Abstract
HP (2-20), a 19-residue peptide derived from the N-terminus of Helicobacter pylori ribosomal protein L1, has antimicrobial activity but is not cytotoxic to human erythrocytes. We synthesized several peptide analogues to investigate the effects of substitutions on structure and antimicrobial activity. Replacement of Gln16 and Asp18 with tryptophan [anal-3 (analogue-3)] caused a dramatic increase in lytic activities against bacteria and fungi. By contrast, a decrease in amphiphilicity caused by replacement of Phe5 or Leu11 with serine was accompanied by a reduction in antimicrobial activity. Analysis of the tertiary structures of the peptides in SDS micelles by NMR spectroscopy revealed that they have a well-defined α-helical structure. Among the analogues, anal-3 has the longest α-helix, from Val4 to Trp18. The enhanced hydrophobicity and increased α-helicity results in enhanced antimicrobial activity in anal-3 without an increase in haemolytic activity. Fluorescence experiments proved that the bacterial-cell selectivity of the anal-3 peptide is due to its high binding affinity for negatively charged phospholipids in bacterial cells. Results showing the effect of spin-labels on the NMR spectra indicated that the side chains in the hydrophobic phase of the amphiphilic α-helix are buried on the surface of the micelle and the tryptophan indole ring is anchored in the membrane surface. Because anal-3 shows high selectivity towards bacterial and fungal cells, it may provide an avenue for the development of new antibiotics.
Keywords: antibiotic, antimicrobial peptide, haemolysis, HP (2-20), membrane, NMR
Abbreviations: anal-3, analogue-3; DPC, dodecylphosphocholine; DQF-COSY, double-quantum-filtered COSY; EYPC, egg yolk L-α-phosphatidylcholine; EYPE, EY L-α-phosphatidylethanolamine; EYPG, EY L-α-phosphatidylglycerol; HP (2-20), N-terminal peptide of Helicobacter pylori ribosomal protein L1; NOE, nuclear Overhauser effect; MIC, minimum inhibitory concentration; PC, phosphatidylcholine; PMAP, porcine myeloid antibacterial peptide; PS, phosphatidylserine; RBC, red blood cell; R.M.S.D., root mean square deviation; SEM, scanning electron microscopy; SUV, small unilamellar vesicle
INTRODUCTION
Antibiotic agents have become indispensable in the modern health care system, assisting and complementing the natural immune system. However, the appearance of resistant bacterial strains and the cytotoxicity of antibiotic agents have necessitated ongoing efforts to identify more potent and safer antibiotic agents. In this respect, endogenous antimicrobial peptides and their derivatives are attractive candidates. Antimicrobial peptides play important roles in the innate host defence mechanisms of most living organisms, including plants, insects, amphibians and mammals [1–4]. In addition, they are known to possess potent antibiotic activity against bacteria, fungi and even certain viruses [5,6].
It has been reported that the bacterium Helicobacter pylori, implicated as a predisposing factor in gastrointestinal illnesses, including gastritis and peptic ulcers, produces the N-terminal peptide of H. pylori ribosomal protein L1 [HP (2-20)] [7–9]. Although H. pylori itself is resistant to this peptide, it has antimicrobial activity similar to that observed in other cecropin-like N-terminal peptides [8,9]. HP (2-20), a 19 amino acid peptide, contains highly positively charged residues, including six lysines and one arginine, as well as seven hydrophobic residues. Chemically synthesized HP (2-20) is microbicidal against representative Gram-negative and Gram-positive bacteria. However, HP (2-20) does not display any haemolytic activity against human erythrocytes, even at a concentration of 100 μM, suggesting that it has a certain degree of cell-membrane target selectivity [8–10].
Tryptophan residues in melittin and mastoparan B are critical for their haemolytic activities, as well as their antimicrobial activities [11–15]. These peptides are thought to form an amphipathic α-helix that inserts in to the membrane, generating an ion channel or pore that disrupts membrane function. However, a tryptophan at the N-terminus of the cecropin A–maganin hybrid peptide mediates partial insertion into, and initial binding of the cell membrane, and this peptide does not have haemolytic activities [16]. Therefore it is important to investigate the role of tryptophan in each antimicrobial peptide in order to develop powerful antimicrobial peptides that do not have haemolytic activity.
To obtain peptides with improved antimicrobial activity and no cytotoxicity, novel analogues have been designed based on the sequence and α-helical wheel-diagram of HP (2-20) [10]. An increase in hydrophobicity has been introduced by replacement of specific residues with tryptophan, and decreases in hydrophobicity have been introduced by replacement with serine.
In the present study, we examined the structures of HP (2-20) and five analogues that have different degrees of hydrophobicity and amphiphilicity. In addition, we examined the effects of tryptophan residues on the structures and activities of these peptides. Interactions between the HP analogues and the membrane were also studied by 1H NMR spectroscopy, SEM (scanning electron microscopy) and fluorescence spectroscopy. Structure-antibiotic activity relationships of these peptides will aid in understanding their target cell selectivity against the bacterial cell membrane.
EXPERIMENTAL
Peptide synthesis
The peptides were synthesized by the solid-phase method using Fmoc (fluoren-9-ylmethoxycarbonyl) chemistry [17]. The peptides were purified by reverse-phase preparative HPLC using a 15 μm Deltapak C18 column (19×30 cm, Waters, Milford, U.S.A.). The purified peptides were greater than 95% pure as judged by analytical HPLC with an Ultrasphere C18 column (4.6×25 cm, Beckman, Fullerton, U.S.A.). All peptides had the correct molecular mass, as determined by matrix-assisted laser-desorption ionization–time-of-flight MS [18].
Microbial strains
Listeria monocytogenes (KCTC 3710), Candida albicans (KCTC 7270), Aspergillus flavus (KCTC 9471), and Aspergillus fumigatus (KCTC 6145) were obtained from the Korean Collection for Type Cultures (Korea Research Institute of Bioscience and Biotechnology, Taejon, Korea). Escherichia coli O157 (ATCC 43895) was obtained from the A.T.C.C.
Antibacterial activity
The bacteria were grown to mid-logarithmic phase in a medium containing 10 g/l bactotryptone, 5 g/l yeast extract and 10 g/l NaCl (pH 7.0). The peptide was serially diluted to concentrations of 100, 50, 25, 12.5, 6.25, 3.125, 1.56, 0.78, 0.39, 0.195 and 0.097 μM in media containing 1% bactopeptone. The tested organism (final bacterial suspension=5×103 colony forming units/ml) suspended in growth medium (100 μl) was mixed with 100 μl of the test peptide solution in a microtitre-plate well. Three replicates were generated for each test condition. Following incubation for 10 h at 37 °C, microbial growth was determined by the increase in absorbance measured at 620 nm.
Measurement of antifungal activity
The fungal strains were grown at 28 °C in potato dextrose broth. Fungal cells (2×103 cells in 100 μl) in potato dextrose broth were seeded on to a 96-well microtiter plate. A 10 μl aliquot of the serially diluted peptide and various concentrations of amphotericin B were added to each well, and the cell suspension was incubated for 24 h at 28 °C. Next, 10 μl of 5 mg/ml 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide in PBS (35 mM phosphate buffer, 150 mM NaCl, pH 7.4) was added to each well, and the plates were incubated for an additional 4 h at 37 °C. A 30 μl quantity of 20% (w/v) SDS containing 0.02 M HCl was then added, and the plates were incubated at 37 °C for 16 h to dissolve the formazan crystals that had formed [19,20]. The absorbance of each well at 570 nm was measured using an Emax microtitre plate reader (Molecular Devices, Sunnyvale, CA, U.S.A.).
Haemolytic activity assay
The haemolytic activities of the peptides were evaluated by determining the amount of Hb released from the 8% suspension of fresh human RBCs (red blood cells). A 100 μl aliquot of the 8% RBC suspension was added to the wells in a 96-well plate. Then, 100 μl of the peptide solution dissolved in PBS was added until the final peptide concentration was 100 μM. Haemolysis was measured by absorbance at 414 nm with an Emax plate reader. Controls for no- and complete-haemolysis were determined in PBS and 0.1% Triton-X 100 respectively. The degree of haemolysis was calculated using the following equation:
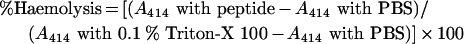 |
The effect of peptides on cellular morphology
RBCs were incubated for 1 h at 37 °C with 60% MIC (minimum inhibitory concentration) of amphotericin B or the peptides anal-3 (analogue-3) or melittin; C. albicans cells were incubated at 28 °C for 4 h with 12.5 μM anal-3; and A. flavus cells were treated at 28 °C for 4 h with 25 μM anal-3. Negative controls contained neither peptides or amphotericin B. The RBCs were fixed with equal volumes of 4% glutaraldehyde and 1% paraformaldehyde in 0.05 M cacodylate buffer (pH 7.2). After fixation for 3 h at 4 °C, the samples were centrifuged at 150 g and washed twice with 0.05 M cacodylate buffer (pH 7.2). The samples were dehydrated with sequential treatments with 50, 70, 90, 95 and 100% ethanol. After freeze drying and gold-coating, the samples were examined by SEM (HITACHI, Tokyo, Japan).
Quenching of tryptophan fluorescence by acrylamide
SUVs (small unilamellar vesicles) composed of EYPE (egg yolk L-α-phosphatidylethanolamine)/EYPG (EY L-α-phosphatidylglycerol) (7:3, w/w) and EYPC (EY L-α-phosphatidylcholine)/cholesterol (10:1, w/w) were prepared for quenching experiments. Briefly, each phospholipid (7.5 mg) was dissolved in chloroform and dried under a nitrogen stream and stored overnight under vacuum to completely remove trace amounts of organic solvent. Dried lipid films were resuspended in 2 ml Tris/HCl buffer (pH 7.4), (10 mM Tris/HCl, 150 mM NaCl and 0.1 mM EDTA) by vortex-mixing. The resulting lipid dispersion was sonicated for 10–20 min with a titanium tip ultrasonicator in an ice-bath under nitrogen flow until the turbidity had cleared. An appropriate amount of a peptide stock solution was added to obtain the desired lipid-to-peptide molar ratio. Excitation of tryptophan was performed at 295 nm instead of 280 nm to decrease absorbance contributed by acrylamide [21]. Tryptophan fluorescence was quenched by the titration of acrylamide, from a 4 M stock solution, to the final concentration of 0.4 M in the presence of liposome at a peptide/lipid molar ratio of 1:200. The quenching data were analysed by the Stern–Volmer plot using the equation, F0/F=1+Ksv[Q], where F0 is the fluorescence of the peptide in the absence of acrylamide, F is the fluorescence of the peptide in the presence of acrylamide, Ksv is the Stern–Volmer quenching constant and [Q] is the concentration of acrylamide.
CD analysis
CD experiments were performed using a J720 spectropolarimeter (Jasco, Tokyo, Japan) using a cell with a 1 mm path length. The CD spectra of the peptides at 100 μM were recorded at 25 °C at 0.1 nm intervals from 190 to 250 nm. To investigate the conformational changes induced by membrane-mimetic environments, DPC (dodecylphosphocholine), and SDS micelles of defined composition were added to the peptides. For each spectrum, the data from four scans were averaged and smoothed using the J720/98 system Version 120C. CD data were expressed as mean residue ellipticity [θ] given in deg·cm2·dmol−1.
NMR experiments
Perdeuterated SDS (SDS-D25) was purchased from Cambridge Isotope Laboratories (Andover, MA, U.S.A.). Peptides were dissolved at 1.0 mM in 0.45 ml of H2O/2H2O (9:1, v/v) (pH 4.0) containing 150 mM SDS micelles. DQF-COSY (double-quantum-filtered COSY), TOCSY and NOESY were performed by time-proportional phase incrementation [22–27]. TOCSY experiments were performed using 20 and 80 ms MLEV-17 spin-lock mixing pulses. Mixing times of 250 and 350 ms were used for NOESY experiments. For DQF-COSY experiments, 512 transients with 4 K complex data points were collected for each increment, and the data along the t1 dimension were zerofilled to 4 K before two-dimensional Fourier transformation. The 3JHNα coupling constants were measured from the DQF-COSY spectra with a spectral width of 4193.24 Hz and a digital resolution of 1.02 Hz/point. Chemical shifts are expressed relative to the 4,4-dimethyl-4-silapentane-1-sulphonate signal at 0 p.p.m. To investigate the intramolecular hydrogen bonding in the peptides, temperature coefficients were calculated from the TOCSY experiments at 298, 308 and 318 K. NMR spectroscopy can give useful information about the interaction between the peptides and micelles [28,29]. TOCSY experiments with 5 mM 5-doxylstearate spin-label and 16-doxylstearate spin-label were performed with a 40 ms mixing-time. All of the NMR spectra were recorded on an Bruker Avance-400 spectrometer at Konkuk University and a Bruker Avance-600 MHz spectrometer at the Korea Basic Science Institute. NMR spectra were processed off-line using the FELIX software package (Accelrys Inc., San Diego, CA, U.S.A.) on an SGI workstation.
Structure calculations
Distance constraints were extracted from the NOESY spectra with mixing-times of 150 and 250 ms. The volumes of the NOEs (nuclear Overhauser effects) between the two beta protons of the phenylalanine residue were used as references. All other volumes were converted into distances by assuming a 1/r6 distance dependence. All of the NOE intensities were divided in to three classes depending on their distance ranges: strong, 1.8 to 2.7 Å (1 Å=0.1 nm); medium, 1.8 to 3.3 Å; and weak, 1.8 to 5.0 Å [30,31]. Structure calculations were carried out using CNS version 1.1 (Yale University, New Haven, U.S.A.) with the topology and parameter sets ‘topallhdg’ and ‘parallhdg’ respectively. Standard pseudo-atom corrections were applied to the non-stereospecifically assigned restraints [32], and an additional 0.5 Å was added to the upper bounds for NOEs involving methyl protons [33]. A hybrid distance geometry-dynamically simulated annealing protocol [34,35] was employed to generate the structures. A total of 50 structures were generated, and the 20 structures with the lowest energies were selected for further analysis.
RESULTS
Antimicrobial activities of HP (2-20) and analogues
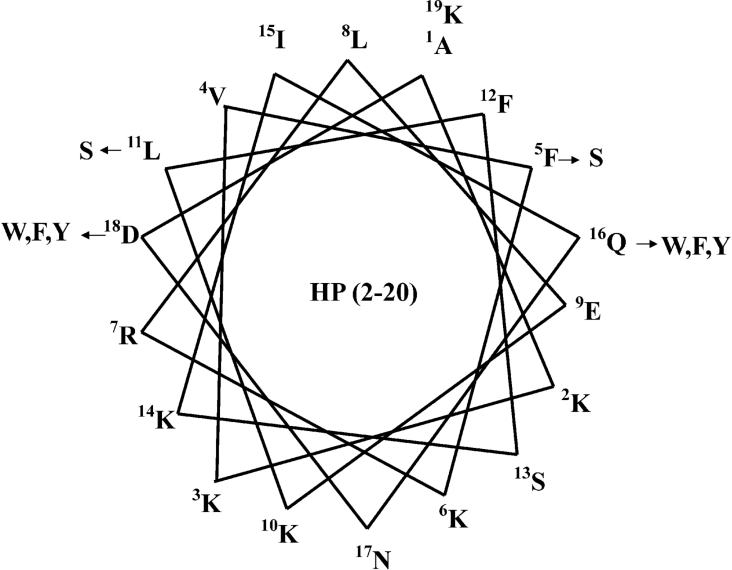
To investigate the relationship between amphiphilicity of antibiotic peptides and antimicrobial peptides, HP analogues were designed with increased or decreased hydrophobicity, by analysis of the α-helical wheel-diagram of HP (2-20) (Figure 1) [10,36–39]. The amino acid sequences and hydrophobic moments for each peptide are summarized in Table 1. Hydrophobicity was increased by replacement of hydrophilic residues such as Gln16 or Asp18 with tryptophan, phenylalanine and tyrosine, and was decreased by replacement of hydrophobic residues such as Phe5 or Leu11 with serine. As shown in Table 1, average hydrophilicity was calculated by CCS parameters, anal-3 and anal-6 show lower hydrophilicities of −1.01 and −0.97 respectively, compared with other peptides [40]. The amphipathicity of peptides was estimated by determining the mean hydrophobic moments (μH) using the Eisenberg equation [41]:
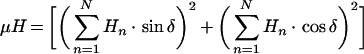 |
Where Hn is the hydrophobicity index value of residue n, and δ=100° for peptides in a helical conformation. According to the wheel-diagram and the hydrophobic moments, anal-3 and anal-6 have an optimum balance between the hydrophobic and hydrophilic phases, resulting in a best amphiphilicity of 0.68 among all analogues.
Figure 1. The α-helical wheel-diagram of HP (2-20).
The arrows indicate the positions of amino acid substitutions in HP (2-20). The HP (2-20) wheel is a perfect amphipathic α-helix with the hydrophobic residues in the upper region and the hydrophilic residues in the lower region.
Table 1. Amino acid sequences and hydrophobic moments of the synthetic antimicrobial peptide HP (2-20) derived from the N-terminus of H. pylori ribosomal protein L1 and its analogues.
| Peptide | Amino acid sequence | Substitution | H* | μH/μHmax† |
|---|---|---|---|---|
| HP (2-20) | AKKVFKRLEKLFSKIQNDK | Native | −2.78 | 0.63 |
| Anal 1 | AKKVFKRLEKLFSKIQNWK | (D18→W18) | −1.83 | 0.66 |
| Anal 2 | AKKVFKRLEKLFSKIWNDK | (Q16→W16) | −1.95 | 0.67 |
| Anal 3 | AKKVFKRLEKLFSKIWNWK | (Q16 D18→W16 W18) | −1.01 | 0.68 |
| Anal 4 | AKKVFKRLEKSFSKIQNDK | (L11→S11) | −3.52 | 0.59 |
| Anal 5 | AKKVSKRLEKLFSKIQNDK | (F5→S5) | −3.53 | 0.57 |
| Anal 6 | AKKVFKRLEKLFSKIFNFK | (Q16 D18→F16 F18) | −0.97 | 0.68 |
| Anal 7 | AKKVFKRLEKLFSKIYNYK | (Q16 D18→Y16 Y18) | −1.76 | 0.66 |
* Average hydrophilicity caculated by CCS parameters.
† Relative amphipathicity, i.e. the hydrophobic moment relative to that of a maximally amphipathic peptide. To facilitate comparisons, the amphipathicity of peptides is given relative to the maximum possible value (μHmax) resulting from a perfectly amphipathic 18-residue peptide composed only of leucine and aspartic acid, which would thus be assigned an μH/μHmax value of 1.
As shown in Table 2, the substitution of Gln16 and Asp18 of HP (2-20) with tryptophan in anal-3 resulted in 2-20 times greater antibacterial activity compared with HP (2-20) (MIC=0.39 and 6.25 μM against L. monocytogenes). The decrease in hydrophobicity caused by replacing Leu5 (anal-4) or Phe11 (anal-5) of HP (2-20) with serine residues produced peptides that were 2- or 16-fold less active than HP(2-20) respectively, against all of the bacterial cells tested. Anal-3 was as effective as melittin against most bacterial cells.
Table 2. Antibacterial and antifungal activities of HP (2-20) and its analogues.
| MIC [μM] | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ME* | HP† | A1 | A2 | A3 | A4 | A5 | A6 | A7 | |
| Bacteria | |||||||||
| B. subtilis | 0.78 | 3.13 | 1.56 | 1.56 | 1.56 | 6.25 | 3.13 | 1.56 | 1.56 |
| S. epidermidis | 3.13 | 6.25 | 3.13 | 3.13 | 3.13 | >12.5 | 12.5 | 6.25 | 3.13 |
| S. aureus | 0.78 | 12.5 | 3.13 | 6.25 | 1.56 | >12.5 | >12.5 | 3.13 | 3.13 |
| E. coli | 0.78 | 3.13 | 0.78 | 3.13 | 1.56 | 12.5 | 6.25 | 3.13 | 3.13 |
| S. typhimurium | 0.39 | 1.56 | 0.39 | 0.78 | 0.39 | 12.5 | 12.5 | 0.39 | 0.39 |
| P. vulgaris | 1.56 | 6.25–3.13 | 3.13 | 3.13 | 3.13 | >12.5 | >12.5 | 3.125 | 6.25–3.13 |
| P. aeruginosa | 3.13 | 6.25 | 3.13 | 6.25 | 3.13 | 12.5 | 12.5 | 6.25 | 3.13 |
| E. coli O157 | 3.13 | 12.5 | 6.25 | 6.25 | 3.13 | >12.5 | 6.25 | 6.25 | 3.13 |
| L. monocytogenes | 0.39 | 6.25 | 0.78 | 1.56 | 0.39 | >12.5 | >12.5 | 0.78 | 0.78 |
| Fungi | |||||||||
| T. beigelii | 1.56 | 3.13 | 1.56 | 1.56 | 1.56 | 12.5 | 12.5 | 3.13 | 1.56 |
| C. albicans | 12.5 | >25 | 25 | 25 | 12.5 | >25 | >25 | >25 | 12.5 |
| S. cerevisiae | 12.5 | >25 | 25 | 25 | 25 | >25 | >25 | 25 | 25 |
| A. flavus | 25 | 50 | 50 | 50 | 25 | >50 | >50 | 50 | 50 |
| A. fumigatus | 12.5 | >50 | 50 | 50 | 25 | >50 | >50 | 50-25 | 50-25 |
| F. moniliforme | 12.5 | 25 | 12.5 | 25 | 12.5 | >50 | >50 | 12.5 | 12.5 |
| B. cinerea | 1.56 | 12.5 | 6.25 | 6.25 | 3.13 | >50 | >50 | 12.5 | 6.25 |
* melittin
† HP (2-20).
We next examined the antifungal activity of the peptides against pathogenic fungi by measuring the MIC as assessed by mitochondrial conversion of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide [42]. The results indicated that anal-3 displayed approx. 2- to 4-fold more potency at inhibiting fungal growth than HP (2-20) (Table 2). Anal-3 was also as effective as melittin.
In order to investigate the effects of various aromatic residues, peptides with systematic substitutions of Gln16 and Asp18 with tryptophan, phenylalanine, or tyrosine were synthesized and named as anal-3, anal-6 and anal-7 respectively. Antimicrobial activities were measured for anal-3, anal-6 and anal-7. Table 2 shows that anal-3, which has replacement of Gln16 and Asp18 with tryptophan shows approx. 2-fold better antimicrobial activities compared with anal-6 or anal-7, which has phenylalanine or tyrosine substitutions, respectively. Also, tryptophan is a good fluorophore to investigate the interactions between the peptide and the membrane. Therefore further experiments were performed only for anal-3 and the structure of the most active anal-3 was studied representatively.
Haemolytic activities of HP (2-20) and its analogues
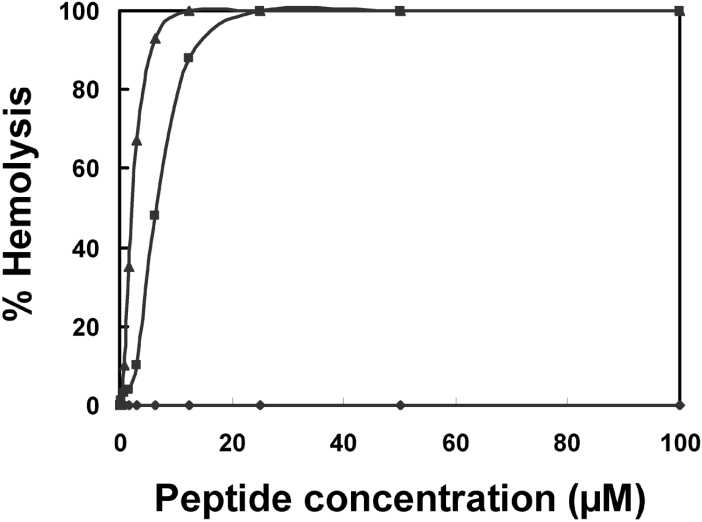
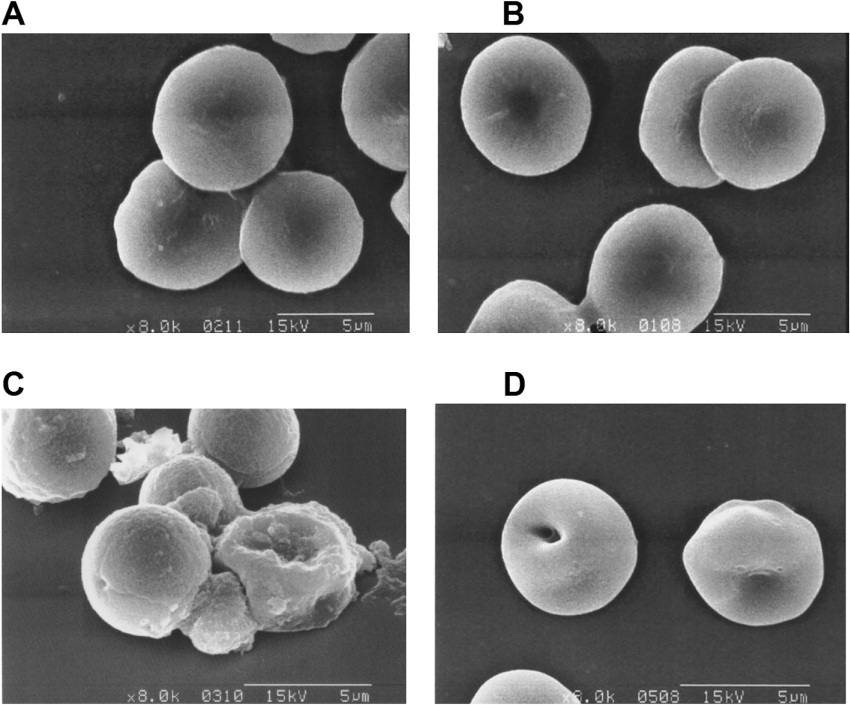
Since anal-3 shows the highest antimicrobial activities among all HP analogues, we further investigated the interactions between the antimicrobial peptide anal-3 and the various types of biological membranes. First, cytotoxicity of anal-3 against mammalian cells was studied. Although anal-3 had remarkable antibacterial and antifungal activity, it did not exhibit haemolytic activity, even at high concentrations, as shown in Figure 2. Both melittin and amphotericin B, which are well known antimicrobial peptide and antibiotic, were potent inducers of haemolysis. At concentrations of melittin or amphotericin B causing 60% haemolysis, SEM showed a significant difference in the morphology of the RBCs, including the frequent observation of pores in the cell surface (Figure 3). Even at concentrations above 100 μM, anal-3 did not cause any difference in the morphology of RBCs or damage to RBC surfaces. HP (2-20) [10] and other analogues (results not shown) also did not show any haemolytic activities.
Figure 2. Haemolytic activities of anal-3.
The haemolytic activities of anal-3 (◆), melittin (▲), and amphotericin B (■) were determined by measuring Hb release at 414 nm from 8% suspensions of fresh human RBCs.
Figure 3. Scanning electron micrographs showing the haemolytic effects of (A) no addition, (B) anal-3, (C) melittin, and (D) amphotericin B on human RBCs.
RBCs were incubated for 1 h at 37 °C with 60% MIC of amphotericin B or the peptides. At MIC melittin and amphotericin B destroyed RBCs completely and it was not possible to examine the morphology of RBCs. Therefore all of the micrographs were taken at 60% MIC. Even at concentrations above 100 μM, anal-3 did not cause any differences in the morphology of RBCs or damage to RBC surfaces.
Morphological changes induced in C. albicans and A. flavus by anal-3
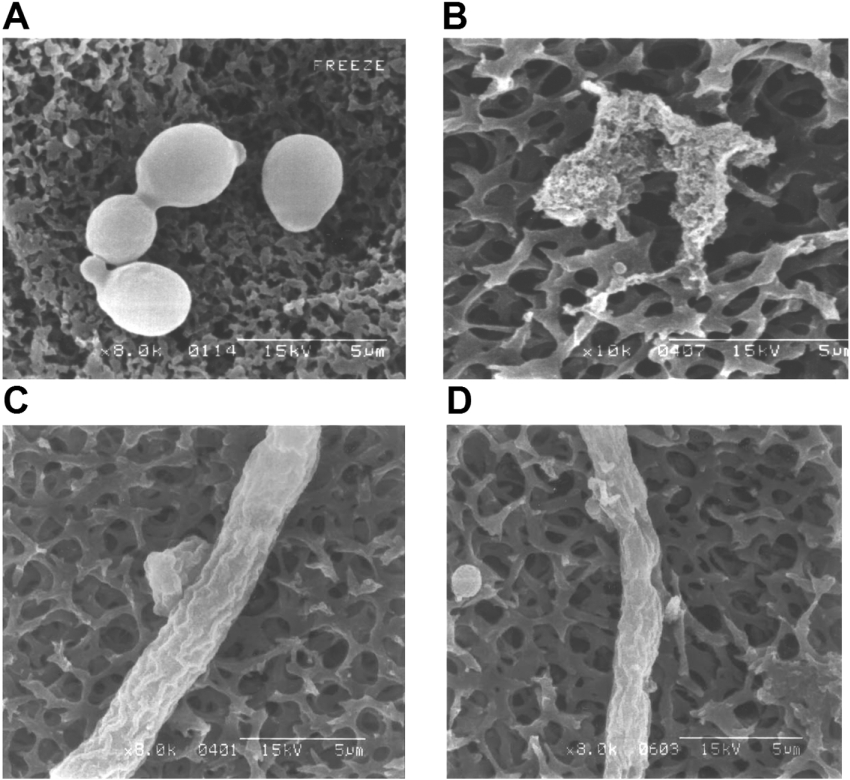
The morphological changes induced in C. albicans by anal-3 were examined by SEM. At peptide concentrations corresponding to the MIC value, we found significant differences in the morphologies of the fungi. Untreated cells had a normal, smooth cell surface (Figure 4A), whereas the cells treated for 4 h with anal-3 showed surface roughening and cellular debris from cell lysis of C. albicans was observed (Figure 4B). Similarly, in A. flavus, untreated cells had a normal cell surface (Figure 4C), whereas cells treated with anal-3 at 60% MIC concentration showed large holes in their surface (Figure 4D). These results provide additional evidence that anal-3 has potent antifungal activity due to disruption of fungal plasma membranes.
Figure 4. Morphological changes in C. albicans and A. flavus induced by anal-3.
Scanning electron micrographs of C. albicans treated for 4 h at 28 °C with (A) no addition and (B) 12.5 μM anal-3. Scanning electron micrographs of A. flavus treated for 4 h at 28 °C with (C) no addition or (D) 25 μM anal-3.
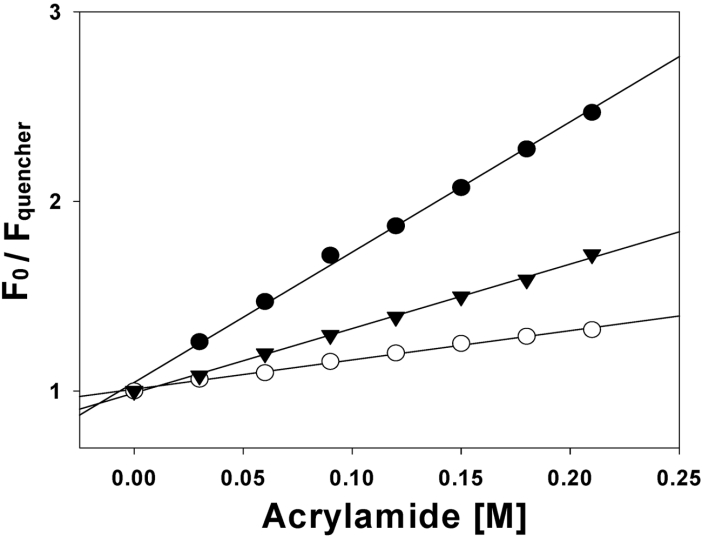
Tryptophan fluorescence quenching by acrylamide
To determine the relative extent of burial of the tryptophan residues of the peptides into model membranes, a fluorescence quenching experiment was performed with the water-soluble fluorescence quencher acrylamide, by determining the Stern–Volmer constants (Ksv). Stern–Volmer plots for the quenching of tryptophan fluorescence in the anal-3 peptide, recorded by acrylamide in the presence of phospholipid vesicles, are depicted in Figure 5. The quenching of tryptophan fluorescence in anal-3 peptide is less effective with negatively charged vesicles than with zwitterionic vesicles, suggesting that the tryptophan residue of anal-3 peptide is buried more extensively in negatively charged phospholipid vesicles than in zwitterionic phospholipid vesicles. This result suggests the bacterial cell selectivity of anal-3 peptide is due to its high binding affinity with negatively charged phospholipids in bacterial cells.
Figure 5. Stern–Volmer plots for the quenching of tryptophan fluorescence of anal-3.
Fluorescence was quenched by aqueous acrylamide, in the presence of Tris buffer (pH 7.4) (●), EYPE/EYPG (7:3, w/w) SUVs (○) and EYPC/cholesterol (10:1, w/w) SUVs (▼). The concentrations of peptides and phospholipid vesicles were 3.0 and 600 μM respectively.
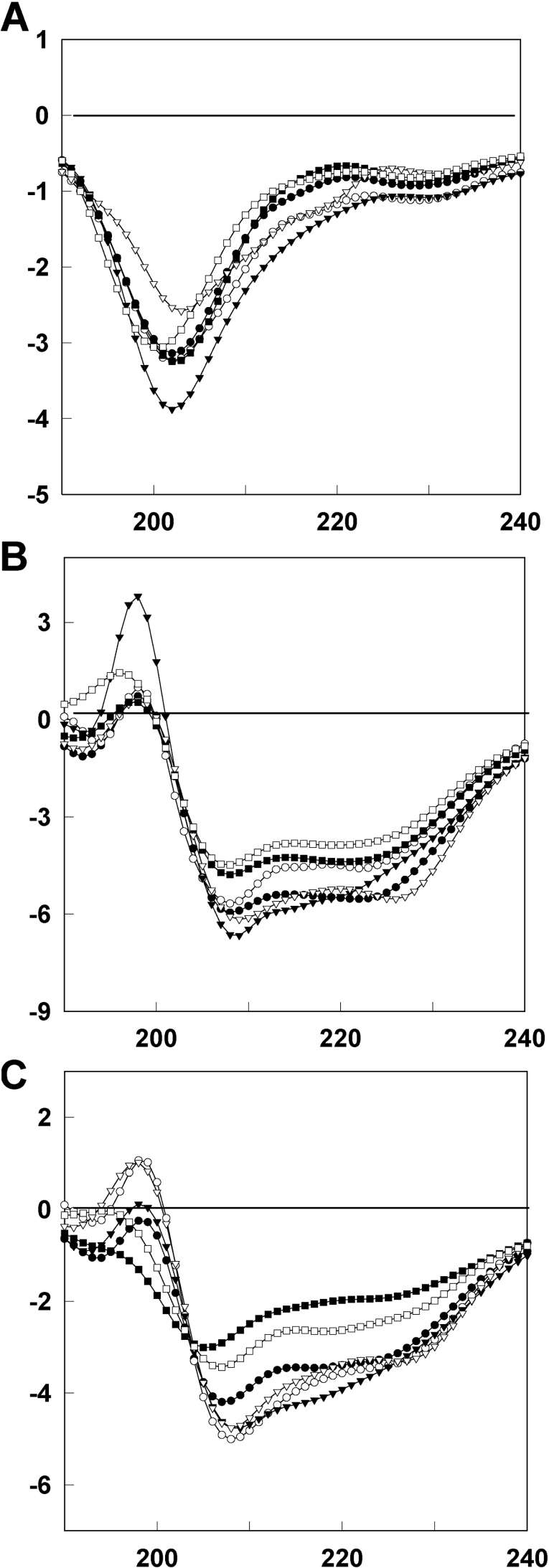
CD measurements of HP (2-20) and its analogues
To investigate the secondary structure of HP (2-20) and its analogues in membrane-mimetic environments, we measured CD spectra of HP (2-20) and its peptide analogues dissolved in an aqueous solution of 100 mM SDS micelles, or 50 mM DPC micelles (Figure 6). The CD spectra of HP (2-20) and its analogues show that they have random-coil structures in aqueous solution, whereas they adopt α-helical conformations in membrane-mimetic environments. Anal-4 and anal-5, which have less antibiotic activity than HP (2-20), have a lower α-helical content than the other peptides. Anal-1, anal-2 and anal-3 have slightly higher α-helicity than HP (2-20) in these membrane-mimetic environments.
Figure 6. CD spectra of HP (2-20) and its analogues in (A) aqueous solution, (B) 100 mM SDS micelles and (C) 50 mM DPC micelles.
Shown are spectra for (●) HP (2-20), (○) anal-1, (▼) anal-2, (▽) anal-3, (■) anal-4 and (□) anal-5. The concentration of peptides was 50 μM.
Resonance assignment of HP (2-20) and its analogues
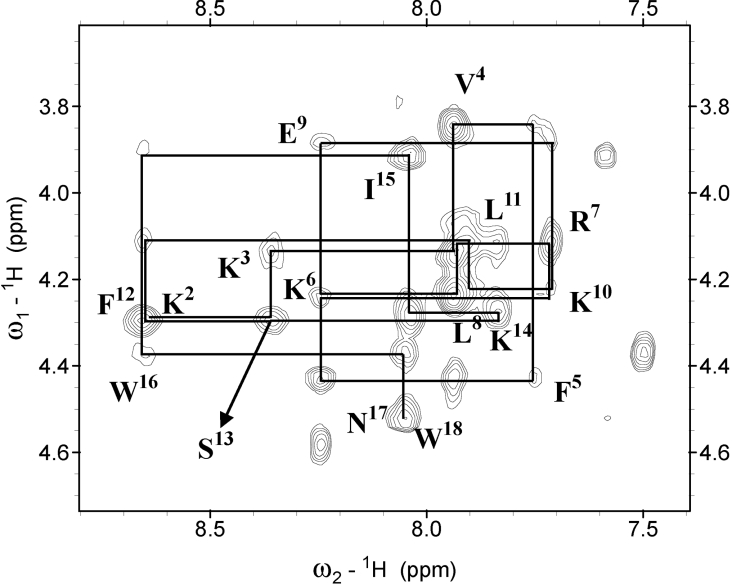
Sequence specific resonance assignments were performed using mainly the DQF-COSY, TOCSY and NOESY data [43]. Figure 7 shows the NOESY spectra with the sequential assignments for anal-3 and its analogues in the NH-CαH region. The chemical shifts of HP (2-20) and its analogues in SDS micelles at 318 K and pH 6.0, were deposited in the BioMagResBank database under accession number 5265. The overall chemical shift of HP (2-20) is similar to those of its analogues, except for the resonances in the regions with substitutions.
Figure 7. NH-CαH region of NOESY spectra with a 250 ms mixing-time for anal-3 in SDS micelles.
For the sake of clarity, only the intra-residual NH-CαH cross-peaks are labelled.
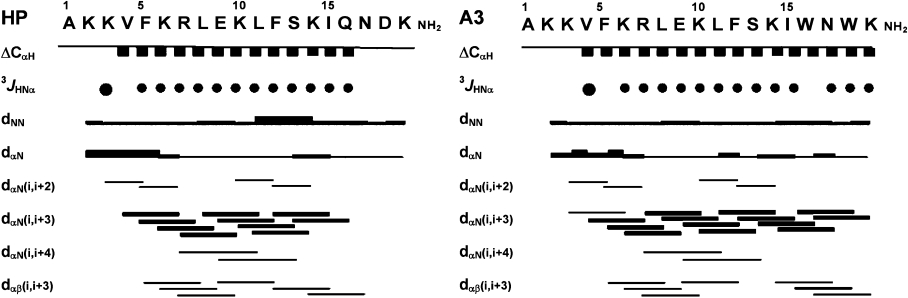
The sequential NOE connectivities and the other NMR data are illustrated in Figure 8. As shown in Figures 7 and 8 a number of non-sequential NOE connectivities that are characteristic of an α-helix, specifically dαβ(i,i+3) and dαN(i,i+3), were observed for all of the peptides. The observed value of the 3JHNα coupling constant for the helical region of all of the peptides, derived using the method of Kim and Prestegard [44], was generally below 6 Hz. The amide-proton temperature coefficient has been used to predict hydrogen bond donors, and values above −4.5 p.p.b./K indicate that the amide proton is involved in intramolecular hydrogen bonding [45]. Temperature coefficients of the amide protons in the α-helical region of the peptides were generally above −4.5 p.p.b./K, indicating that the mid-regions of these peptides form α-helices.
Figure 8. Summary of the NOE connectivities, JHNα coupling constants (JHNα< 6 Hz) and CαH chemical shift indices for HP (2-20) (HP) and anal-3 (A3) in SDS micelles.
The line thickness for the NOEs reflects the intensity of the NOE connectivities.
Structure of HP (2-20) and its analogues
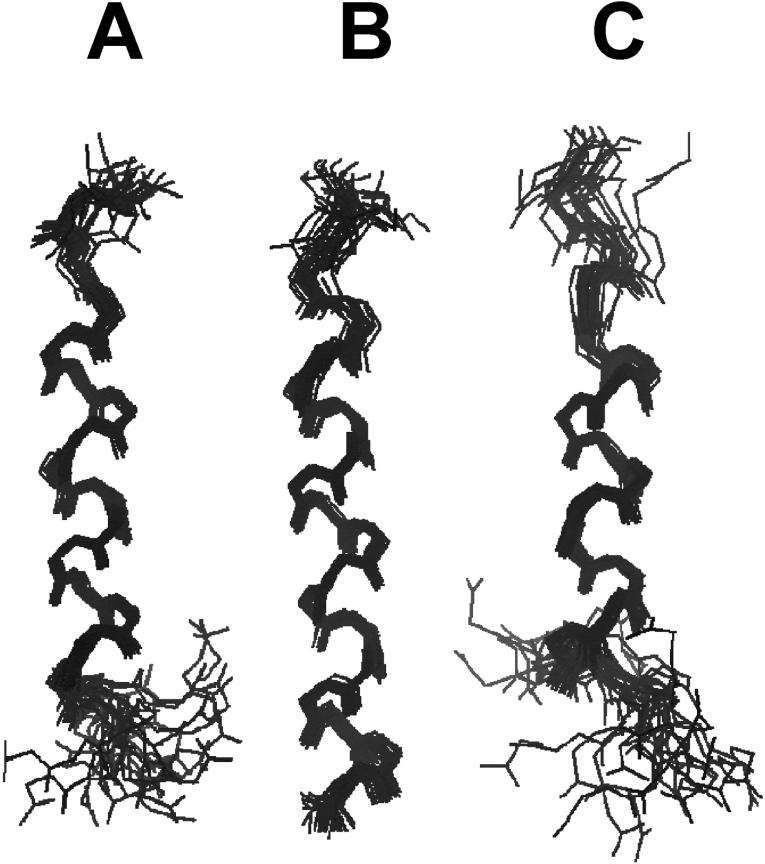
To calculate the tertiary structures of HP (2-20) and its analogues, we used experimental restraints, such as sequential (|i-j|=1), medium-range (1<|i-j|≤5), long-range (|i-j|>5), intraresidual distance and torsion-angle restraints. They are listed in the Supplementary data (http://www.BiochemJ.org/bj/394/bj3940105add.htm). Of the structures that were accepted with small deviations from the idealized covalent geometry and the experimental restraints (≤0.05 Å for bonds, ≤5° for angles, ≤5° for chirality, ≤0.3 Å from NOE restraints and ≤3° from torsion angle restraints), we analysed 20 output structures with the lowest energy for each peptide. None of the structures had violations over 0.5 Å from the NOE distance restraints or 3° from dihedral angle restraints, and all the structures exhibited good covalent geometry. The statistics for the 20 final simulated annealing structures of HP (2-20) and its analogues are also given in the Supplementary data (http://www.BiochemJ.org/bj/394/bj3940105add.htm). When we superimposed the 20 lowest energy structures of HP (2-20), anal-4 and anal-5 over the backbone atoms (from Val4 to Gln16), their R.M.S.Ds (root mean squared deviations) from the mean structures were 0.35±0.15 Å, 0.38±0.18 and 0.35±0.11 for the backbone atoms (N, Cα, C′, O) and 1.11±0.14 Å, 1.12±0.19 and 1.06±0.20 for all heavy atoms respectively. When the low-energy structures of anal-1, anal-2 and anal-3 were superimposed over the backbone atoms from Val4 to Trp18, their R.M.S.Ds from the mean structures were 0.35±0.12 Å, 0.30±0.09 and 0.26±0.08 for the backbone atoms and 1.21±0.1 Å, 1.10±0.09 and 1.22±0.12 for all heavy atoms respectively. Figure 9 shows the superposition of the 20 lowest energy structures of HP (2-20), and anal-3 and anal-5 over the backbone atoms in 150 mM SDS micelles. HP (2-20) peptides have a stable amphiphilic helix from Val4 to Gln16. Because of the increase in hydrophobicity caused by substitution of tryptophan at the C-terminus, anal-1, anal-2 and anal-3 have longer amphiphilic helical-structures from Val4 to Trp18 than HP (2-20). Because anal-4, and anal-5 do not have tryptophan residues at the C-terminus and anal-4 and anal-5 have lower amphiphilicity due to replacement of leucine or phenylalanine with serine compared with HP (2-20), they have helical structures only from Val4 to Gln16. These results show that increasing the hydrophobicity by the addition of tryptophan residues at the C-terminus stabilizes the overall α-helical structures in anal-1, anal-2 anal-3 and results in longer amphiphilic helical structures and higher antimicrobial activity.
Figure 9. The superpositions of the 20 lowest energy structures of (A) HP(2-20), (B) anal-3 and (C) anal-5 in SDS micelles.
Backbone atoms of residues 5–19 were superimposed. Other structures are shown as Supplementary data (http://www.BiochemJ.org/bj/394/bj3940105add.htm). This figure was generated using MOLSCRIPT.
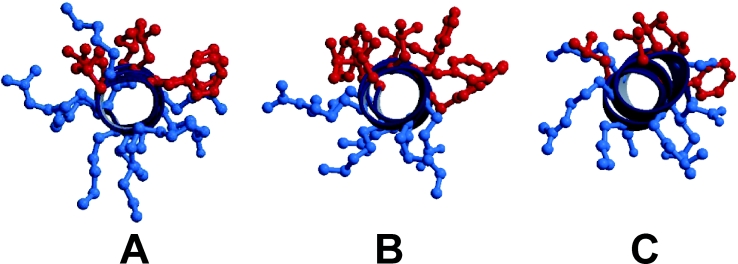
Figure 10 shows the orientation of the hydrophobic and hydrophilic side chains of HP (2-20), anal-3 and anal-5 from Val4 to Trp18. It is well known that when an amphipathic peptide forms an ion channel, the hydrophilic residues face inwards to contact the solvent and the hydrophobic side-chains face towards the acyl chains of the hydrophobic lipid [16,46–48]. The hydrophobic side-chains in these peptides, shown in red in the Figure, protrude towards one side, and the hydrophilic side-chains, shown in blue, protrude towards the other side. Among the HP (2-20) analogues, anal-3 has the highest hydrophobic moment and has a good balance between the hydrophobic residues and the hydrophilic residues, as shown in Figure 10(B).
Figure 10. Head-on view of (A) HP(2-20), (B) anal-3, and (C) anal-5 generated using MOLSCRIPT.
The hydrophobic side-chains are indicated in red, and the hydrophilic side-chains are shown in blue.
The position of anal-3 relative to the SDS micelles
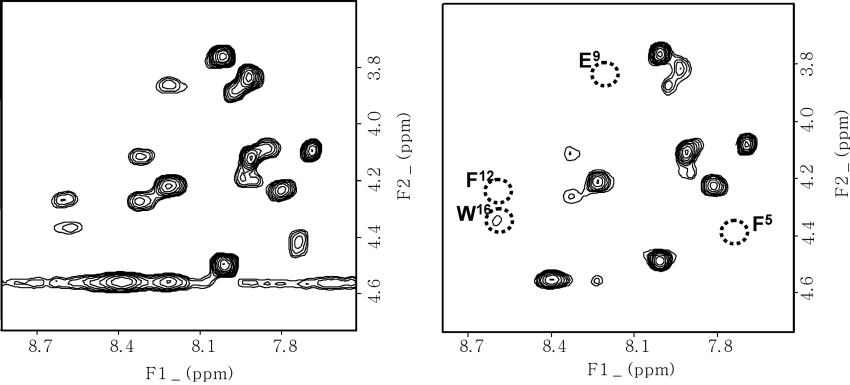
To probe the position of anal-3 relative to the SDS micelles, we added stearate which was spin-labelled at the 5 or 16 position with a doxyl group containing a stable nitroxide radical. The average position of the spin-labels in the SDS micelles was derived from the line-broadening of the resonances in the 13C spectrum of SDS, as reported previously [28,49]. The effects of the paramagnetic probes depend upon the distance between them and the protons in the peptide, and they can affect the chemical shifts and the signal intensities in the two-dimensional TOCSY spectrum. As shown in Figure 11, addition of 5-doxylstearate caused the intensities of the resonances from the Phe5, Leu8, Glu9, Phe12 and Trp16 residues to substantially weaken and disappear whereas 16-doxylstearate did not cause any changes in the spectrum. Because the resonance of Trp18 overlapped with that of Asn17, it was not possible to monitor the effects on Trp18. These results imply that these residues are buried on the surface of the SDS micelles and have hydrophobic interactions in the micelle with the acyl chains of the phospholipid near the spin-labels.
Figure 11. TOCSY spectra of anal-3.
Spectra in the absence of spin-labelled stearate is shown on the left and spectra in the presence of 3.0 mM 5-doxylstearate at 318 K is shown on the right. Concentrations of the peptides for all NMR experiments were 1 mM in 150 mM SDS micelles.
DISCUSSION AND CONCLUSIONS
Analysis of tertiary structures by NMR spectroscopy revealed that the tryptophan residue at the C-terminus of anal-1, anal-2 and anal-3 stabilizes the amphipathic α-helical structure of the C-terminus. By contrast, a decrease in the hydrophobicity of anal-1, anal-2 and anal-3, by replacing leucine (anal-4) or phenylalanine (anal-5) with serine decreased antibacterial and antifungal activities. Anal-3 shows only slightly better antimicrobial activities than anal-6 and anal-7 which carry replacement of Gln16 and Asp18 with phenylalanine and tyrosine residues respectively. Anal-3 and anal-6 have the highest hydrophobic moments among all the peptides. Therefore the hydrophobicity at the positions where the tryptophan residues are located in the sequence is important and it can be tryptophan or phenylalanine. According to the helical wheel-diagram, anal-3 shows the optimum balance between the hydrophobic and hydrophilic amino acids, and amphipathicity of the α-helix plays an important role in antibiotic activity.
Anal-3 interacts more strongly with anionic than neutral phospholipid vesicles because it has seven positively charged amino acids (six lysine residues and one arginine). This suggests that the positively charged residues in anal-3 preferentially interact with the head groups of anionic phospholipids rather than those of neutral phospholipids. This possibility is supported by the tryptophan fluorescence quenching experiments. These suggested that the tryptophan residue of anal-3 peptide is buried more extensively in negatively charged phospholipid vesicles than in zwitterionic phospholipid vesicles. These results imply that the bacterial cell selectivity of the anal-3 peptide is due to its high binding affinity with negatively charged phospholipids in bacterial cells.
The differing cytotoxic effects of the peptides on bacterial cells and RBCs may be due to differences in their cellular membrane lipid compositions. In the RBC membrane, the lipids in the two leaflets are distributed so that zwitterionic phospholipids, such as phosphatidylcholine and sphingomyelin, are concentrated in the outer leaflet of the bilayer, whereas the negatively charged phosphatidylserine residue is present exclusively in the inner leaflet [14,46]. However, in bacterial or fungal cells, negatively charged lipids are present exclusively in the outer leaflet. Therefore it is not surprising that highly positively charged peptides with high amphiphilicity show strong lytic activity against bacterial or fungal cells. Indeed, in the current studies, SEM showed that anal-3 causes severe damage to the surface of C. albicans and A. flavus but no damage to RBCs.
It is well known that aromatic rings in peptide play an important role in hydrophobic membrane interactions [16,46–48,50]. Tryptophan residues, in particular, are known to have an important role in the interaction between peptides and biological membranes [16] and tryptophan is critical for the haemolytic activity of melittin [14]. In melittin, tryptophan residues are located in the hydrophobic core of the peptide, restricting its movement in the membrane, and are involved in hydrophobic interactions with the acyl chains of phospholipids [11–14]. These interactions help to allow melittin to form a channel in the membrane. By contrast, the cecropin A–magainin hybrid peptide and PMAP (porcine myeloid antibacterial peptide) 23, which lack haemolytic activity against RBCs even at concentrations up to 100 μM, have tryptophan residues at their termini and have helix-bend-helix structures [16,51]. The partial insertion of their tryptophan residues in to the membrane, as well as the electrostatic interactions between the positively charged lysine residues and the anionic phospholipid head groups, mediate the primary binding to the cell membrane. Thereafter, the flexibility or bending potential induced by the hinge regions in the central part of cecropin A–magainin hybrid peptide and PMAP23 may allow the α-helix to span the lipid bilayer [16,51]. Thus the different cell specificities of these two peptides and melittin are probably due to their different modes of interaction with and insertion into biological membranes.
In the case of anal-3, the tryptophan residue is located at a rather flexible region in the C-terminus, and the peptide does not show haemolytic activity. Furthermore, anal-3 has a linear amphiphilic α-helical structure. A model of the relative position of anal-3 in the SDS micelle was derived from the effects of the spin-labels on anal-3 resonances. The spin-label of 5-doxylstearate is located near the surface of the SDS micelles whereas that of 16-doxylstearate is located in the middle of the SDS micelles. Since only 5-doxylstearate affected the intensity of the resonances of anal-3, the side-chains of the hydrophobic phase of the amphiphilic α-helix must be buried in the surface of the micelles. Also, Lys2 and Lys3 at the N-terminus and Lys19 at C-terminus may produce an optimum arrangement for electrostatic interactions between the sulphate head groups of SDS and the positively charged lysyl NH3+.
The Asp18 and Gln16 side-chains in HP (2-20) are partially negatively charged and are located at the boundary of the hydrophobic and hydrophilic phases in the helical wheel-diagram. Therefore they may repel the negatively charged head group in the bacterial membrane. Replacement of these residues with tryptophan in anal-3 resulted in the highest hydrophobic moments and good amphiphilicity, which may explain why anal-3 has the highest antibacterial activity among the HP (2-20) analogues. Positively charged lysine and arginine residues, in addition to interactions between the indole ring of tryptophan in anal-3 and the membrane, mediate the initial binding to the negatively charged membrane surface of the bacteria, whereupon the peptides form amphipathic α-helical structures. Hydrophobic regions in the amphipathic structures in anal-3 are buried in the surface of the membrane and result in disruption of the microbial cell membrane. Overall, the results of this study help to clarify the structure-activity relationship of HP (2-20) and its analogues and should assist in our efforts to design novel antimicrobial peptides that have potent antibiotic activity but no haemolytic effects.
Online data
Acknowledgments
This work was supported by a grant from the Molecular and Cellular BioDiscovery Research Program (M10301030000) of the Ministry of Science and Technology, and by grants from the Ministry of Science and Technology, Korea and the Korea Science and Engineering Foundation through the Research Center for Proteineous Materials.
References
- 1.Koczulla A. R., Bals R. Antimicrobial peptides-current status and therapeutic potentials. Drugs. 2003;63:389–406. doi: 10.2165/00003495-200363040-00005. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrovial peptides of multicellular organisms. Nature (London) 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Barra D., Simmaco M. Amphibian skin: a promising resource for antimicrobial peptides. Trends Biotechnol. 1995;13:205–209. doi: 10.1016/S0167-7799(00)88947-7. [DOI] [PubMed] [Google Scholar]
- 4.Lehrer R. I., Lichtenstein A. K., Ganz T. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 5.Boman H. G., Faye I., Gudmundsson G. H., Lee J. Y., Lidholm D. A. Cell-free immunity in Cecropia. A model system for antibacterial proteins. Eur. J. Biochem. 1991;201:23–31. doi: 10.1111/j.1432-1033.1991.tb16252.x. [DOI] [PubMed] [Google Scholar]
- 6.De Lucca A. J., Walsh T. J. Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 1999;43:1–11. doi: 10.1128/aac.43.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser M. J. The versatility of Helicobacter pylori in the adaptation to the human stomach. J. Physiol. Pharmacol. 1997;48:307–314. [PubMed] [Google Scholar]
- 8.Putsep K., Branden C.-I., Boman H. G., Normark S. Antibacterial peptide from H. pylori. Nature (London) 1999;398:671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- 9.Putsep K., Normark S., Boman H. G. The origin of cecropins: implications from synthetic peptides derived from ribosomal protein L1. FEBS Lett. 1999;451:249–252. doi: 10.1016/s0014-5793(99)00582-7. [DOI] [PubMed] [Google Scholar]
- 10.Lee D. G., Kim P. I., Park Y., Jang S.-H., Park S.-C., Woo E.-R., Hahm K.-S. HP (2–20) derived from the amino terminal region of Helicobacter pylori ribosomal protein L1 exerts its antifungal effects by damaging the plasma membranes of Candida albicans. J. Peptide Sci. 2002;8:453–460. doi: 10.1002/psc.405. [DOI] [PubMed] [Google Scholar]
- 11.Tosteson M. T., Holmes S. J., Razin M., Tosteson D. C. Melittin lysis of red cells. J. Membr. Biol. 1985;87:35–44. doi: 10.1007/BF01870697. [DOI] [PubMed] [Google Scholar]
- 12.Blondelle S. E., Simpkins L. R., Perez-Paya E., Houghten R. A. Influence of tryptophan residues on melittin's hemolytic activity. Biochim. Biophys. Acta. 1993;1202:331–336. doi: 10.1016/0167-4838(93)90024-l. [DOI] [PubMed] [Google Scholar]
- 13.Hancock R. E., Chapple D. S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh A. K., Rukmini R., Chattopadhyay A. Modulation of tryptophan environment in membrane-bound melittin by negatively charged phospholipids: implications in membrane organization and function. Biochemistry. 1997;36:14291–14305. doi: 10.1021/bi971933j. [DOI] [PubMed] [Google Scholar]
- 15.Yu K., Kang S., Park N., Shin J., Kim Y. Relationship between the tertiary structures of mastoparan B and its analogs and their lytic activities studied by NMR spectroscopy. J. Peptide Res. 2000;55:51–62. doi: 10.1034/j.1399-3011.2000.00146.x. [DOI] [PubMed] [Google Scholar]
- 16.Oh D., Shin S-Y., Lee S., Kang J. H., Kim S. D., Ryu P. D., Hahm K.-S., Kim K., Kim Y. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin a (1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures. Biochemistry. 2000;39:11855–11864. doi: 10.1021/bi000453g. [DOI] [PubMed] [Google Scholar]
- 17.Merrifield R. B. Solid phase synthesis. Science (Washington, D.C.) 1986;232:341–347. doi: 10.1126/science.3961484. [DOI] [PubMed] [Google Scholar]
- 18.Duncan M. W., Matanovic G., Cerpa-Poljak A. Quantitative analysis of low molecular weight compounds of biological interest by matrix-assisted laser desorption ionization. Rapid Commun. Mass Spectrom. 1993;7:1090–1094. doi: 10.1002/rcm.1290071207. [DOI] [PubMed] [Google Scholar]
- 19.Jahn B., Martin E., Stueben A., Bhakdi S. Susceptibility testing of Candida albicans and Aspergillus species by a simple microtiter menadione-augmented 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay. J. Clin. Microbiol. 1995;33:661–667. doi: 10.1128/jcm.33.3.661-667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H., Kinnunen P. K. Binding of the antimicrobial peptide temporin L to liposomes assessed by Trp fluorescence. J. Biol. Chem. 2002;277:25170–25177. doi: 10.1074/jbc.M203186200. [DOI] [PubMed] [Google Scholar]
- 22.Derome A. E., Williamson M. P. Rapid-pulsing artifacts in double-quantum-filtered COSY. J. Magn. Reson. 1990;88:177–185. [Google Scholar]
- 23.Bax A., Davis D. G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 24.Macura S., Ernst R. R. Elucidation of cross-relaxation in liquids by two-dimensional NMR spectroscopy. Mol. Phys. 1980;41:95–117. [Google Scholar]
- 25.Bax A., Davis D. G. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 1985;63:207–213. [Google Scholar]
- 26.Marion D., Wuthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 27.Mueller L. P.E.COSY, a simple alternative to E.COSY. J. Magn. Reson. 1987;72:191–197. [Google Scholar]
- 28.Papavoine C. H., Konings R. N., Hilbers C. W., van de Ven F. J. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 1994;33:12990–12997. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- 29.Lee S., Kim Y. Solution structure of neuromedin B by 1H nuclear magnetic resonance spectroscopy. FEBS Lett. 1999;460:263–269. doi: 10.1016/s0014-5793(99)01346-0. [DOI] [PubMed] [Google Scholar]
- 30.Clore G. M., Gronenborn A. M. Determination of three-dimensional structures of proteins and nucleic acids in solution by nuclear magnetic resonance spectroscopy. Crit. Rev. Biochem. Mol. Biol. 1989;24:479–564. doi: 10.3109/10409238909086962. [DOI] [PubMed] [Google Scholar]
- 31.Clore G. M., Gronenborn A. M. Structures of larger proteins, protein-ligand and protein-DNA complexes by multi-dimensional heteronuclear NMR. Protein Sci. 1994;3:372–390. doi: 10.1002/pro.5560030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wuthrich K., Billeter M., Braun W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constrains with nuclear magnetic resonance. J. Mol. Biol. 1983;169:949–961. doi: 10.1016/s0022-2836(83)80144-2. [DOI] [PubMed] [Google Scholar]
- 33.Clore G. M., Gronenborn A. M., Nilges M., Ryan C. A. Three-dimensional structure of potato carboxypeptidase inhibitor in solution. A study using nuclear magnetic resonance, distance geometry, and restrained molecular dynamics. Biochemistry. 1987;26:8012–8023. doi: 10.1021/bi00398a069. [DOI] [PubMed] [Google Scholar]
- 34.Nilges M., Clore G. M., Gronenborn A. M. Determination of three-dimensional structures of proteins from interprotein distance data by hybrid distance geometry-dynamical simulated annealing calculations. FEBS Lett. 1988;229:317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- 35.Kuszewski J., Nilges M., Brunger A. T. Sampling and efficiency of metric matrix distance geometry: a novel partial metization algorithm. J. Biomol. NMR. 1992;2:33–56. doi: 10.1007/BF02192799. [DOI] [PubMed] [Google Scholar]
- 36.Blondelle S. E., Houghten R. A. Design of model amphipathic peptides having potent antimicrobial activities. Biochemistry. 1992;31:12688–12694. doi: 10.1021/bi00165a020. [DOI] [PubMed] [Google Scholar]
- 37.Saberwal G., Nagaraj R. Cell-lytic and antibacterial peptides that act by perturbing the barrier function of membranes: facets of their conformational features, structure-function correlations and membrane-perturbing abilities. Biochim. Biophys. Acta. 1994;1197:109–131. doi: 10.1016/0304-4157(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 38.Kiyota T., Lee S., Sugihara G. Design and synthesis of amphiphilic alpha-helical model peptides with systematically varied hydrophobic-hydrophilic balance and their interaction with lipid- and bio-membranes. Biochemistry. 1996;35:13196–13204. doi: 10.1021/bi961289t. [DOI] [PubMed] [Google Scholar]
- 39.Lee D. G., Park J.-H., Shin S. Y., Lee S. G., Lee M. K., Kim K. L., Hahm K.-S. Design of novel analogue peptides with potent fungicidal but low hemolytic activity based on the cecropin A-melittin hybrid structure. Biochem. Mol. Biol. Int. 1997;43:489–498. doi: 10.1080/15216549700204281. [DOI] [PubMed] [Google Scholar]
- 40.Tossi A., Sandri L., Giangaspero A. Peptides 2002: Proceedings of the Twenty-Seventh European Peptide Symposium. Napoli, Italy: Edizioni Ziino; 2002. New consensus hydrophobicity scale extended to non-proteinogenic amino acids; pp. 416–417. [Google Scholar]
- 41.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 1984;53:595–623.. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 42.Jahn B., Martin E., Stueben A., Bhakdi S. Susceptibility testing of Candida albicans and Aspergillus species by a simple microtiter menadione-augmented 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay. J. Clin. Microbiol. 1995;33:661–667. doi: 10.1128/jcm.33.3.661-667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wuthrich K. NY: Wiley-Interscience; 1986. NMR of protein and nucleic acid. [Google Scholar]
- 44.Kim Y., Prestegard J. H. Measurement of vicinal coupling from cross peaks in COSY spectra. J. Magn. Reson. 1989;84:9–13. [Google Scholar]
- 45.Baxter N. J., Williamson M. P. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR. 1997;9:359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 46.Gennis R. B. NY: Springer Verlag; 1989. Biomembranes: Molecular Structure and Function; pp. 155–156. [Google Scholar]
- 47.Holak T. A., Engstrom A., Kraulis P. J., Lindeberg G., Bennich H., Jones T. A., Gronenborn A. M., Clore G. M. The solution conformation of the antibacterial peptide cecropin A: a nuclear magnetic resonance and dynamical simulated annealing study. Biochemistry. 1988;27:7620–7629. doi: 10.1021/bi00420a008. [DOI] [PubMed] [Google Scholar]
- 48.Katsu T., Kuroko M., Morikawa T., Sanchika K., Yamanaka H., Shinoda S., Fujita Y. Interaction of wasp venom mastoparan with biomembranes. Biochim. Biophys. Acta. 1990;1027:185–190. doi: 10.1016/0005-2736(90)90083-z. [DOI] [PubMed] [Google Scholar]
- 49.Ohman A., Lycksell P. O., Jureus A., Langel U., Bartfai T., Graslund A. NMR study of the conformation and localization of porcine galanin in SDS micelles. Comparison with an inactive analog and a galanin receptor antagonist. Biochemistry. 1998;37:9169–9178. doi: 10.1021/bi980153n. [DOI] [PubMed] [Google Scholar]
- 50.Fujita K., Kimura S., Imanishi K. Self-assembly of mastoparan X derivative having fluorescence probe in lipid bilayer membrane. Biochim. Biophys. Acta. 1994;1195:157–163. doi: 10.1016/0005-2736(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 51.Park K., Oh D., Shin S. Y., Hahm K.-S., Kim Y. Structural studies of porcine myeloid antibacterial peptide PMAP-23 and its analogues in DPC micelles by NMR spectroscopy. Biochem. Biophys. Res. Commun. 2002;290:204–212. doi: 10.1006/bbrc.2001.6173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.