedes aegypti midgut cells. Two (a 100 kDa and a 65 kDa) GPI-anchored proteins that bound Cry11Aa toxin were preferentially extracted after treatment of BBMV (brush boder membrane vesicles) from Ae. aegypti midgut epithelia with phospholipase C. The 65 kDa protein was further purified by toxin affinity chromatography. The 65 kDa protein showed ALP activity. The peptide-displaying phages (P1.BBMV and P8.BBMV) that bound to the 65 kDa GPI–ALP (GPI-anchored ALP) and competed with the Cry11Aa toxin to bind to BBMV were isolated by selecting BBMV-binding peptide-phages by biopanning. GPI–ALP was shown to be preferentially distributed in Ae. aegypti in the posterior part of the midgut and in the caeca, by using P1.BBMV binding to fixed midgut tissue sections to determine the location of GPI–ALP. Cry11Aa binds to the same regions of the midgut and competed with P1.BBMV and P8.BBMV to bind to BBMV. The importance of this interaction was demonstrated by the in vivo attenuation of Cry11Aa toxicity in the presence of these phages. Our results shows that GPI–ALP is an important receptor molecule involved in Cry11Aa interaction with midgut cells and toxicity to Ae. aegypti larvae.
Keywords: alkaline phosphatase, Cry-receptor, GPI-anchor, mosquitocidal toxins, phage display
Abbreviations: ALP, alkaline phosphatase; APN, aminopeptidase-N; BBMV, brush border membrane vesicles; Cry, crystal protein; GPI, glycosylphosphatidyl-inositol; pfu, plaque-forming units; PIPLC, phospholipase-C
INTRODUCTION
The occurrence of arthropod-borne human diseases continues to pose significant health risks in many parts of the world. The difficulties of controlling insect vectors for these diseases is compounded by the resistance of some vector populations, not only to chemical insecticides, but more recently also to Bacillus sphaericus. An important alternative method for control of these insect vectors is through the use of Bacillus thuringiensis subsp. israelensis, which is toxic to different mosquito and black fly species [1]. Consequently, formulations using this bacterial strain are being increasingly used worldwide for the control of insect vector populations.
B. thuringiensis subsp. israelensis produces crystal inclusions during sporulation that are composed principally of six toxins: Cry4Aa, Cry4Ba, Cry10Aa, Cry11Aa, Cyt1Aa and Cyt2Ba [2]. The crystal toxins when ingested by susceptible larvae dissolve in the alkaline environment of the larval midgut and release soluble proteins. The inactive Cry (crystal protein) protoxins are then cleaved at specific sites by midgut proteases yielding active 60 kDa protease resistant fragments [3]. In the case of the 70 kDa Cry11Aa protoxin, proteolytic activation involves both cleavage of 28 amino-terminal residues and intra-molecular cleavage, this results in the formation of two fragments of 36 and 32 kDa. These fragments, however, remain associated and retain insect toxicity [4–7]. The active toxin then binds to specific membrane receptors on the apical brush border of the midgut epithelium columnar cells, leading to oligomerization, membrane insertion and pore formation [3].
Receptor binding is a key factor in determining the specificity of Cry toxins. For Cry1A lepidopteran toxins, at least four different protein receptors have been characterized; a CADP (cadherin-like protein), a GPI (glycosylphosphatidyl-inositol)-anchored APN (aminopeptidase-N), a GPI-anchored ALP (alkaline phosphatase) and a 270 kDa glycoconjugate [8–12]. Previously, we provided evidence that, in Manduca sexta, binding of monomeric Cry1Ab toxin to the cadherin receptor promotes complete proteolytic activation of the toxin, facilitating the formation of a pre-pore oligomeric structure that is the membrane insertion competent [13]. The pre-pore oligomer then binds to a second receptor, GPI-anchored APN, leading to toxin insertion into membrane lipid rafts [13].
In the case of Cry11Aa and Cry4Ba mosquitocidal toxins, two toxin binding proteins of 65 and 62 kDa were identified on BBMV (brush border membrane vesicles) from Ae. aegypti larvae by toxin overlay assays and the 62 kDa was shown to be a degradation product of the 65 kDa protein [14]. The identity of these binding molecules and their role as receptors of the Cry11Aa and Cry4Ba toxins, however, is still unknown. Also, two midgut APNs were identified in Aedes aegypti larvae, although no evidence for their role in Cry11Aa or Cry4Ba binding was provided [15].
The crystal structures of the chymotrypsin-, trypsin-, or papain- activated Cry4Ba (dipteran specific), Cry1Aa (lepidopteran specific) and Cry3A (coleopteran specific) toxins, and the protoxin of Cry2Aa (dipteran–lepidopteran specific), have been solved [16–19]. The structures have common features: the-N-terminal domain I is formed of seven α-helices that have been characterized as the pore-forming domain, domain II consists of three anti-parallel β-sheets with exposed loop regions and domain III is a β-sandwich [16–19]. Domains II and III are important for receptor recognition and, in particular, loop regions of domain II are involved in receptor binding (for a review see [20]). In the case of Cry11Aa toxin, the predicted loop α8 (and probably β4–β5 region and loop 3 of domain II) was shown to be important binding epitopes involved in interaction with mosquito midgut receptors [21].
The identity of the Cry11Aa receptor(s) remains unknown. GPI-anchored proteins have been shown to be important receptor proteins of other insecticidal toxins. For the Cry1A toxins both the APN and ALP receptors are GPI-anchored proteins [10,11]. Also, a GPI-anchored α-glucosidase [Cpm1(Culex pipiens maltase 1)] is the functional receptor in C. pipiens for the mosquitocidal Bin toxin produced by B. sphaericus [22]. In the present study we analysed the role of GPI-anchored proteins in the binding of Cry11Aa to Ae. aegypti BBMV and report the purification and partial characterization of a Cry11Aa receptor. We identified two GPI-anchored proteins of 65 kDa and 62 kDa that bound to the Cry11Aa toxin and had ALP activity. In addition, we identified two peptide-displaying phages (P1.BBMV and P8.BBMV) that bound this 65 kDa ALP protein. The P1.BBMV phage interfered with the interaction of Cry11Aa with Ae. aegypti BBMV and also interfered with the toxicity of Cry11Aa toxin to mosquito larvae. Immunofluorescence experiments using P1.BBMV revealed that the GPI–ALP is located in the same midgut sites that Cry11Aa toxin binds, which are in the caeca and posterior midgut. Our results show that GPI–ALP is a functional Cry11Aa receptor in Ae. aegypti midgut.
EXPERIMENTAL
Toxin Cry11Aa purification and activation
The Bt (B. thuringiensis) CG6 strain harbouring the pCG6 plasmid that encodes the cry11Aa gene [23] was used for Cry11Aa production. The CG6 strain was grown for 72 h in sporulation medium [24] plus erythromycin (25 μg/ml) and was shaken at 200 r.p.m. at 30 °C until complete sporulation. The spores and crystal were harvested and washed three times with buffer containing 1 mM EDTA, 0.1 mM PMSF, 1 μg/ml pepstatin, 5 μg/ml leupeptin in PBS [150 mM NaCl, 2.8 mM NaH2PO4, 4 mM Na2HPO4. 7H2O (pH 7.2)]. Crystals were isolated using sucrose gradients as previously described in [25]. The crystals were recovered in 50 mM Tris/HCl (pH 8.0), 1 mM PMSF, solubilized for 1 h at 4 °C with 0.1 M NaOH and activated with trypsin (1:50 dilution; w/w) for 2 h at 25 °C. The molecular size and the integrity of proteins were determined using SDS/15% PAGE.
Preparation of BBMV
Ae. aegypti larvae, hatched from eggs kindly supplied by Dr J. Ibarra (Departamento de Biotecnología y Bioquímica, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico), were fed cat food pellets. BBMV were prepared from dissected midguts of fourth instar larvae by differential precipitation using MgCl2, as previously reported in [26], and stored at −70 °C until used.
Qualitative binding assays of Cry11Aa toxin to BBMV
The binding of Cry11Aa toxin to Ae. aegypti BBMV was performed as described in [27]. Binding was performed in 100 μl binding buffer [PBS (pH 7.6), 0.1% (w/v) BSA, 0.1% (v/v) Tween 20]. BBMV protein (10 μg) was incubated with biotinylated toxin (10 nM) in the presence or absence of a several-fold excess of either unlabelled toxin, different phages or synthetic peptides for 1 h. The unbound toxin was removed by centrifugation (10 min at 14000 g). BBMV were washed three times in binding buffer and suspended in 15 μl of PBS, 5 μl of Laemmli sample loading buffer 4X [0.125 M Tris/HCl (pH 6.8), 4% (w/v) SDS, 20% (v/v) glycerol, 10% (v/v) 2-mercaptoethanol, 0.01% (w/v) Bromophenol Blue. The samples were boiled for 5 min, separated by SDS/10% PAGE, and electroblotted on to Hybond nitrocellulose membranes (Amersham Biosciences). The biotinylated-Cry11Aa toxin was visualized by incubating membranes with streptavidin–peroxidase conjugate (1:6000 dilution; Amersham Biosciences) for 1 h followed by incubation with SuperSignal chemiluminescence substrate (Pierce) as described in the manufacturer's protocol. Synthetic peptides were synthesized by Invitrogene and their sequence is shown in Table 1.
Table 1. Synthetic peptide sequences.
| Name | Sequence | Description |
|---|---|---|
| Loopα8 | GVSIPVNYNEWY | Amino acid sequence of residues 257–268 Cry11Aa |
| β4 | GNGRTNNFNFADNN | Amino acid sequence of residues 323–336 Cry11Aa |
| Loop3 | LTYNRIEYDSPTTEN | Amino acid sequence of residues 447–461 Cry11Aa |
| P1.BBMV | AAKTMDTLRPPR | Sequence of peptide displayed in P1.BBMV |
Toxin overlay assay
Analysis of BBMV proteins recognized by Cry11Aa or by peptide-displaying phages was as follows. BBMV proteins (10 μg) were separated through SDS/10% PAGE gels and electroblotted on to Hybond nitrocellulose membranes as described above. After blocking, the membranes were incubated for 1 h with 30 nM Cry11A or 1×1010 pfu (plaque-forming units)/ml phages in blocking buffer [0.1M NaHCO3 (pH 9.6), 5 mg/ml BSA, 0.02% (w/v) NaN3] at 25 °C. Unbound toxin or phages were removed by washing six times for 10 min in washing buffer [PBS, 0.1% (v/v) Tween 20] at 25 °C. The bound toxin was identified by incubation with anti-Cry11A rabbit antibody and anti-rabbit HRP (horseradish peroxidase) antibody conjugate (1:2000 dilution), while the bound phages were identified by incubation with anti-M13 mouse antibody and anti-mouse peroxidase conjugate (1:2000 dilution), for 1 h at 25 °C. Binding was visualized using SuperSignal chemiluminescence substrate as described above.
Purification of GPI-anchored proteins that interact with Cry11A toxin
Ae. aegypti BBMV (300 μg) were suspended in 500 μl of PBS2 [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 (pH 7.2)]. PIPLC (phospholipase-C) (3 units) of from Bacillus cereus (Boehringer Mannheim) was added to the BBMV suspension and incubated for 90 min at 30 °C, as previously reported in [28]. Membranes were recovered by centrifugation (11000 g, for 20 min), and the supernatant was analysed for the presence of GPI-anchored proteins by SDS/10% PAGE and Cry11A toxin overlay as described above. Then Cry11Aa was biotinylated using biotinyl-N-hydroxysuccinimide ester (catalogue number RPN28; Amersham Biosciences) according to the manufacturer's directions. A matrix of agarose–streptavidin (200 μl; Sigma–Aldrich) was incubated with 7 μg of biotinylated-Cry11A toxin overnight at 4 °C. After incubation, the matrix was washed ten times in 1X PBS2 to remove any unbound protein. The solution containing GPI-anchored proteins (42 μg) was incubated with the 200 μl of Cry11Aa–agarose matrix. The matrix was washed four times with 1X PBS2. The binding proteins were then eluted using 200 μl of 1X PBS2, pH 9.5. The pH was adjusted to 7.2 and the samples were stored at 4 °C in the presence of protease inhibitors.
ALP and APN specific activities
Specific ALP and APN enzymatic activities were measured using 1 mg/ml of p-nitrophenyl phosphate and leucine-p-nitroanilide (Sigma–Aldrich) as substrates respectively. BBMV proteins (5 μg) were mixed with ALP buffer [0.5 mM MgCl2, 100 mM Tris/HCl (pH 9.5)] containing 1.25 mM of p-nitrophenyl phosphate or APN buffer [0.2 M Tris/HCl (pH 8), 0.25 M NaCl] containing 1 mM of leucine-p-nitroanilide. Enzyme activities were monitored as the change in the absorbance at 450 nm for 3 min at 25 °C.
Phage display library
The phage-peptide library (Ph.D.-12 phage display peptide library kit) used in the present study was obtained from New England BioLabs. This library has a complexity of 2.7×109 transformants, and is based on a combinatorial library of random 12-mers fused to the minor coat protein (pIII) of the M13 phage.
Selection and sequencing of peptide-displaying phage clones
BBMV-binding phages were selected by a panning procedure performed in solution. Briefly, BBMVs (185 μg total protein) were incubated with the phage-library (1×1011 pfu) in washing buffer for 1 h at 25 °C. Bound phages were harvested by centrifugation (11000 g for 20 min) and washed six times in washing buffer. The phages were obtained by incubation with elution buffer [0.2 M glycine/HCl (pH 2.2), 1 mg/ml BSA] for 10 min at 25 °C. Phages were neutralized with 1M Tris/HCl (pH 9.1) and used to perform subsequent rounds of selection. To increase the specificity of the selected phages, increasing concentrations of Tween 20 [0.1 to 0.4%(v/v)], were employed in each panning round. There were four rounds of panning against BBMV performed.
The DNA from independent phages, selected after the third and fourth rounds in both panning procedures, was purified and analysed by sequencing using the primer 5′-GCCCTCATAGTTAGCGTAACG-3′ provided in the Ph.D.-12 phage display peptide library kit (New England Biolabs).
Binding assay of phages to BBMV
Phages (103) were incubated in 100 μl of binding buffer with 10 μg of BBMV, in the presence or absence of competitors, for 1 h at 25 °C. The competitors used were 10 nM Cry11Aa toxin or 1000 nM synthetic peptides. After incubation, the unbound phages and competitors were removed by centrifugation (10 min at 11000 g). The BBMV pellet was washed six times in PBS with 0.1% (v/v) Tween 20 by centrifugation at 11000 g and 25 °C for 10 min. The phage–BBMV complex was separated by acidic treatment with 0.2 M glycine/HCl (pH 2.2), 1 mg/ml BSA and neutralized with 0.15 M Tris/HCl (pH 9.1). The pfu were quantified by infection of an ER2738 Escherichia coli culture in the exponential phase of growth (0.7 D600).
Immunohistochemistry on paraffin sections
Early fourth-instar larvae were fixed in 4% (v/v) paraformaldehyde in PBS overnight at 4 °C. Fixed larvae were washed three times in PBS and dehydrated in an ethanol series [20, 40, 70, 100% (v/v)] in PBS for 20 min per step at 25 °C and kept overnight at 25 °C in 100% ethanol to ensure complete dehydration. Larvae were then incubated in ethanol/xylene mixtures [70/30 and then 30/70 (v/v)] for 3 h per mixture and then in 100% xylene overnight. Paraffin chips were added, intially at 20–50% of the total xylene volume, first at 25 °C for 3 h and then at 55 °C overnight. Finally, samples were incubated four times for 12 h per incubation with 100% paraffin at 55 °C. The larvae were embedded in paraffin blocks. The blocks were sectioned, 8–10 μm thick, and the sections placed on glass slides coated with gelatin. The slides were washed twice in 100% xylene for 10 min to eliminate the paraffin, re-hydrated by serial ethanol washes [100, 70, 40, 20% (v/v)] and washed in double distilled water for 10 min. The slides were treated with blocking solution [2% (w/v) BSA, 0.1% (v/v) Triton X-100 in PBS] for 2 h at 25 °C. The slides were then incubated with Cry11Aa toxin (10 nM) or phages (1×103 pfu), in the absence or presence of different competitors, for 1 h in blocking solution at 25 °C. The slides were washed six times in washing solution 1 [0.1% (w/v) BSA, 0.1% (v/v) Triton X-100 in PBS] to eliminate unbound toxin or phages, and then incubated overnight at 4 °C with primary antibody (anti-Cry11A antibody or anti-M13 antiody respectively) diluted in washing solution 1. The slides were washed three times with washing solution 2 [0.1% (w/v) BSA, 0.1% (v/v) Triton X-100, 2% (w/v) goat serum in PBS] and incubated with the corresponding secondary antibody (1:1000 dilution of Cy3-conjugated goat-anti-rabbit antibody for slides incubated with Cry11A toxin, or with a 1:1000 dilution of Cy3-conjugated goat-anti-mouse antibody for slides incubated with phage) together with Alexa Fluor 488 (1:100 dilution; Molecular Probes). The slides were washed three times in washing solution 2, mounted in 90% (v/v) glycerol, 4% (v/v) N-propyl gallate in PBS and analysed using a Confocal Zeiss LSM 510 Axioplan2 microscope with a helium/neon laser. All images were imported into, and assembled and labelled in, Adobe PhotoShop (V6).
Insect bioassays
Ae. aegypti mosquitoes were reared at 28 °C, 87% humidity and a 12:12, light/dark, photoperiod. Early fourth-instar larvae (20) were placed in 100 ml of dechlorinated water. The effects of different concentrations of Cry11Aa crystal proteins (150–600 ng/ml) were tested in the absence (positive control) or presence of competitors (selected phages) during a 24 h period at 28 °C. The toxic effect was determined by counting dead larvae.
RESULTS
Analysis of GPI-anchored proteins that interact with Cry11Aa toxin
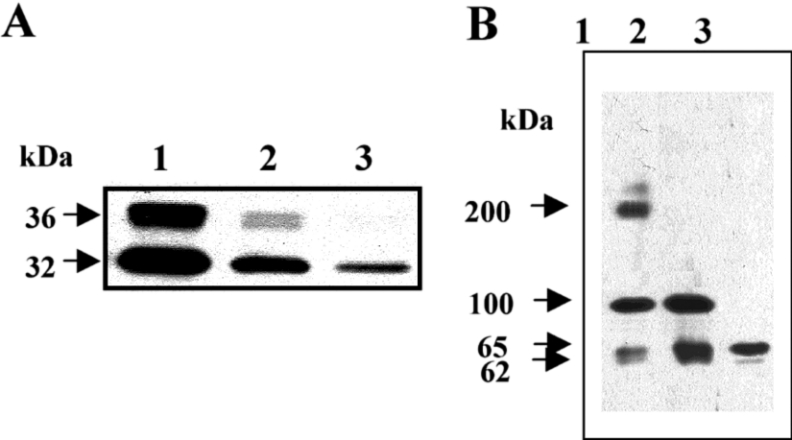
GPI-anchored proteins have been shown to be important molecules for the interaction with Cry1 toxins in lepidopteran insects and also with the Bin toxin in mosquitoes [10,11,22]. To determine if GPI-anchored proteins are also involved in the binding of Cry11Aa to BBMV, we compared the binding of biotinylated-Cry11Aa toxin to BBMV or BBMV treated with PIPLC. PIPLC removes GPI-anchored proteins from membranes by cleaving the GPI anchor. Figure 1(A) shows that the binding of biotinylated-Cry11Aa was greatly reduced after treatment of BBMV with PIPLC. Figure 1(A), lane 3, shows a complete loss of BBMV binding to the 36 kDa band (corresponding to the N-terminus of the toxin Cry11A) and a substantial reduction in binding to the 32 kDa band (corresponding to the C-terminus that is involved in binding) [7]. This result suggests that a GPI-anchored protein is involved in binding of Cry11Aa to BBMV.
Figure 1. Characterization of Ae. aegypti GPI-anchored proteins involved in binding Cry11A toxin.
(A) SDS/PAGE showing binding of biotinylated Cry11A toxin (10 nM) to BBMV (10 μg), in solution. Lane 1, biotinylated Cry11Aa; lane 2, biotinylated Cry11Aa bound to BBMV; lane 3, biotinylated Cry11Aa to bound to BBMV, previously treated with PIPLC. (B) Toxin overlay assay of Cry11Aa toxin binding to BBMV proteins, visualised with anti-Cry11Aa antibody. Lane 1, binding of Cry11Aa to BBMV proteins; lane 2, binding of Cry11Aa to proteins in supernantants obtained after treatment of BBMV with PIPLC; lane 3, binding of Cry11Aa to proteins obtained from Cry11Aa ligand chromatography.
To characterize the GPI-proteins that bound Cry11Aa toxin, a ligand blot of BBMV was performed. Figure 1(B), lane 1, demonstrates that Cry11Aa binds to the BBMV 200 kDa, 100 kDa, 65 kDa (as well as traces of 62 kDa) proteins. In contrast, the ligand blot of the proteins released by PIPLC treatment, revealed three GPI-anchored proteins of 100 kDa, 65 kDa, and traces of a 62 kDa that bound the Cry11Aa toxin (Figure 1B, lane 2). To further characterize the proteins involved in Cry11Aa binding, a purification protocol using biotinylated-Cry11Aa toxin was performed. Biotinylated-Cry11Aa was bound to a Sepharose–agarose–streptavidin matrix and the proteins from the supernatants of PIPLC-treated BBMV were applied to the column. After elution of Cry11Aa-binding proteins by an alkaline buffer treatment, the protein samples were separated by SDS/PAGE and the binding of Cry11Aa to the eluted proteins was visualized using the anti-Cry11Aa polyclonal antibody. The 65 kDa and trace amounts of the 62 kDa protein were purified when proteins obtained from treatment of BBMV with PIPLC were applied to the biotinylated-Cry11Aa coupled agarose–streptavidin (Figure 1B). The presence of traces of a 62 kDa protein (in addition to the 65 kDa protein) in the sample after Cry11Aa affinity chromatography was confirmed using silver stained gels (results not shown). This result shows that the 65 kDa protein, in contrast to the 100 kDa protein, binds the Cry11Aa toxin in its native conformation.
In the case of the Cry1A toxins, two GPI-anchored proteins, APN and ALP have been shown to be functional receptor molecules [10,11]. To determine if the 65 kDa protein could be an APN or ALP enzyme, the specific activity of these enzymes was determined in BBMV, in PIPLC supernatants and in the eluants from Cry11Aa affinity chromatography. Table 2 shows that both APN and ALP activities were enriched in the supernatants of PIPLC-treated BBMV. Nevertheless, only ALP activity was present in the eluant obtained after Cry11Aa affinity chromatography. Table 2 shows that there was a 5.7-fold enrichment of ALP activity after the 65 kDa protein was purified, by Cry11Aa affinity chromatography, from BBMV. This result demonstrates that the 65 kDa protein is a GPI-anchored ALP.
Table 2. Specific ALP and APN activities of BBMV proteins.
The results are expressed as the means±S.D. (n=3). ND, not detected.
| Sample | ALP activity (nM·ml−1·min−1/μg) | APN activity (μM·ml−1·min−1/μg) |
|---|---|---|
| BBMV | 717±94 | 1380±45 |
| BBMV/PIPLC | 2241±67 | 9557±347 |
| Cry11Aa ligand chromatography | 4081±115 | ND |
In the case of the lepidopteran insects Heliothis. virescens and M. sexta, the interaction of 1 μM of Cry1Ac toxin with native ALP resulted in a 50% inhibition of the phosphatase activity [29,30]. We determined the specific ALP activity in the PIPLC extracted proteins in the presence and absence of 1 μM of Cry11Aa toxin. The ALP activity was reduced by 40% in the presence of Cry11Aa toxin.
Isolation and characterization of ALP-binding peptides that interfere with Cry11Aa–BBMV interaction and toxicity
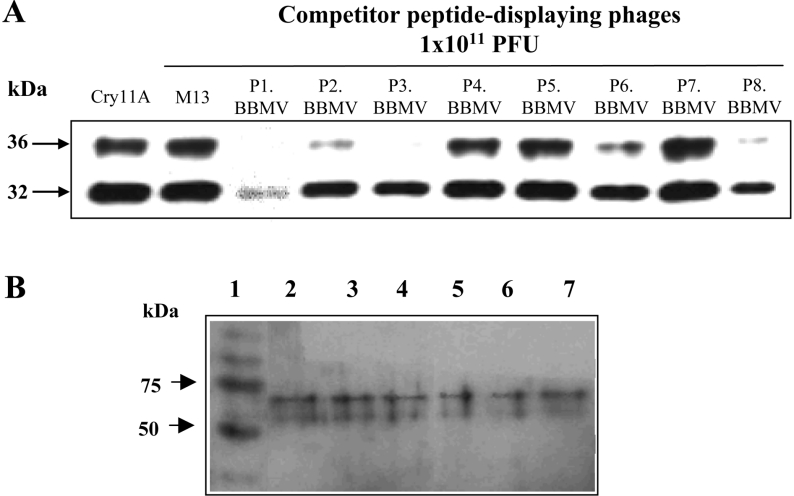
In order to identify peptides that bind to the Cry11Aa receptor and that interfere with toxin-receptor interaction, a library of M13 phages displaying random 12-residue peptides was panned against Ae. aegypti BBMV in solution for four consecutive rounds of selection. Ten phages from the third and ten from the fourth round of panning were randomly selected for further analysis. Sequence analyses revealed eight different peptide sequences. Competition-binding analysis of Cry11Aa toxin to BBMV in the presence of each of the selected phages revealed that four of these phages interfered with the binding of Cry11Aa to Ae. aegypti BBMV (Figure 2A). The deduced amino acid sequences of the displayed peptides in these phages are as follows: P1.BBMV, AAKTMDTLRPPR; P2.BBMV, STTQQHRYLNPA; P3.BBMV, WHWLPSTSVLRR and P8.BBMV, AHLKDWFLPRIP.
Figure 2. Identification of peptide-displaying phages selected against Aedes aegypti BBMV that inhibit Cy11Aa binding to BBMV.
(A) Qualitative binding assay of biotinylated Cry11Aa toxin to BBMV in solution, performed in the presence of different phages (1×1011 pfu) selected from panning against Ae. aegypti BBMV. The results are representative of three independent experiments. (B) Peptide-phage overlay assays to BBMV proteins. Lane 1, molecular mass marker; lanes 2, 3 and 4, binding of peptide-phage P1.BBMV and lanes 5, 6 and 7, binding of P8.BBMV, visualised with anti-M13 antibody. Lanes 2 and 5 correspond to total BBMV proteins. Lanes 3 and 6 to proteins recovered after PIPLC treatment of BBMV. Lanes 4 and 7 proteins recovered after Cry11Aa ligand chromatography.
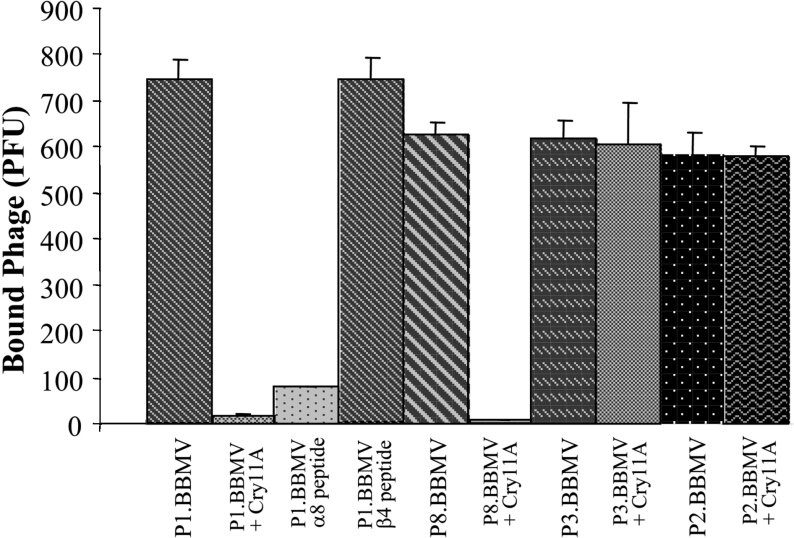
To further characterize the interaction of the peptide phages with Ae. aegypti BBMVs, a competition binding assay of the selected phages to BBMV, in the presence of Cry11Aa toxin, was performed. In this assay, 1000 peptide-phages were incubated with BBMV and the bound phages were scored after centrifugation washing and elution of phage particles. Figure 3 shows that binding of P1.BBMV and P8.BBMV to BBMV was competed for by Cry11Aa toxin, in contrast the binding of P2.BBMV and P3.BBMV to BBMV were not competed for by Cry11Aa. This result suggests that P1.BBMV and P8.BBMV bind to a specific Cry11Aa receptor molecule.
Figure 3. Binding of P1.BBMV and P8.BBMV to Ae. aegypti BBMV competed for by Cry11Aa toxin.
Binding of different peptide-phages (103 pfu) to 10 μg of BBMV and competition in the presence of 10 nM Cry11Aa toxin or 1000 nM synthetic peptides. Phages bound to BBMV were recovered after washing steps and pfus were determined.
To identify the BBMV proteins that were recognized by P1.BBMV and P8.BBMV, we analysed the proteins recognized by these phages using ligand blot assays. Figure 2(B) shows that P1.BBMV and P8.BBMV recognized the 65 kDa GPI–ALP, and also the 62 kDa protein in total BBMV, in eluants of PIPLC treated BBMV and in the purified proteins obtained after Cry11Aa affinity-chromatography. These results indicate that P1.BBMV and P8.BBMV peptide phages recognize the GPI–ALP Cry11Aa-receptor.
Localization of Cry11Aa and ALP in Ae. aegypti midgut tissue sections
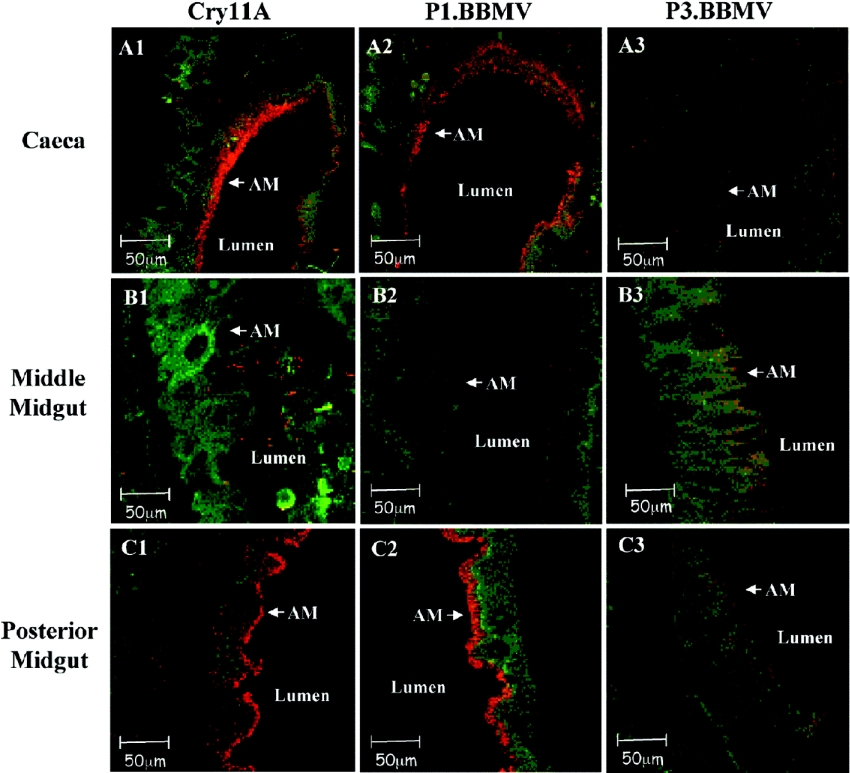
We analysed the binding of the Cry11Aa toxin to midgut tissue sections by immunofluorescence localization to determine to which regions of the midgut this toxin binds. In addition, we analysed the binding of P1.BBMV phage to the midgut tissue sections to determine the localization of ALP in the different regions of the midgut epithelium. Figure 4 shows that Cry11Aa toxin binds preferentially to the microvilli membranes of caeca and the posterior midgut. A similar binding pattern was observed with P1.BBMV, in contrast P3.BBMV, used as control phage, does not affect toxin-receptor interaction and did not bind to tissue sections (Figure 4). Furthermore, the binding of P1.BBMV phages to midgut epithelial cells was competed for by the Cry11Aa toxin, showing that this peptide-phage recognizes the same binding sites as the Cry11Aa in the midgut epithelial cells (Figure 5, panel B5, shows Cry11Aa binding competition of P1.BBMV). P8.BBMV showed a similar binding pattern to P1.BBMV (results not shown).
Figure 4. ALP is located in the same regions as Cry11Aa binding sites in the Ae. aegypti midgut.
Binding of Cry11A, P1.BBMV and P3.BBMV to different regions of Ae. aegypti fourth instar larvae midgut tissue sections (A1–A3, caeca tissue sections; B1–B3, anterior midgut tissue sections and C1–C3, posterior midgut tissue sections). Cry11Aa toxin was detected with anti-Cry11Aa primary antibody raised in mice and a Cy3-conjugated goat-anti-mouse secondary antibody (red colour). The peptide-phages were detected with anti-M13 primary antibody raised in rabbits and a secondary antibody Cy3-conjugated goat-anti-rabbit. (also red colour). Actin was detected with Alexa Fluor 488 (green colour). AM, apical microvilli.
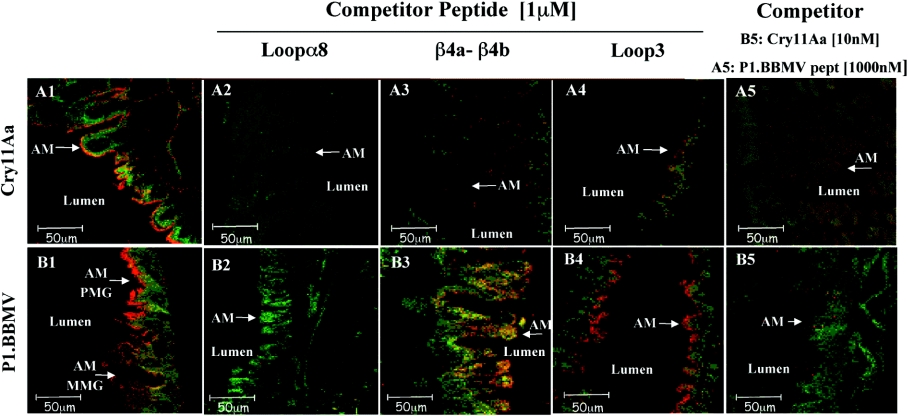
Figure 5. Domain II loop α8 is involved in Cry11Aa–ALP interaction.
Competition binding assay of Cry11Aa (panels A1–A5) and P1.BBMV phage (panels B1–B5). to Ae. aegypti fixed midgut tissue sections from the posterior region using synthetic peptides or Cry11A toxin as competitors. P1.BBMV and Cry11Aa were detected as indicated in Figure 4.
Analysis of epitopes in Cry11Aa toxin-ALP interaction
Previous work demonstrated that Cry11Aa interacts with its receptor through three domain II regions, loopα8, β4a–β4b region and loop 3 [21]. To determine if the P1.BBMV phage bound ALP in similar regions to the Cry11Aa receptor-binding epitopes, binding of P1.BBMV phage to BBMV in solution was analysed in the presence of synthetic peptides with amino acid sequences corresponding to the domain II loop α8, β4–β5 region or loop 3 of Cry11Aa. We observed that the binding of phage P1.BBMV to Ae. aegypti BBMV was inhibited in the presence of Cry11Aa toxin or in the presence of peptide loop α-8. In contrast, peptide β4a–β4b that had no effect on this interaction (Figure 3). To confirm these results, we also analysed the binding of P1.BBMV to midgut tissue sections of Ae. aegypti larvae in the presence of synthetic peptides corresponding to exposed regions of Cry11Aa toxin. Cry11Aa binding to fixed midgut tissue sections showed that loop α-8, β-4 and loop 3 peptides competed with this toxin to bind to the microvilli of the posterior part of the midgut (Figure 5, panels A2, A3 and A4). Figure 5, panel B2 shows that loop α-8 peptide, in contrast to β-4 or loop 3 synthetic peptides (panels B3 and B4), competed with the P1.BBMV phage to bind to midgut tissue. These results confirm that P1.BBMV recognizes the ALP receptor through an amino acid region that is also recognized by the loop α-8 region of Cry11Aa. The P8.BBMV binding to BBMV was not competed for by any of the loop synthetic peptides (results not shown).
Involvement of GPI–ALP in Cry11Aa toxicity
To determine if ALP is involved in Cry11Aa toxicity, Ae. aegypti larvae were fed with Cry11Aa toxin alone or in combination with the P1.BBMV or P8.BBMV. Table 3 shows that the toxicity of Cry11Aa toxin was reduced by 68% when the toxin was previously incubated with P1.BBMV. The P8.BBMV phage also reduced the toxicity of Cry11Aa, in contrast to treatment with P3.BBMV which did not affect toxin-receptor interaction or toxicity. None of the selected phages was toxic to Ae. aegypti larvae when assayed alone (results not shown). This result suggests that interaction of Cry11Aa toxin with ALP is important for Cry11Aa toxicity.
Table 3. Toxicity of Cry11Aa toxin to Ae. aegypti fourth instar larvae in the presence of peptide-phage competitors.
Mortalities are expressed as the means±S.E.M. (n=3), using 45 larvae per treatment. For each treatment 300 ng/ml of Cry11A toxin plus 1011 colony forming units of peptide-displaying phages/cup as competitor were used.
| Treatment | Mortality (%) |
|---|---|
| Cry11A | 72±2.9 |
| Cry11A/P1.BBMV | 23±7.4 |
| Cry11A/P8.BBMV | 37±2.0 |
| Cry11A/P3.BBMV | 70±1.5 |
| Water | 0 |
DISCUSSION
Several Cry1A receptors are attached to the membrane by GPI anchors, including APN and ALP [10,11]. Also, as in the case of the Bin toxin produced by B. sphaericus, a GPI-anchored α-glucosidase is the functional receptor in C. pipiens [22]. These and other GPI-anchored proteins are proposed to be selectively included in lipid rafts that are considered to be spatially differentiated, liquid-ordered microdomains in cell membranes, although the existence of these membrane microdomains in living cells is still in doubt [31]. Lipid rafts are enriched in glycosphingolipids, cholesterol and GPI-anchored proteins and are proposed to be involved in signal transduction, sorting and trafficking of plasma membrane proteins in mammalian cells [31]. Also, they may function as pathogen portals for different viruses, bacteria and toxins [32,33]. Recently, we demonstrated that APNs in M. sexta and H. virescens, in contrast with the 210 kDa cadherin receptors, are probably located in lipid rafts and that the integrity of these microdomains is essential for Cry1Ab pore activity [34]. A model of the sequential participation of cadherin receptor and APN, leading to Cry1A toxin insertion into lipid rafts was recently proposed [13,34]. Therefore it seems that targeting GPI-anchored proteins may be a general strategy by which pore-forming toxins to interact with its target cells. The interaction of pore-forming toxins with lipid rafts could result in additional cellular events, including toxin internalization, signal transduction and cellular response.
In this work we analysed the GPI-anchored proteins of Ae. aegypti midgut that interact with Cry11Aa toxin. Ligand blot experiments identified three proteins of 200 kDa, 100 kDa and 65 kDa that bound Cry11Aa toxin. Of these proteins, the 100 and 65 kDa (as well as traces of 62 kDa) proteins were shown to be anchored to the membrane by GPI. However, proteins purified after Cry11Aa affinity chromatography revealed only two proteins of 65 kDa and small amounts of 62 kDa that bound Cry11Aa toxin (Figure 1B), suggesting that the 100 kDa protein did not interact with Cry11Aa toxin in its native conformation. The role of the 200 kDa protein as a Cry11Aa receptor remains to be characterized. Previously, the 65 kDa protein was identified by Cry11Aa affinity chromatography and was also shown to bind to the Cry4B toxin, while the 62 kDa protein was characterized as degradation product of the 65 kDa protein [14]. The present study has provided evidence showing that the 65 kDa protein is a GPI-anchored ALP enzyme. In different insect species, the midgut GPI–ALPs are of similar sizes, 65–68 kDa [11]. The ALP specific activity was enriched 6-fold after PIPLC treatment of BBMV and Cry11Aa affinity chromatography (Table 2). Since the 65 kDa ALP was purified almost to homogeneity (only trace amounts of the 62 kDa protein were also present), the 6-fold increase in ALP activity suggests that this enzyme is an abundant protein in the BBMV accounting for approx. 15 to 20% of total protein in the BBMV. Enrichment of specific activity was reported for the purification of the APN Cry1A receptor in M. sexta larvae, using a similar experimental protocol, suggesting that APN is also a very abundant protein in the midgut cells of M. sexta [10].
Several lines of evidence indicate that ALP is also an important Cry receptor in lepidopteran insects. In a proteomic analysis of M. sexta BBMV proteins it was shown that the Cry1Ac toxin binds to a 65 kDa ALP protein [35]. Reduced levels of a 65 kDa ALP correlated with resistance of H. virescens larvae to Cry1Ac toxin [11]. In this work we show that the Ae. aegypti GPI–ALP protein binds Cry11Aa toxin and is involved in the toxicity of Cry11Aa. We identified two peptide displaying phages, P1.BBMV and P8.BBMV, that specifically bound the ALP, competed with the binding of the toxin to BBMV, and interfered with the toxicity of Cry11Aa toxin. The P1.BBMV and P8.BBMV were identified among eight different peptide-phages that bound BBMV. The abundance (see above) of the 65 kDa ALP may explain our success in isolating peptide-phages that bind this protein when selecting for proteins that bound to whole BBMV vesicles.
Interestingly, we found that Cry11Aa diminished ALP specific activity. As mentioned previously, in the case of the lepidopteran insects H. virescens and M. sexta, the interaction of Cry1Ac with native ALP resulted in the inhibition of the phosphatase activity at similar ALP levels as those reported here [29,30]. Therefore it could be possible that reducing ALP activity is a mechanism by which Cry-toxin-related toxicity occurs, leading to the impairment of the enzymatic machinery of midgut cells. Nevertheless, it should be mentioned that lowering the levels of ALP is not a major determinant in toxicity, since in H. virescens mutations that lower the ALP activity to 30% of wild-type levels correlate with resistance to Cry1Ac toxin [11], suggesting that ALP could have an important role as a binding protein in H. virescens.
In the present study we also provided evidence that the domain II loop α8 of Cry11Aa is involved in the interaction with the GPI–ALP, since binding of Cry11Aa and P1.BBMV to BBMV or to midgut tissue sections was specifically inhibited by a synthetic peptide with a sequence corresponding to loop α8. In previous work, we identified a peptide phage (P5.tox) that bound Cry11Aa in domain II loop α8 and inhibited the interaction of the toxin with BBMV [22]. The identification of the cognate binding epitope in ALP will require the cloning of the gene encoding this protein, followed by a comparison of the deduced amino acid sequence with that of the P5.tox peptide. Binding of P8.BBMV to BBMV was not competed for by any of the Cry11Aa loop synthetic peptides suggesting that P8.BBMV recognizes the 65 kDa ALP at a different epitope from P1.BBMV.
Finally, we determined the distribution of the ALP enzyme in different regions of Ae. aegypti larvae midgut by analysing the binding of P1.BBMV phage to midgut tissue sections. ALP was preferentially localized in the caeca and in the posterior part of the midgut. This distribution is similar to that observed by immunolocalization of the Cry11Aa toxin. These regions coincide with the different pH regions of the Ae. aegypti gut, where the caeca and posterior midgut have a pH 8.0, whereas the anterior midgut has a pH of up to 11 [36]. Furthermore, the distribution of ALP is very similar to that of a GPI-carbonic anhydrase, which may be involved in maintaining the alkaline pH homoeostasis in the insect gut [37]. Overall our results show that the GPI–ALP is an important receptor molecule that mediates Cry11Aa toxicity in Ae. aegypti larvae.
Acknowledgments
Our thanks to Dr Isabel Gomez (Departamento de Microbiología Molecular, Instituto de Biotecnología Universidad Nacional Autónoma de México, Mexico) for helpful discussions, and to Mrs Lizbeth Cabrera (Departamento de Microbiología Molecular, Instituto de Biotecnología Universidad Nacional Autónoma de México, Mexico) for technical assistance. This research was supported in part by DGAPA/UNAM (Dirección General del Personal Académico/Universidad Nacional Autónoma de México) IN207503-3, IN206503-3 and IX217404, CONACyT (Consejo Nacional de Ciencia y Tecnología) 36505-N, USDA (United States Department of Agriculture) 2002-35302-12539 and NIH (National Institutes of Health, U.S.A.) 1R01 AI066014-01. Luisa E. Fernandez received a DGEP UNAM (Dirección General de Estudios de Postgrado Universidad Nacional Autónoma de México) PhD fellowship.
References
- 1.Margalith Y., Ben-Dov E. Techniques for environmental protection. In: Rechcigl J. E., Rechcigl N. A., editors. Insect Pest Management. Boca Raton: CRC Press; 2000. p. 243. [Google Scholar]
- 2.Porter A. G., Davidson E. W., Liu J. W. Mosquitocidal toxins of bacilli and their genetic manipulation for effective biological control of mosquitoes. Microbiol. Rev. 1993;57:838–861. doi: 10.1128/mr.57.4.838-861.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Maagd R. A., Bravo A., Crickmore N. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 2001;17:193–199. doi: 10.1016/s0168-9525(01)02237-5. [DOI] [PubMed] [Google Scholar]
- 4.Yamahiwa M., Ogawa R., Yasuda K., Natsuyama H., Sen K., Sakai H. Active form of dipteran-specific insecticidal protein Cry11A produced by Bacillus thuringiensis subsp. israelensis. Biosci., Biotechnol., Biochem. 2002;66:516–522. doi: 10.1271/bbb.66.516. [DOI] [PubMed] [Google Scholar]
- 5.Revina L. P., Kostina L. I., Ganushkina L. A., Zalunin I. A., Chestukina G. G. Reconstruction of Bacillus thuringiensis ssp. israelensis Cry11A endotoxin from fragments corresponding to its N- and C-moieties restores its original biological activity. Biochemistry (Moscow) 2004;69:181–187. doi: 10.1023/b:biry.0000018949.70836.dc. [DOI] [PubMed] [Google Scholar]
- 6.Dai S. M., Gill S. S. In vitro and in vivo proteolysis of the Bacillus thuringiensis subsp. israelensis CryIVD protein by Culex quinquefasciatus larval midgut proteases. Insect Biochem. Mol. Biol. 1993;23:273–283. doi: 10.1016/0965-1748(93)90008-g. [DOI] [PubMed] [Google Scholar]
- 7.Yamagiwa M., Sakagawa K., Sakai H. Functional analysis of two processed fragments of Bacillus thuringiensis Cry11A toxin. Biosci., Biotechnol., Biochem. 2004;68:523–528. doi: 10.1271/bbb.68.523. [DOI] [PubMed] [Google Scholar]
- 8.Vadlamudi R. K., Weber E., Ji I., Ji T. H., Bulla L. A., Jr Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J. Biol. Chem. 1995;270:5490–5494. doi: 10.1074/jbc.270.10.5490. [DOI] [PubMed] [Google Scholar]
- 9.Nagamatsu Y., Koike T., Sasaki K., Yoshimoto A., Furukawa Y. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal. FEBS Lett. 1999;460:385–390. doi: 10.1016/s0014-5793(99)01327-7. [DOI] [PubMed] [Google Scholar]
- 10.Knight P., Crickmore N., Ellar D. J. The receptor for Bacillus thuringiensis CryIA(c) δ-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 1994;11:429–436. doi: 10.1111/j.1365-2958.1994.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurat-Fuentes J. L., Adang M. J. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 2004;271:3127–3135. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 12.Valaitis A. P., Jenkins J. L., Lee M. K., Dean D. H., Garner K. J. Isolation and partial characterization of Gypsy moth BTR-270 an anionic brush border membrane glycoconjugate that binds Bacillus thuringiensis Cry1A toxins with high affinity. Arch. Insect Biochem. Physiol. 2001;46:186–200. doi: 10.1002/arch.1028. [DOI] [PubMed] [Google Scholar]
- 13.Bravo A., Gómez I., Conde J., Muñoz-Garay C., Sánchez J., Miranda R., Zhuang M., Gill S. S., Soberón M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim. Biophys. Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Buzdin A. A., Revina L. P., Kostina L. I., Zalunin I. A., Chestukhina G. G. Interaction of 65- and 62-kD proteins from the apical membranes of the Aedes aegypti larvae midgut epithelium with Cry4B and Cry11A endotoxins of Bacillus thuringiensis. Biochemistry (Moscow) 2002;67:540–546. doi: 10.1023/a:1015594127636. [DOI] [PubMed] [Google Scholar]
- 15.Pootanakit K., Angsuthanasombat C., Panyin S. Identification of two isoforms of aminopeptidase N in Aedes aegypti larval midgut. J. Biochem. Mol. Biol. 2003;36:508–513. doi: 10.5483/bmbrep.2003.36.5.508. [DOI] [PubMed] [Google Scholar]
- 16.Boonserm P., Davis P., Ellar D. J., Li J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J. Mol. Biol. 2005;348:363–382. doi: 10.1016/j.jmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Grochulski P., Masson L., Borisova S., Pusztai-Carey M., Schwartz J. L., Brousseau R., Cygler M. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. 1995;254:447–464. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Carroll J., Ellar D. J. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature (London) 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 19.Morse R. J., Yamamoto T., Stroud R. M. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure. 2001;9:409–417. doi: 10.1016/s0969-2126(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 20.Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J. R., Feitelson J., Zeigler D., Dean D. H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998;62:705–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández L. E., Pérez C., Segovia L., Rodríguez M. H., Gill S. S., Bravo A., Soberón M. Cry11Aa toxin from Bacillus thuringiensis binds its receptor in Aedes aegypti mosquito larvae trough loop α-8 of domain II. FEBS Lett. 2005;579:3508–3514. doi: 10.1016/j.febslet.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 22.Darboux I., Nielsen-LeRoux C., Charles J. F., Pauron D. The receptor of Bacillus sphaericus binary toxin in Culex pipiens (Diptera: Culicidae) midgut: molecular cloning and expression. Insect Biochem. Mol. Biol. 2001;31:981–990. doi: 10.1016/s0965-1748(01)00046-7. [DOI] [PubMed] [Google Scholar]
- 23.Chang C., Yu Y. M., Dai S. M., Law S. K., Gill S. S. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl. Environ. Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lereclus D., Agaisse H., Gominet M., Chaufaux J. Overproduction encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spoOA mutant. Bio/Technology. 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- 25.Thomas W. E., Ellar D. J. Bacillus thuringiensis var israelensis crystal δ-endotoxin: effects on insect and mammalian cells in vitro and in vivo. J. Cell Sci. 1983;60:181–197. doi: 10.1242/jcs.60.1.181. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen-LeRoux C., Charles J. F. Binding of Bacillus sphaericus binary toxin to a specific receptor on midgut brush-border membranes from mosquito larvae. Eur. J. Biochem. 1992;210:585–590. doi: 10.1111/j.1432-1033.1992.tb17458.x. [DOI] [PubMed] [Google Scholar]
- 27.Aranda E., Sanchez J., Perferoen M., Güereca L., Bravo A. Interactions of Bacillus thuringiensis crystal proteins with the midgut epithelial cells of Spodoptera frugiperda (Lepidoptera: Noctuidae) J. Invertebr. Pathol. 1996;68:203–212. doi: 10.1006/jipa.1996.0087. [DOI] [PubMed] [Google Scholar]
- 28.Lorence A., Darszon A., Bravo A. Aminopeptidase dependent pore formation of Bacillus thuringiensis Cry1Ac toxin on Trichoplusia ni membranes. FEBS Lett. 1997;414:303–307. doi: 10.1016/s0014-5793(97)01014-4. [DOI] [PubMed] [Google Scholar]
- 29.Sangadala S., Walters F. S., English L. H., Adang M. J. A mixture of Manduca sexta aminopetidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)- K+ efflux in vitro. J. Biol. Chem. 1994;269:10088–10092. [PubMed] [Google Scholar]
- 30.English L., Readdy T. L. δ endotoxin inhibits a phosphatase in midgut epithelial membranes of Heliothis virescens. Insect Biochem. 1989;19:145–152. [Google Scholar]
- 31.Munro S. Lipid rafts: elusive or illusive? Cell (Cambridge, Mass.) 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberger C. M., Brumell J. H., Finlay B. B. Microbial pathogenesis: lipid rafts as pathogen portals. Current Biol. 2000;10:823–825. doi: 10.1016/s0960-9822(00)00788-0. [DOI] [PubMed] [Google Scholar]
- 33.Cabiaux V., Wolff Ch., Ruysschaert J. M. Interaction with a lipid membrane: a key step in bacterial toxins virulence. Int. J. Biol. Macromol. 1997;21:285–298. doi: 10.1016/s0141-8130(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang M., Oltean D. I., Gómez I., Pullikuth A. K., Soberón M., Bravo A., Gill S. S. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J. Biol. Chem. 2002;277:13863–13872. doi: 10.1074/jbc.M110057200. [DOI] [PubMed] [Google Scholar]
- 35.McNall R. J., Adang M. J. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem. Mol. Biol. 2003;33:999–1010. doi: 10.1016/s0965-1748(03)00114-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang Z., Linser P. J., Harvey W. R. Antibody to H(+) V-ATPase subunit E colocalizes with portasomes in alkaline larval midgut of fresh water mosquito (Aedes aegypti) J. Exp. Biol. 1999;202:2449–2460. doi: 10.1242/jeb.202.18.2449. [DOI] [PubMed] [Google Scholar]
- 37.Seron T., Hill J., Linser P. J. A GPI-linked carbonic anhydrase expressed in the larval mosquito midgut. J. Exp. Biol. 2004;207:4559–4572. doi: 10.1242/jeb.01287. [DOI] [PubMed] [Google Scholar]