Abstract
The cyclotide family of plant proteins is of interest because of their unique topology, which combines a head-to-tail cyclic backbone with an embedded cystine knot, and because their remarkable chemical and biological properties make them ideal candidates as grafting templates for biologically active peptide epitopes. The present study describes the first steps towards exploiting the cyclotide framework by synthesizing and structurally characterizing two grafted analogues of the cyclotide kalata B1. The modified peptides have polar or charged residues substituted for residues that form part of a surface-exposed hydrophobic patch that plays a significant role in the folding and biological activity of kalata B1. Both analogues retain the native cyclotide fold, but lack the undesired haemolytic activity of their parent molecule, kalata B1. This finding confirms the tolerance of the cyclotide framework to residue substitutions and opens up possibilities for the substitution of biologically active peptide epitopes into the framework.
Keywords: cyclic protein, cyclotide, haemolytic activity, kalata, NMR, protein engineering
Abbreviations: Boc, t-butoxycarbonyl; CCK, cyclic cystine knot; DQF-COSY, double-quantum-filtered COSY; ECOSY, exclusive COSY; ES-MS, electrospray MS; HBTU, 2-(1-H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate; HD50, peptide concentration causing 50% haemolysis; NOE, nuclear Overhauser effect; RMSD, root-mean-square deviation; RP, reversed-phase; TFA, trifluoroacetic acid
INTRODUCTION
The cyclotides [1] are a fascinating family of plant proteins that are distinguished by a head-to-tail peptide backbone and six conserved cysteine residues that are paired to form a knotted network of three disulphide bonds within the molecular core. The combination of a circular peptide backbone and a tightly knotted disulphide network forms a CCK (cyclic cystine knot) motif and makes the cyclotides exceptionally stable. In general, the limited stability of peptides in vivo makes them poor candidates for drug development. However, the cyclotides are resistant to thermal unfolding, chemical denaturants and proteolytic degradation and their use in native medicine as a uterotonic tea suggests that they are very stable in biological systems [2]. This inherent stability makes the cyclotides an attractive scaffold for the design of peptide-based therapeutics [3].
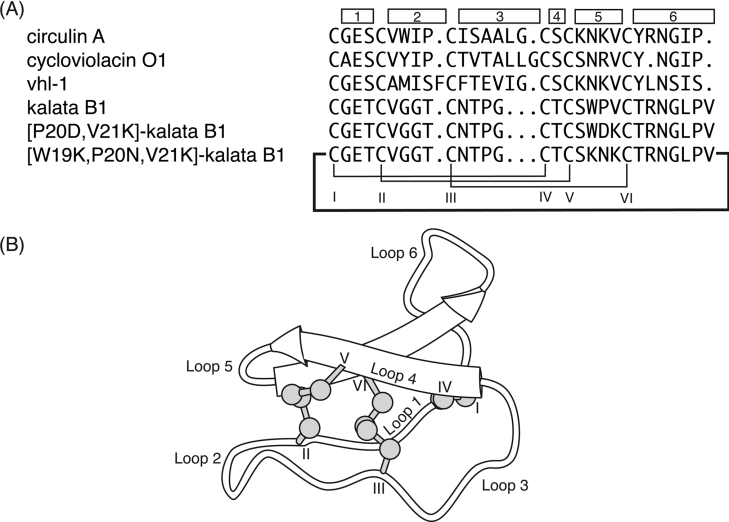
Over 80 cyclotides containing between 28 and 37 amino acids have been reported to date, with example sequences shown in Figure 1(A) [4]. The backbone segments between the conserved cysteine residues, referred to as loops, have various levels of sequence diversity, as illustrated in Figure 1(B). Loops 1 and 4 are highly conserved across all the cyclotides and, together with the three disulphide bonds, form the cystine knot. Loop 6 has recently been identified as the point of cyclization in the biosynthesis of the cyclotides and, although it varies in size between different cyclotides, it has a number of highly conserved residues that are believed to be critical in the processing of the cyclotide precursor protein [5,6]. The remaining loops (2, 3 and 5) are more variable in size and composition, but several residues are still highly conserved. On the basis of the presence or absence of a cis-proline residue in loop 5, the cyclotides have been divided broadly into two subfamilies, namely the bracelet and Möbius respectively.
Figure 1. Selected cyclotide sequences and a schematic diagram of the CCK framework.
(A) Cyclotide sequences showing the disulphide connectivity between cysteine residues (labelled with Roman numerals), with the individual loops indicated by the boxes. The original papers reporting the peptides are as follows: circulin A [16]; cycloviolacin O1 [1]; vhl-1 [13]; and kalata B1 [48,49]. Circulin A, cycloviolacin O1 and vhl-1 are members of the bracelet subfamily, whereas kalata B1 is a Möbius cyclotide, as it contains a cis-proline in loop 5. (B) The CCK framework illustrating the cyclic backbone and cystine-knot motif. The β-sheet region is shown and the cysteine residues and loops are numbered.
A number of three-dimensional structures of cyclotides have been determined by NMR, including kalata B1 and B2 [7,8], cycloviolacin O1 [7], circulins A and B [9,10], palicourein [11], tricyclon A [12], vhl-1 [13] and vhr-1 [14]. The conserved structural elements of the cyclotides include a β-hairpin that is part of a triple-stranded β-sheet formed by loops 4, 5 and 6. The third strand is distorted from ideal geometry and contains a β-bulge. In addition, the bracelet cyclotides have a short section of helical structure in loop 3.
The cyclotides exhibit a diverse range of biological activities. These include uterotonic [15], anti-HIV [16], haemolytic [17–19], antimicrobial [19], antitumour [20] and cardiotoxic activity [15], as well as inhibition of trypsin [21] and neurotensin binding [22]. In addition, they exhibit insecticidal properties, and it is believed that the natural role of cyclotides in plants is as a defence against predation [5,8].
A predominant feature of the Möbius cyclotides, exemplified by the prototypical kalata B1, is the large number of hydrophobic residues that are exposed on a localized region of the protein surface. This large hydrophobic patch in kalata B1 is formed predominantly by the residues of loops 5 and 6 and has a major influence on the folding of the peptide. Previously it has been shown that addition of a hydrophobic solvent to the folding buffer in vitro significantly improves the yield of correctly folded kalata B1 [18]. Furthermore, this hydrophobic patch has been implicated as providing a key contribution to the biological activity of these peptides [7].
The CCK framework is an attractive scaffold on which to graft biologically active peptide epitopes. In the present paper we describe the synthesis of two ‘grafted’ kalata B1 variants and examine their structural and biological properties with the aim of exploring the plasticity of the CCK framework. An understanding of the tolerance of the CCK framework to changes in the variable loops is crucial if we are to utilize it as a scaffold for drug design. Our first step towards this understanding is to investigate the role of the hydrophobic patch in both folding and structure of the cyclotides. Both of the analogues have polar or charged residues introduced into loop 5 to replace residues that are part of the hydrophobic patch. [W19K/P20N/V21K]kalata B1 has three residues (KNK) from a bracelet cyclotide substituted into loop 5 of a Möbius cyclotide while [P20D/V21K]kalata B1 has two charged residues (DK) substituted into loop 5 (Figure 1A). The former analogue is thus a chimaera and the latter a charge mutant. The KNK sequence was chosen as it is a conserved sequence in loop 5 of the bracelet cyclotides and it was of interest to see if it would be tolerated in the Möbius cyclotides. In the second mutant the DK residues were chosen to see if the interaction between neighbouring positive and negative charges would counteract any destabilization caused by the disruption of the hydrophobic patch. By introducing polar and charged residues into loop 5 the size of the hydrophobic patch is reduced and consequently we could examine the effect on folding and biological activity. This will provide an understanding of the tolerance of the CCK framework of non-native epitopes and hence establish its viability as a structural scaffold for the development of peptide-based pharmaceuticals.
EXPERIMENTAL
Peptide synthesis
All peptides were assembled on PAM (phenylacetamidomethyl) resin (Applied Biosystems, Foster City, CA, U.S.A.) by manual solid-phase peptide synthesis using the in situ neutralization/HBTU [2-(1-H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate] protocol for Boc (t-butoxycarbonyl) chemistry [23]. The peptide chain was attached to the resin via a linker which generates a C-terminal thioester on HF cleavage [24]. Amino acid side chain protection was as follows: Arg(Tos), Asn(Xan), Asp(OChx) Glu(OChx), Ser(Bzl), Thr(Bzl) and Trp-(CHO) (where Tos is tosyl, Xan is xanthyl, OChx is cyclohexyl ester, Bzl is benzoyl and CHO is formyl). The CHO protecting group on the tryptophan residue was not removed prior to cleavage as the conditions required for deprotection are not compatible with the TAMPAL (trityl-associated mercaptopropionic acid leucine) linker. Cleavage of the peptide from the resin was achieved using HF with p-cresol and p-thiocresol as scavengers (HF/p-cresol/thiocresol, 9:0.8:0.2, by vol.). The reaction was allowed to proceed at −5 to 0 °C for 1 h; HF was removed under vacuum and the peptide precipitated with diethyl ether. Following cleavage, the peptide was dissolved in 50% acetonitrile containing 0.05% TFA (trifluoroacetic acid) and freeze-dried. The crude peptide was purified by RP (reversed-phase)-HPLC on a Phenomenex C18 column using a gradient of 0–80% B (Buffer A: water/0.05% TFA; Buffer B: 90% acetonitrile/10% water/0.045% TFA) in 80 min and the eluent was monitored at 230 nm. These conditions were used in subsequent purification steps. Analytical RP-HPLC and ES-MS (electrospray MS) confirmed the purity and molecular mass of the synthesized peptide.
The linear reduced peptides were cyclized and oxidized ‘in one pot’ by incubating in 0.1 M NH4HCO3 (pH 8.5)/propan-2-ol (50:50, v/v) with 1 mM GSSG overnight at room temperature. The mixture was then purified by RP-HPLC to yield the cyclic/oxidized peptide. Analytical RP-HPLC and ES-MS confirmed the purity of the final product and 1H NMR was used to confirm that the peptide had correctly folded.
NMR spectroscopy
Samples for NMR spectroscopy were prepared by dissolving the peptide in either 90% 1H2O/10% 2H2O or 100% 2H2O to a final concentration of 1 mM and a pH of approx. 4. All NMR experiments were carried out on Bruker 500 MHz and 750 MHz spectrometers with sample temperatures of 290 and 295 K. The two-dimensional NMR experiments performed included DQF (double-quantum-filtered)-COSY, TOCSY [25], ECOSY (exclusive COSY) [26] and NOESY with mixing times of 100 and 200 ms. All spectra were acquired as previously described [7] and spectra were processed using XWINNMR software (Bruker). To identify slowly exchanging amide protons, a series of one-dimensional and TOCSY spectra were obtained from the fully protonated peptide immediately after dissolving the sample in 2H2O. Chemical shifts were referenced to DSS (dimethyl-2-silapentane-5-sulphonate) at 0 p.p.m. Processed spectra were analysed and assigned within the program XEASY [27] using the sequential assignment technique [28].
Structural restraints
Cross-peaks primarily from the 200 ms NOESY spectrum at 290 K were integrated in XEASY and distance restraints were derived using the CALIBA function of DYANA [29] with appropriate pseudoatom corrections. Backbone dihedral restraints were derived from 3JNH-αH coupling constants measured both from the one-dimensional spectrum and from the lineshape analysis of HN–Hα peaks in the COSY spectrum. The angles were restrained to −120±30° for 3JNH-αH couplings of 8–9.5 Hz, −120±15° for 3JNH-αH couplings of >9.5 Hz and 50±40° where 3JNH-αH couplings were ≈7.0 Hz and strong intra-residue HN–Hα NOEs (nuclear Overhauser effects) were observed. In addition, backbone dihedral angles were restrained to −100±80° where sequential HN–Hα NOEs were significantly stronger than intra-residue HN–Hα NOEs.
3JαH-βH coupling constants derived from ECOSY spectra together with NOE intensities from a 100 ms NOESY spectrum were used to obtain stereospecific assignments and φ1 dihedral angles of 60, −180 or −60°.
Structure calculations
Preliminary three-dimensional structures were calculated using a torsion-angle simulated-annealing protocol within DYANA [29] to check NOE restraints for violations and resolve ambiguous cross-peaks. The dihedral angle restraints were included in these structure calculations, and where hydrogen bonds could be determined and assigned unambiguously, these were included in subsequent calculations. Final structures were calculated using simulated-annealing and energy-minimization protocols in explicit water [30] within CNS version 1.1 [31] as previously described [7]. Structures were analysed using PROMOTIF [32] and PROCHECK-NMR [33].
Haemolytic assay
Human type A erythrocytes were washed with PBS and centrifuged at 150 g for 30 s in a microcentrifuge several times until a clear supernatant was obtained. The assay was performed by adding 20 μl of sample to 80 μl of a 1% suspension of erythrocytes in PBS. The concentration of the sample stock was determined by measurement of the UV absorbance at 215 nm. Test concentrations ranged from 0.5 to 1400 μM. Synthetic melittin (Sigma) was used as a standard. The mixtures were incubated at room temperature (23 °C) for 1 h and then centrifuged at 150 g for 1 min. The supernatant (35 μl) was diluted 1:30 in deionized (Milli-Q) water, and the absorbance was measured at 415 nm. The peptide concentration causing 50% haemolysis (HD50) was calculated.
RESULTS
Design and synthesis
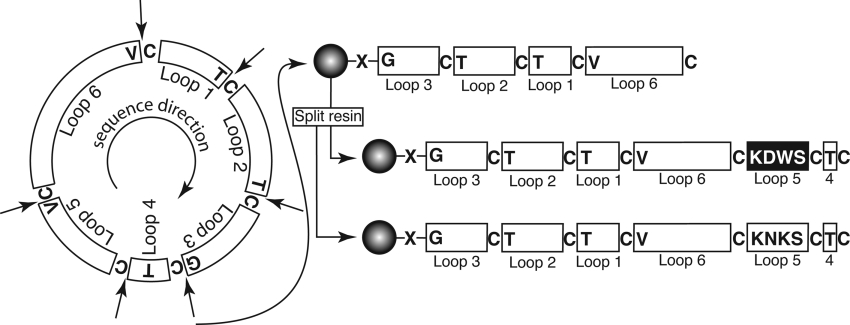
Both [W19K/P20N/V21K]kalata B1 and [P20D/V21K]kalata B1 were synthesized using solid-phase peptide synthesis. The strategy for synthesis of the grafted cyclotide analogues is shown in Figure 2. The linear precursor sequences were designed to have cysteine residues at their N-termini to facilitate cyclization via an intramolecular thioester-mediated reaction as described previously [34]. In the present study, the two linear sequences synthesized corresponded to CTCSWDKCTRNGLPVCGETCVGGTCNTPG for [P20D/V21K]kalata B1 and CTCSKNKCTRNGLPVCGETCVGGTCNTPG for [W19K/P20N/V21K]kalata B1. The most efficient method of synthesizing multiple loop 5 analogues is to begin the synthesis at the end of loop 4 (i.e. Thr16) as the sequence common to both peptides can be synthesized in one pot and the resin then divided to complete the individual peptides. However, we chose to begin the synthesis at loop 3, rather than loop 4, as the intramolecular cyclization reaction proceeds more rapidly when the less bulky glycine is at the C-terminus [35]. If the synthesis had started from loop 4, then the C-terminal residue would have been a more bulky threonine residue, which would have probably reduced the rate of the cyclization reaction.
Figure 2. Schematic illustration of the synthetic strategy utilized for grafted cyclotide analogues.
There are six possible starting points for the synthesis, at the N-terminal side of each of the cysteine residues, which are indicated by arrows; the amino acid that would become the C-terminal residue in the linear precursor is also shown. The two modified kalata B1 analogues described in the present study were synthesized using solid-phase methods by starting at the glycine residue in loop 3, as shown on the right, and attached to the resin via a thioester linker (X). The peptide chain was built up on-resin (starting at the C-terminus) until loop 5 was reached, when the resin was split and the synthesis of the two analogues completed separately. After cleavage of the peptide chain from the resin, cyclization and disulphide formation was done simultaneously by incubating the peptides in aqueous buffer at pH 8.5.
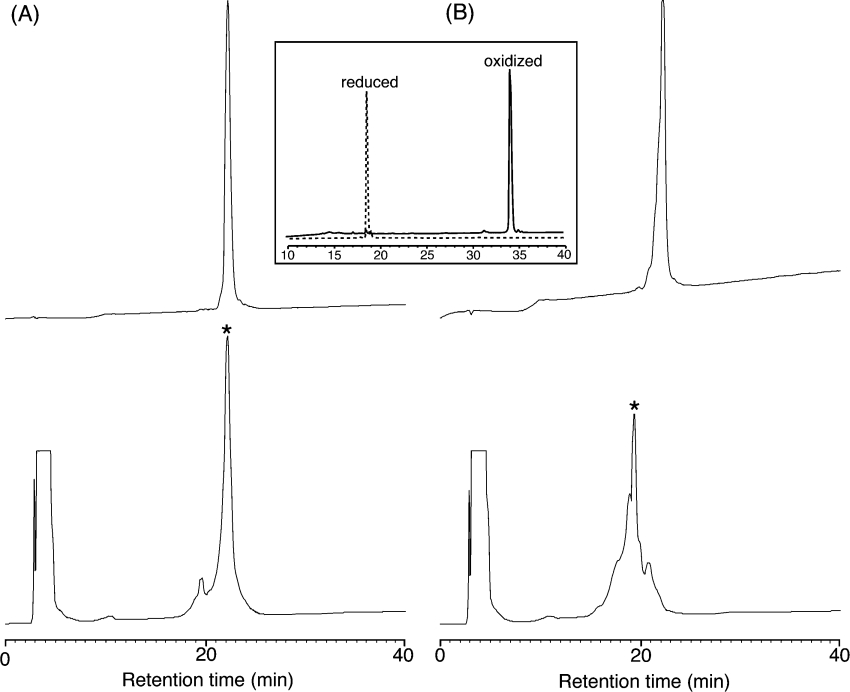
After assembly and cleavage from the resin using HF, the peptides were cyclized and oxidized in a buffer containing ammonium bicarbonate in aq. 50% (v/v) propan-2-ol to form the cyclic backbone and three disulphide bonds. The identity and purity of both folded peptides was confirmed by analytical RP-HPLC and ES-MS. The correct disulphide connectivity was subsequently confirmed from NMR experiments. The retention time of oxidized kalata B1 is significantly longer than that of its reduced form, owing to the formation of the hydrophobic patch on folding. The oxidation profiles of the two cyclotide analogues (Figure 3) clearly show that the retention times of the oxidized peptides are very similar to that of the corresponding reduced forms. This reflects the substitution of the hydrophobic residues in kalata B1 with the polar and charged residues in the two analogues.
Figure 3. RP-HPLC oxidation profiles of [P20D/V21K]kalata B and [W19K/P20N/V21K]kalata B1.
(A) Linear, reduced [P20D/V21K]kalata B1 (top) and the oxidation profile (below) with the correctly folded major isomer marked with an asterisk. (B) Linear, reduced [W19K/P20N/V21K]kalata B1 (top) and the oxidation profile (below) with the correctly folded major isomer marked with an asterisk. A typical oxidation for kalata B1 (inset) illustrates the large difference in retention times between the reduced and oxidized forms.
NMR assignment and structure determination
NMR spectra were recorded at 500 and 750 MHz and used to determine the three-dimensional structure of the two analogues. Assignments of the NMR spectra of the analogues were made using two-dimensional homonuclear methods [28]. Chemical shifts in the amide region are generally well dispersed, and the fingerprint region in the NOESY spectrum of each peptide shows a complete cycle of αH–NH sequential connectivities for the whole protein, unbroken except at proline residues. Both proline residues in [P20D/V21K]kalata B1 and [W19K/P20N/V21K]kalata B1 were determined to be in the trans conformation, on the basis of the presence of Hα(i−1)–Hδ(i) NOEs and the absence of Hα(i−1)–Hα(i) NOEs in the 2H2O NOESY spectrum. The presence of sequential NOEs between residues Gly14 and Cys15, corresponding to the N- and C-termini of the linear precursors, provided confirmatory evidence for the cyclic nature of the peptides.
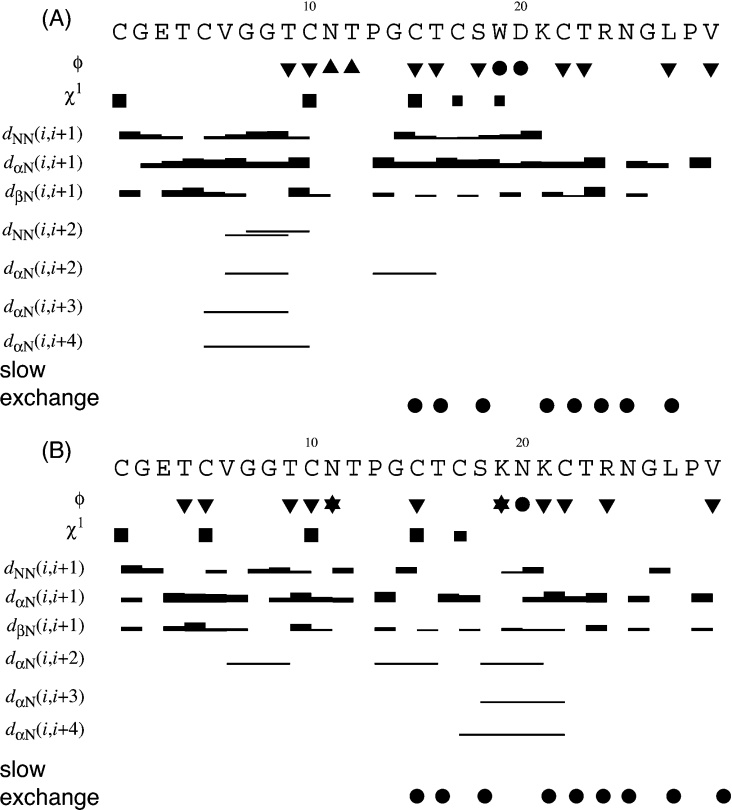
From a consideration of the experimental NOE, slow exchange and coupling patterns illustrated in Figure 4, together with some longer-range NOEs between backbone protons, an indication of the likely secondary structure was derived and is schematically illustrated in Figure 5. The predominant secondary-structural feature in both peptides is a β-hairpin that is loosely associated with a third β-strand. The position of the two strands that make up the β-hairpin is consistent with those found in native kalata B1.
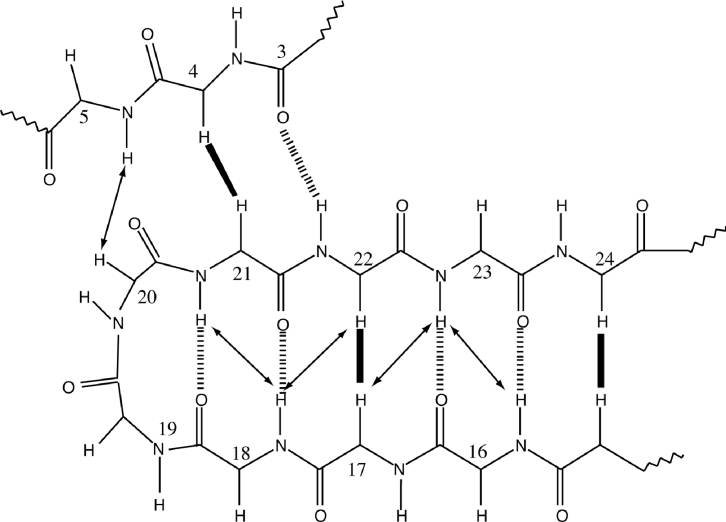
Figure 4. Summary of coupling constants, sequential, and medium-range NOE connectivities and amide exchange for (A) [P20D/V21K]kalata B1 and (B) [W19K/P20N/V21K]kalata B1.
The height of the bar represents the strength of the NOE cross-peak, classified as either strong, medium or weak. Dihedral angle restraints are shown at the top of the diagram: φ angle restraints are represented as ▼ for restraints of −120° ±30°, and represented as ▲ for restraints of −60°±30°. Filled circles (●) indicate positive phi angles (50°±40°). Stars (★) indicate negative phi angles (−100°±80°). χ1 angle restraints are represented as large squares for restraints of 180°±30° and smaller squares for restraints of −60°±30°. Slow-exchanging amide protons still present 16 h after dissolution in 2H2O at 298 K are shown at the bottom of the diagram.
Figure 5. Schematic representation of the secondary structure of the analogues [P20D/V21K]kalata B1 and [W19K/P20N/V21K]kalata B1.
Long-range NOE data from NOESY spectra (in 2H2O or 1H2O) are summarized, with intrastrand NOEs presented at arrows or solid lines for the αH–αH NOEs and hydrogen bonds presented as broken lines.
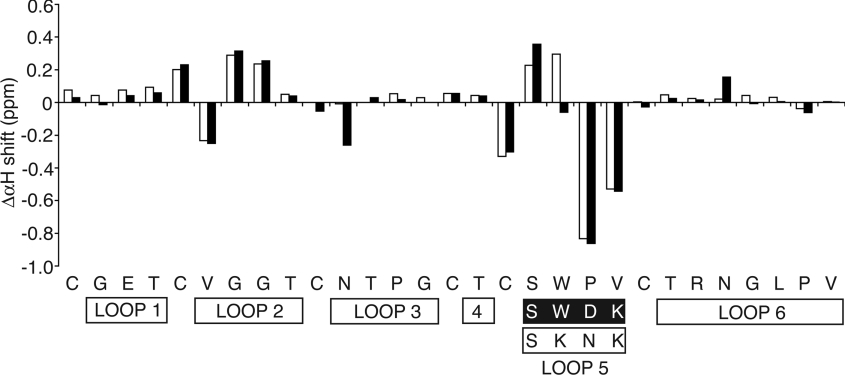
A comparison of the αH chemical shifts of [P20D/V21K] kalata B1 and [W19K/P20N/V21K]kalata B1 with the native peptide is shown in Figure 6. In general, the chemical shifts do not differ substantially from the native, indicating that the overall fold is retained in both mutant peptides. As expected, the most significant changes occur in loop 5, but additional differences are also observed in loop 2, indicating that mutations in loop 5 have an influence on the structure of loop 2.
Figure 6. Comparison of the secondary αH chemical shifts of [P20D/V21K]kalata B1 (black bars) and [W19K/P20N/V21K]kalata B1 (white bars) with those of native kalata B1.
The values shown are the chemical-shift values of the analogues subtracted from the values for kalata B1.
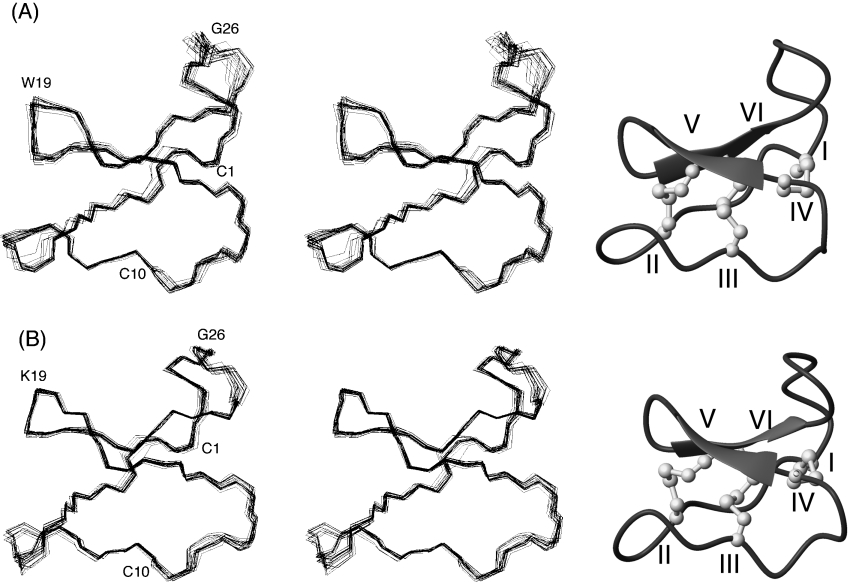
The three-dimensional structure of [P20D/V21K]kalata B1 was calculated with 228 distance restraints and 26 angle restraints using a simulated annealing protocol in CNS. A total of 100 sequential, 47 medium-range and 81 long-range distance restraints were derived from NOESY spectra with a mixing time of 200 ms. A total of 16 φ angles were obtained from 3JHN-Hα coupling constants and ten χ1 angles were derived from the 3JHα-Hβ coupling constants and NOESY cross-peaks intensities. The disulphide connectivity was assumed to be identical with that of kalata B1 on the basis of the similarities in chemical shifts. Eight hydrogen bonds were established through the slow-amide-exchange data and were included in the structure calculations. The 20 lowest-energy structures were chosen to represent the structure of [P20D/V21K]kalata B1, and a summary of the structural statistics is shown in Table 1. An overlay of the lowest-energy structures is given in Figure 7(A).
Table 1. Structural statistics for the 20 lowest-energy structures of [P20D,V21K]-kalata B1 and [W19K, P20N, V21K]-kalata B1.
The values in the Table are the means±S.D. Note 1 kcal=4.184 kJ.
| Parameter | [P20D/V21K] kalata B1 | [W19K/P20N/V21K] kalata B1 |
|---|---|---|
| Experimental restraints | ||
| Sequential NOEs | 101 | 89 |
| Medium-range NOEs | 47 | 48 |
| Long-range NOEs | 81 | 56 |
| Hydrogen bonds | 8 | 8 |
| Dihedral angles | 24 | 21 |
| Mean RMSDs from experimental restraints | ||
| NOE distances (Å) | 0.033±0.001 | 0.05±0.003 |
| Dihedral angles (°) | 0.72±0.18 | 0.64±0.26 |
| Mean RMSDs from idealized covalent geometry | ||
| Bonds (Å) | 0.004±0.0002 | 0.005±0.0002 |
| Angles (°) | 0.53±0.03 | 0.58±0.03 |
| Impropers (°) | 0.39±0.04 | 0.48±0.03 |
| Mean energies (kcal·mol−1) | ||
| ENOE | 12.9±0.8 | 29.9±3.3 |
| EcDihedral | 0.79±0.4 | 0.6±0.52 |
| Ebond | 5.9±0.66 | 8.2±0.63 |
| Eangle | 29.7±3.9 | 35.9±3.7 |
| Eimproper | 4.5±0.93 | 6.5±0.87 |
| Evan der Waals | −75.6±6.68 | −55.5±4.1 |
| Atomic RMSDs (Å)* | ||
| Backbone atoms | 0.4±0.12 | 0.38±0.09 |
| Heavy atoms | 1.29±0.23 | 1.18±0.19 |
| Ramachandran statistics (%)† | ||
| Residues in most favoured regions | 83.2 | 71.8 |
| Residues in additional allowed regions | 16.5 | 28.2 |
* Atomic RMSDs are the pairwise RMS difference for the family of structures.
† Procheck_NMR was used to calculate the Ramachandran statistics.
Figure 7. Stereoview of the ensemble of 20 low-energy structures representing the solution structures superimposed over all the backbone N, Cα and C atoms and corresponding ribbon diagrams of (A) [P20D/V21K]kalata B1 and (B) [W19K/P20N/V21K]kalata B1.
An analysis of the three-dimensional structure of [P20D/V21K]kalata B1 with PROMOTIF_NMR reveals that the major secondary structural element is a β-hairpin comprising residues 16–23. Residues 16–18 and 21–23 form the two β-strands and a type I′ β-turn between residues 18 and 21 connects these two strands. A third β-strand is not formally recognized, but is loosely associated with residues 3 and 4. The structure also contains a number of tight turns, including type I′ β-turns between residues 6 and 9 and between residues 24 and 27, and a type II β-turn between residues 12 and 15. Two inverse gamma turns were also identified in [P20D/V21K]kalata B1, located between residues 5 and 7 and between residues 27 and 29. The disulphide bond between Cys5 and Cys17 is characterized as a right-handed spiral by PROMOTIF_NMR, whereas those between Cys1 and Cys15 and between Cys10 and Cys22 are recognized as a left-handed spirals, and these disulphide-bond conformations agree with the native structure. Generally the overall three-dimensional structure of [P20D/V21K]kalata B1 is well-defined [global backbone RMSD (root-mean-square deviation) 0.40±0.12 Å; 1 Å=0.1 nm] and, as is evident from Figure 7(A), the general fold closely resembles that of the native kalata B1.
The three-dimensional structure of [W19K/P20N/V21K]kalata B1 was calculated with 278 NOE distance restraints and 21 dihedral angle restraints using a simulated annealing protocol in CNS. The 20 lowest-energy structures consistent with experimental data were chosen to represent the family of structures for the KNK chimaera of kalata B1, as shown in Figure 7(B). A summary of the energy and geometric statistics for these structures is given in Table 1.
Analysis of the structures with PROMOTIF and PROCHECK-NMR revealed structural elements very similar to those of [P20D/V21K]kalata B1. The β-hairpin encompasses residues 16–23, but is connected by a type II β-turn rather than the type I′ turn observed in [P20D/V21K]kalata B1. Turn types identical with those of [P20D/V21K]kalata B1 were observed for residues 12–15 and 24–27, which are type II and type I′ β-turns respectively. The β-turn observed between residues 6 and 9 is classified as a type IV β-turn in [W19K/P20N/V21K]kalata B1. In addition, two inverse γ-turns are present between residues 5 and 7 and between residues 27 and 29. The disulphide bonds between Cys1 and Cys15 and between Cys10 and Cys22 are characterized as left-hand spirals, whereas the disulphide bond between Cys5 and Cys17 is a right-hand spiral.
The NMR structural co-ordinates of [P20D/V21K]kalata B1 (PDB ID 2F2I) and [W19K/P20N/V21K]kalata B1 (PDB ID 2F2J) have been submitted to the Protein Data Bank at the Research Collaboratory for Structural Bioinformatics (http://www.rcsb.org/pdb/) [36].
Haemolytic activity
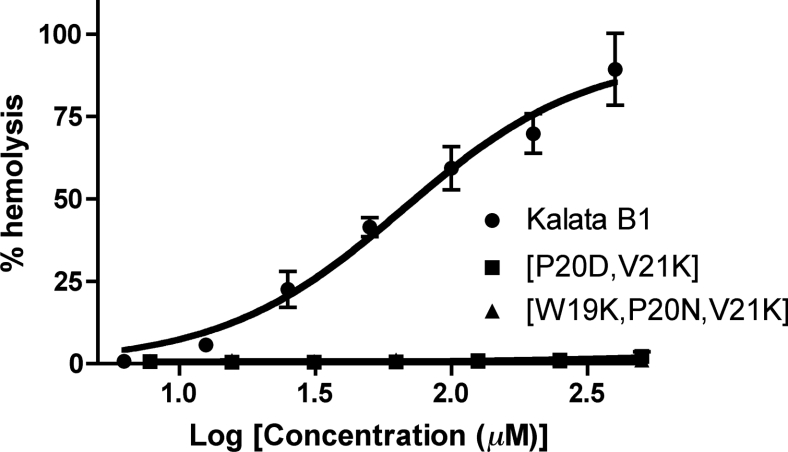
Some of the various biological activities exhibited by cyclotides may be related to membrane disruption, which in turn is related to their hydrophobicity. Kalata B1 has mild haemolytic activity [37], and so a haemolytic assay was used to compare the biological activity of the cyclotide analogues with that of kalata B1. The results of this assay are shown in Figure 8 and support our hypothesis that haemolytic activity could be removed via a decrease in the hydrophobic nature of the cyclotides. Both [P20D/V21K]kalata B1 and [W19K/P20N/V21K]kalata B1 exhibited no haemolytic activity, in contrast with kalata B1, which exhibited mild haemolytic activity consistent with literature values.
Figure 8. Haemolytic activity of [P20D/V21K]kalata B1, [W19K/P20N/V21K]kalata B1 and kalata B1.
It can clearly be seen that the haemolytic activity is abolished in the grafted cyclotide analogues.
DISCUSSION
In the present study we have synthesized and characterized two cyclotide analogues in which residues comprising part of the hydrophobic patch in native kalata B1 have been replaced with grafted epitopes containing polar or charged residues. Both grafted peptides, [W19K/P20N/V21K]kalata B1 and [P20D/V21K]-kalata B1, exhibited a native-like fold, despite the substantial change in residue type and alteration in hydrophobic properties due to the grafting of the new epitopes into loop 5. This work thus confirms that ‘foreign’ epitopes can successfully be grafted into the cyclotide framework without disrupting the folding of the CCK. Importantly, increasing the polarity and charge on the surface of the grafted peptides abolished the haemolytic activity of the native peptide, showing that the cyclotide framework is readily amenable to mutations that enhance its biopharmaceutical properties.
The major aim of the present study was to see if the cyclotide framework was amenable to the grafting of new sequences and whether the replacement of key hydrophobic residues would affect the folding and structure. To address this aim it was necessary to develop a general strategy for the synthesis of grafted cyclotide analogues. Native chemical ligation is a methodology for linking two peptide fragments via a peptide bond utilizing a C-terminal thioester moiety in one peptide and a N-terminal cysteine in the other [24]. If this reaction is performed intramolecularly, then the product has a cyclic peptide backbone. This strategy has been utilized successfully to synthesize both native cyclotides and disulphide-deficient mutants [18,19,34] and was applied here for the grafted mutants. The presence of six cysteine residues in the cyclotide sequences means that there are potentially six starting points for the synthesis of the linear peptide precursor. The possibility of multiple potential starting points is advantageous when synthesizing multiple analogues that contain a range of substitutions in the same loop. By selecting an appropriate starting point, the majority of the sequence can be synthesized before splitting the resin into aliquots and completing each analogue individually. This splitting strategy results in a substantial saving of time relative to the separate synthesis of each mutant and is applicable for the efficient future production of a range of grafted cyclotide analogues.
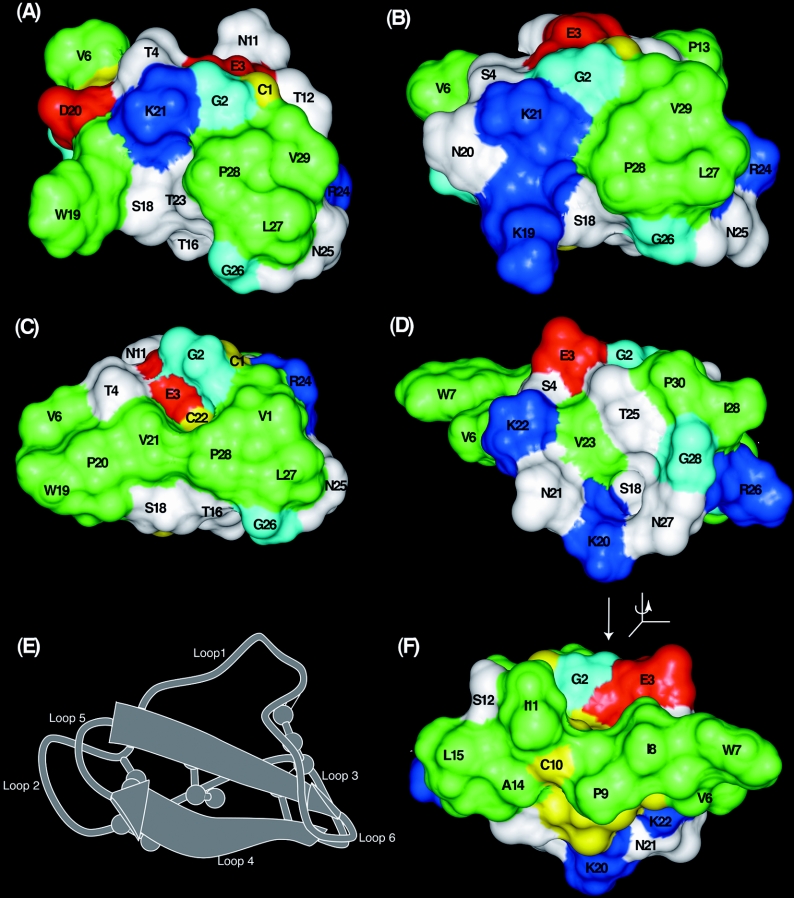
Having designed and tested an appropriate synthetic strategy, we turned to a structural examination of the grafted cyclotides. Figure 9 shows surface representations of the two grafted cyclotides and compares them with kalata B1, a prototypic Möbius cyclotide and circulin A, a prototypic bracelet cyclotide. A comparison of Figures 9(A) and 9(B) for the mutants with Figure 9(C) for kalata B1 shows that the surface-exposed hydrophobic patch in the latter is clearly disrupted in the two mutants. The hydrophobic patch in kalata B1 comprises Val6 from loop 2, Trp19, Pro20 and Val21 from loop 5 and Leu27, Pro28 and Val29 from loop 6. The hydrophobic patch in [P20D/V21K]kalata B1, shown in Figure 9(A), is decreased in size compared with the native peptide, with Leu27, Pro28 and Val29 forming a small patch, while Val6 and Trp19 are isolated owing to the two introduced charged residues. In [W19K/P20N/V21K]kalata B1 (Figure 9B), the corresponding molecular face is more similar to that of circulin A than kalata B1, with only a small hydrophobic patch formed by Leu27, Pro28 and Val29. Figure 9(D) shows the corresponding face of circulin A, which has only a small hydrophobic patch made up of Ile29 and Pro30. The results clearly show that the hydrophobic patch of kalata B1 can be disrupted, but still produce folded cyclotides.
Figure 9. Surface features of (A) [P20D/V21K]kalata B1, (B) [W19K/P20N/V21K]kalata B1 (C) kalata B1 and (D and F) circulin A.
The ribbon diagram in (E) shows the orientation of the views (A)–(D), with (F) flipped 180° about the vertical axis. Hydrophobic residues (alanine, leucine, isoleucine, proline, tryptophan and valine) are green, polar residues (asparagine, serine, threonine and tyropsine) are white, basic residues (arginine and lysine) are blue, acidic residues (aspartic acid and glutamic acid) are red, glycine is cyan and cysteine residues are yellow. (F) Illustrates the hydrophobic patch in circulin A, which is on the opposite face of the molecule to the patch in kalata B1.
In previous studies, the formation of the hydrophobic patch of kalata B1 has been associated with the correct folding of the peptide in vitro [18,34,38]. The yield of the correctly folded form of kalata B1 was significantly enhanced by the inclusion of propan-2-ol in the folding buffer to stabilize the hydrophobic patch. The folding efficiency of the two grafted cyclotides in the presence of propan-2-ol was slightly reduced compared with kalata B1, but reasonable yields were still produced. There was a small decrease in the yield of correctly folded peptide for [W19K/P20N/V21K]kalata B1 when compared with [P20D/V21K]kalata B1 and this may be a reflection of the smaller hydrophobic patch in [W19K/P20N/V21K]kalata B1. The retention times of folded [P20D/V21K]kalata B1 and [W19K/P20N/V21K]kalata B1 were significantly shorter than that of kalata B1 and were almost identical with those of their respective reduced forms. This contrasts with kalata B1, where the retention time increases substantially on folding. The shorter retention time of the two analogues is clearly due to the reduction in the hydrophobic surface character of the molecules rather than a change in the global fold, as the NMR structures are comparable with those of kalata B1 and other members of the cyclotide family.
The ability to modulate the hydrophobic patch on cyclotides as demonstrated here has important consequences for their use as molecular templates. A number of native cyclotides have mild haemolytic activity, which is undesirable in a pharmaceutical framework. Previously we have shown that the haemolytic activity of kalata B1 can be abolished when a break is introduced into the backbone via the synthesis of acyclic permutants [37,39,40]. The anti-HIV activity of kalata B1 is likewise lost in acyclic permutants [41]. In addition it has been proposed that an increase in net positive charge of a cyclotide is directly correlated with both antimicrobial activity and cytotoxicity [19,20]. The present study demonstrates that disruption of the hydrophobic patch in kalata B1, by the addition of polar and charged residues, also abolishes haemolytic activity, but offers the advantage of retention of the stable cyclic backbone. The loss of haemolytic activity in the mutants is consistent with findings for other peptides, with haemolytic activity closely correlated with hydrophobicity in a range of bioactive peptides [42–46]. Following up further on this theme it is noteworthy that although the surfaces of [W19K/P20N/21K]kalata B1 and circulin A are similar for the molecular face shown in Figure 9, circulin A is haemolytic [19], but [W19K/P20N/V21K]kalata B1 is not. However, as shown in Figure 9(F), circulin A has a large hydrophobic surface on the opposite face of the molecule, comprising residues from loops 2 and 3, in contrast with the hydrophobic surface in kalata B1 comprising residues primarily from loops 5 and 6. The fact that loops 2 and 3 are responsible for haemolytic activity in circulin A, whereas loops 5 and 6 facilitate this activity in kalata B1, reinforces the proposal that the cystine knot core of the cyclotides forces exposure of hydrophobic residues, and that this can result in different locations for hydrophobic patches depending on the sequence. In other words, one of the strengths of the CCK framework is that the cystine knot core is a reproducible structural unit that can be ‘decorated’ with residues that can be placed in different, yet precisely defined, locations determined by their position in the amino acid sequence.
In conclusion, the CCK framework, owing to its inherent thermal and biological stability [47], is an attractive molecule for use as a scaffold for the development of peptide-based pharmaceuticals. However, it is desirable for any drug scaffold to be biologically inert, and the mildly haemolytic activity of many native cyclotides represents a potential limitation to their use as pharmaceuticals. The present study has shown that it is possible to ‘engineer out’ this haemolytic activity without any significant effects on the folding efficiency or overall structure of the molecule. This is a vital step towards utilizing cyclotides as drug templates. The CCK scaffold potentially has four sites for the grafting of biologically active epitopes, namely loops 2, 3, 5 and 6. These loops show substantial sequence variation in native cyclotides and, unlike loops 1 and 4, are not an integral part of the cystine knot. On the basis of the structures of the naturally occurring cyclotides it can be envisaged that loops 2 and 5 could accommodate epitopes comprising a β-turn, and helical sequences could be grafted into loop 3 and long extended sequences into loop 6. In the present study we have established that one of these loops, loop 5, has sufficient plasticity to accommodate sequence changes, including non-native sequences, while maintaining the structural integrity of the scaffold.
It is clear from the results presented here that individual cyclotides are able to accommodate changes to their primary structure while maintaining a native-like fold. This finding paves the way for more extensive grafting studies on the CCK framework. The next logical step is to transfer entire non-native sequences on to the framework to see if the overall fold is still maintained. Peptides generally bind with high affinity and selectivity to their target, but are of limited therapeutic value owing to their low stability and poor bioavailability. Therefore, the ability to graft biologically active peptide epitopes on to the remarkably stable CCK framework has the potential to develop new leads for peptide-based therapeutics. The present study represents a first step towards this goal.
Acknowledgments
N. L. D. is a NHMRC (National Health and Medical Research Council) Industry Fellow. D. J. C. is an ARC (Australian Research Council) Professorial Fellow. We thank Ms Lillian Sando for performing the haemolytic assays and Ms Emryn MacLachlan and Ms Nhi Tran for their assistance with the NMR structure determination.
References
- 1.Craik D. J., Daly N. L., Bond T., Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 2.Gran L., Sandberg F., Sletten K. Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J. Ethnopharmacol. 2000;70:197–203. doi: 10.1016/s0378-8741(99)00175-0. [DOI] [PubMed] [Google Scholar]
- 3.Craik D. J., Simonsen S., Daly N. L. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discovery Dev. 2002;5:251–260. [PubMed] [Google Scholar]
- 4.Craik D. J., Daly N. L., Mulvenna J., Plan M. R., Trabi M. Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 5.Jennings C., West J., Waine C., Craik D., Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutton J. L., Renda R. F., Waine C., Clark R. J., Daly N. L., Jennings C. V., Anderson M. A., Craik D. J. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 2004;279:46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 7.Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J. Biol. Chem. 2003;278:8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- 8.Jennings C. V., Rosengren K. J., Daly N. L., Plan M., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Mobius strips exist in Nature? Biochemistry. 2005;44:851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 9.Koltay A., Daly N. L., Gustafson K. R., Craik D. J. Structure of circulin B and implications for antimicrobial activity of the cyclotides. Int. J. Pept. Res. Ther. 2005;11:99–106. [Google Scholar]
- 10.Daly N. L., Koltay A., Gustafson K. R., Boyd M. R., Casas-Finet J. R., Craik D. J. Solution structure by NMR of circulin A: a macrocyclic knotted peptide having anti-HIV activity. J. Mol. Biol. 1999;285:333–345. doi: 10.1006/jmbi.1998.2276. [DOI] [PubMed] [Google Scholar]
- 11.Barry D. G., Daly N. L., Bokesch H. R., Gustafson K. R., Craik D. J. Solution structure of the cyclotide palicourein: implications for the development of a pharmaceutical framework. Structure. 2004;12:85–94. doi: 10.1016/j.str.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Mulvenna J. R., Sando L., Craik D. J. Processing of a 22 kDa precursor protein to produce the circular protein tricyclon A. Structure. 2005;13:691–701. doi: 10.1016/j.str.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Chen B., Colgrave M. L., Daly N. L., Rosengren K. J., Gustafson K. R., Craik D. J. Isolation and characterization of novel cyclotides from Viola hederacea: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide. J. Biol. Chem. 2005;280:22395–22405. doi: 10.1074/jbc.M501737200. [DOI] [PubMed] [Google Scholar]
- 14.Trabi M., Craik D. J. Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell. 2004;16:2204–2216. doi: 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gran L. On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol. Toxicol. 1973;33:400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 16.Gustafson K. R., Sowder R. C., II, Henderson L. E., Parsons I. C., Kashman Y., Cardellina J. H., II, McMahon J. B., Buckheit R. W., Jr, Pannell L. K., Boyd M. R. Circulins A and B: Novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 1994;116:9337–9338. [Google Scholar]
- 17.Schöpke T., Hasan Agha M. I., Kraft R., Otto A., Hiller K. Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci. Pharm. 1993;61:145–153. [Google Scholar]
- 18.Daly N. L., Love S., Alewood P. F., Craik D. J. Chemical synthesis and folding of large cyclic polypeptides: Studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 19.Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svangard E., Goransson U., Hocaoglu Z., Gullbo J., Larsson R., Claeson P., Bohlin L. Cytotoxic cyclotides from Viola tricolor. J. Nat. Prod. 2004;67:144–147. doi: 10.1021/np030101l. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez J. F., Gagnon J., Chiche L., Nguyen T. M., Andrieu J. P., Heitz A., Trinh Hong T., Pham T. T., Le Nguyen D. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 22.Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria Longipes. J. Nat. Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 23.Schnölzer M., Alewood P., Jones A., Alewood D., Kent S. B. H. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Int. J. Pept. Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 24.Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 25.Braunschweiler L., Ernst R. R. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 26.Griesinger C., Sørensen O. W., Ernst R. R. Practical aspects of the E.COSY technique, measurement of scalar spin–spin coupling constants in peptides. J. Magn. Reson. 1987;75:474–492. [Google Scholar]
- 27.Eccles C., Guntert P., Billeter M., Wüthrich K. Efficient analysis of protein 2D NMR spectra using the software package EASY. J. Biomol. NMR. 1991;1:111–130. doi: 10.1007/BF01877224. [DOI] [PubMed] [Google Scholar]
- 28.Wüthrich K. New York: Wiley-Interscience; 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 29.Guntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 30.Linge J. P., Nilges M. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR. 1999;13:51–59. doi: 10.1023/a:1008365802830. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A. T., Adams P. D., Rice L. M. New applications of simulated annealing in X-ray crystallography and solution NMR. Structure. 1997;5:325–336. doi: 10.1016/s0969-2126(97)00190-1. [DOI] [PubMed] [Google Scholar]
- 32.Hutchinson E. G., Thornton J. M. PROMOTIF – A program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 34.Daly N. L., Clark R. J., Craik D. J. Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides. J. Biol. Chem. 2003;278:6314–6322. doi: 10.1074/jbc.M210492200. [DOI] [PubMed] [Google Scholar]
- 35.Hackeng T. M., Griffin J. H., Dawson P. E. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barry D. G., Daly N. L., Clark R. J., Sando L., Craik D. J. Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity. Biochemistry. 2003;42:6688–6695. doi: 10.1021/bi027323n. [DOI] [PubMed] [Google Scholar]
- 38.Daly N. L., Clark R. J., Goransson U., Craik D. J. Diversity in the disulfide folding pathways of cystine knot peptides. Lett. Pept. Sci. 2003;10:523–531. [Google Scholar]
- 39.Daly N. L., Craik D. J. Acyclic permutants of naturally occurring cyclic proteins. Characterization of cystine knot and β-sheet formation in the macrocyclic polypeptide kalata B1. J. Biol. Chem. 2000;275:19068–19075. doi: 10.1074/jbc.M000450200. [DOI] [PubMed] [Google Scholar]
- 40.Simonsen S. M., Daly N. L., Craik D. J. Capped acyclic permutants of the circular protein kalata B1. FEBS Lett. 2004;577:399–402. doi: 10.1016/j.febslet.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 41.Daly N. L., Gustafson K. R., Craik D. J. The role of the cyclic peptide backbone in the anti-HIV activity of the cyclotide kalata B1. FEBS Lett. 2004;574:69–72. doi: 10.1016/j.febslet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Blondelle S. E., Houghten R. A. Hemolytic and antimicrobial activities of the twenty-four individual omission analogues of melittin. Biochemistry. 1991;30:4671–4678. doi: 10.1021/bi00233a006. [DOI] [PubMed] [Google Scholar]
- 43.Blondelle S. E., Houghten R. A. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry. 1992;31:12688–12694. doi: 10.1021/bi00165a020. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y., Mant C. T., Farmer S. W., Hancock R. E., Vasil M. L., Hodges R. S. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiyota T., Lee S., Sugihara G. Design and synthesis of amphiphilic α-helical model peptides with systematically varied hydrophobic-hydrophilic balance and their interaction with lipid- and bio-membranes. Biochemistry. 1996;35:13196–13204. doi: 10.1021/bi961289t. [DOI] [PubMed] [Google Scholar]
- 46.Oren Z., Shai Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: structure–function study. Biochemistry. 1997;36:1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 47.Colgrave M. L., Craik D. J. Thermal, chemical and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 48.Gran L. Oxytocic principles of Oldenlandia affinis. Lloydia. 1973;36:174–178. [PubMed] [Google Scholar]
- 49.Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]