Abstract
Many experiments in the past have demonstrated the requirement of de novo gene expression during the long-term retention of learning and memory. Although previous studies implicated individual genes or genetic pathways in learning and memory, they did not uncover the collective behaviors or patterns of the genes. We have used genome-scale screening to analyze gene expression during spatial learning of rats in the Morris water maze. Our results show distinct temporal gene expression profiles associated with learning and memory. Exogenous administration of one peptide whose sustained increase during memory retention was implicated by microarray analysis, fibroblast growth factor (FGF)-18, improved spatial learning behavior, suggesting that pharmacological modulation of pathways and targets identified may allow new therapeutic approaches for improving learning and memory. Results of this study also suggest that while learning and physical activity involve common groups of genes, the behavior of learning and memory emerges from unique patterns of gene expression across time.
For more than a century, two general forms of memory have been classified by their duration: short-term memory (STM), which is rapidly formed and can outlast training for minutes or hours, and long-term memory (LTM), which lasts from hours to days, weeks, or even years (1). STM involves posttranslational modifications of preexisting molecules that alter the efficiency of synaptic transmission. In contrast, LTM can be blocked by inhibitors of transcription or translation, indicating that it depends on de novo gene expression (2, 3). Proteins that are newly synthesized during memory consolidation may contribute to restructuring processes at the synapse and thereby alter the efficiency of synaptic transmission beyond the duration of STM. Revealing the dependence of LTM on protein synthesis, however, provides no information about the identity and specificity of the required proteins. Because the quantity of a particular protein is often reflected by the abundance of its mRNA, a variety of methods have been used to describe a limited number of differentially expressed mRNAs during LTM. The increased or, less often, decreased expression of genes has been demonstrated during specific time windows after learning (3).
In the past we have used RNA fingerprinting to identify genes that were up-regulated in the hippocampus of water maze-trained rats (4). Spatial learning-induced changes in the expression of some of these genes occur at selective times and in specific hippocampal subfields (4, 5), indicating distinct contributions to learning and memory. Increased expression of one of these genes, the ryanodine receptor type-2, could result in the increased mobilization of Ca2+ that may participate in the synaptic changes underlying associative memory storage (6–8). In these past studies, however, we screened only a small fraction of the genes that may have been differentially expressed during LTM. Thus, the questions remain how many genes are involved in memory and how do they interact functionally to effect memory storage. In addition, each of the identified genes may not act in a linear sequence but in complex networks. Successive screening in different time domains, therefore, may be needed to uncover the networks of genes involved in distinct steps of memory storage.
To begin a comprehensive survey of the gene-based molecular mechanisms that underlie LTM, we have recently used the unprecedented experimental opportunities that genome sequences and the development of cDNA array technology now provide to perform genome-wide expression analysis in classical conditioning of the rabbit nictitating membrane response (9). In the present study we have extended these experiments to analyze the time dependence of patterns of gene regulation during the acquisition and consolidation of spatial learning.
Materials and Methods
Water Maze Learning.
The subjects were 36 adult male Wistar rats, each weighing 200–300 g. Rats were given access to food and water, and were maintained on a 12:12 light/dark cycle in a constant temperature (23°C). All behavioral tests were performed as described (4), carried out in the light phase, and were in accordance with National Institutes of Health guidelines. To reduce stress in the experimental day, the first day was dedicated to swimming training, in the absence of an island. Each rat was placed in the pool for 2 min and was returned to its home cage. In the next day, half of the rats were placed again in the pool for a 2.5-min swimming session and were used as swimming controls. The other half were given four consecutive trials to locate the platform, each trial lasting up to 2 min. Rats were required to spend 30 sec of an intertrial interval on the platform. The rats' escape latency was measured by using a HVS2020 video tracking system (HVS Image, San Diego). One, 6, and 24 h after training, swimming control and water maze-trained rats were killed and their hippocampi were rapidly dissected and frozen on dry ice. To verify that indeed the rats that were used had learned the spatial location of the island, a set of six rats was trained to find the island, and 24 h later they were tested on a quadrant analysis as described (4).
Microarray Analysis.
Hippocampal RNA from untrained animals (naive), swimming control, and water maze-trained individual animals was extracted as described (4). Total RNA samples from each experimental condition were pooled into two groups, reverse transcribed, biotinylated, and hybridized to two GeneChip Rat Neurobiology U34 arrays with the protocol outlined in the GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). The arrays were washed and stained by using a fluidics system with streptavidin-phycoerythrin (Molecular Probes), amplified with biotinylated anti-streptavidin antibody (Vector Laboratories), and then scanned with a GeneArray Scanner (Affymetrix). To determine the quality of labeled targets before analysis on GeneChip Rat Neurobiology U34 arrays, each sample was hybridized to one GeneChip test 3 array. The image data were analyzed by MICROARRAY SUITE 4.0 gene expression analysis program (Affymetrix). Normalization, filtering, and cluster analysis of the data were performed with the GENESPRING 4.2 software (Silicon Genetics, Redwood City, CA). The raw data from each array were normalized as follows: Each measurement for each gene was divided by the 50th percentile of all measurements. Each gene was then normalized to itself by making a synthetic positive control for that gene, and dividing all measurements for that gene by this positive control. This synthetic control was the median of the gene's expression values over all of the samples. Average difference values of less than zero represent probe sets where the intensity of the mismatched probe is, on average, greater than the perfect matched probe and, thus, the probe set is performing poorly. For this reason, normalized values below 0 were set to 0. Data derived from replicates (n = 2) in experimental groups were used to perform pairwise comparisons. An average fold change, derived from all possible pairwise comparisons, greater than 2 and at least one raw average difference value above 100 was used as the cutoff for significant differences in gene expression.
Real-Time Quantitative RT-PCR.
To further confirm the reliability of the array data, the mRNA levels of 15 genes were quantified by real-time quantitative RT-PCR. Aliquots of cDNA (0.1 and 0.2 μg) from naive, swimming control, and water maze-trained rats (six animals per group), and known amounts of external standard (purified PCR product, 102 to 108 copies) were amplified in parallel reactions using specific primers (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). PCR amplifications were performed as described (9, 10). Specificity of PCR products obtained was characterized by melting curve analysis, followed by gel electrophoresis and DNA sequencing.
Behavioral Pharmacology.
Thirteen male Wistar rats (250–300 g) were implanted strereotaxically with stainless steel guide cannulae in the right and left lateral ventricles (AP, −0.80 mm; ML, 1.5 mm; DV, 3.6 mm) (11). On day 1, 1 week after surgery, animals were subjected to a 2-min swimming training session. A water maze training session was then performed on days 2 and 3, which measured the ability of the animals to find a submerged platform to escape from the water. Two trials were given to each animal for each session. The escape latency and distance to find the platform were monitored as described above. Ten minutes after the second trial on day 2, an intracerebroventricular administration of drug or vehicle was performed in both lateral ventricles by introducing stainless steel injection cannulae into the implanted guide cannulae. Each injection cannula was connected to a 25-μl Hamilton syringe fastened onto a pump through polyethylene tubing filled with distilled water. Infusions were performed at a rate of 2 μl/min for 1 min in each side. Six animals received 0.94 pmol of FGF-18 (PeproTech, Rocky Hill, NJ) and the other seven received a control injection of vehicle (saline).
RNA preparation, microarray analyses, quantitative RT-PCR and pharmacological studies were performed in a double-blind manner.
Results
To relate mRNA induction to a learning task we trained the rats for four consecutive trials to locate a submerged island in the water maze. The rats completed the task within 2.56 ± 0.49 min (mean ± SD) and their latency time to find the island was reduced from 47.8 ± 11.3 sec to 26.3 ± 6.9 sec, indicating that the rats indeed learned the task. Swimming control rats were allowed to swim in the pool in the absence of the island for 2.5 min. To verify that the trained rats in fact learned the spatial location of the island, a group of six rats was trained to find the island and tested 24 h later on a quadrant analysis. The trained rats swam significantly longer in the quadrant where the island was located (36.5 ± 3.2% of the total distance compared with 22.5 ± 2% and 21.8 ± 2.9% in the two adjacent quadrants and 19.1 ± 4.1% in the opposite quadrant, ANOVA, P < 0.01).
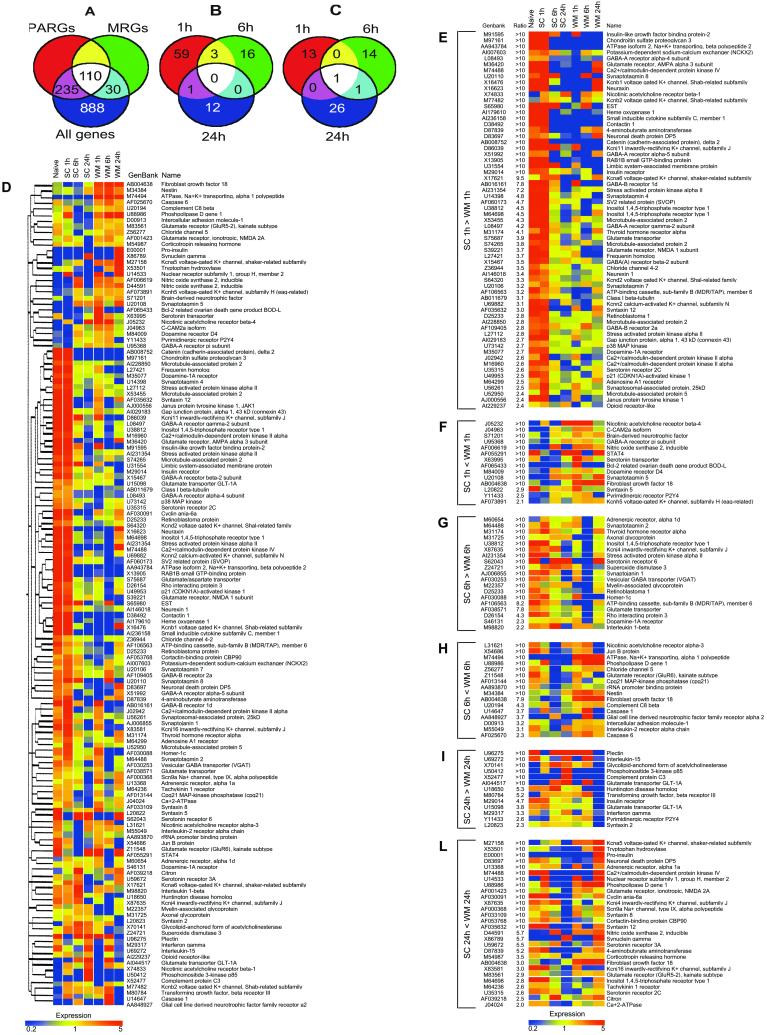
Hippocampal gene expression profiles in naive, swimming control, and water maze-trained animals were measured by using microarrays containing more than 1,200 genes relevant to neurobiology. The complete set of data is available in Table 3, which is published as supporting information on the PNAS web site. When gene expression profiles in the naive and swimming control animals 1, 6, and 24 h after swimming sessions were compared, 345 genes (27.3%) were found to be differentially expressed more than 2-fold in at least two of the four conditions (Fig. 1A). These genes, operationally defined as “physical activity-related genes” (PARGs) indicate that physical activity and mild stress associated with behavioral training has a significant impact on hippocampal gene expression.
Fig. 1.
Differential gene expression during water maze learning. (A and B) Venn diagrams of differentially expressed hippocampal genes. Genes differentially expressed in naive and swimming control animals 1, 6, and 24 h after training were operationally defined as “physical activity-related genes” (PARGs), whereas genes differentially expressed in water maze-trained animals compared with swimming controls were operationally defined as “memory-related genes” (MRGs) (A). Among these, 91 genes were down-regulated (B), whereas 55 genes were up-regulated (C) in at least one of three time points examined. (D) Hierarchical clustering of memory-related genes. A hierarchical clustering algorithm (Pearson correlation, separation ratio 0.5, minimum distance 0.001) was used to order memory-related genes in a dendrogram in which the pattern and length of the branches reflect the relatedness of the samples. Data are presented in a matrix format. Each row represent a single gene and each column represents an experimental condition. The averaged normalized intensity from two replicates is represented by the color of the corresponding cell in the matrix. Blue, yellow, and red cells, respectively, represent transcript levels below, equal to, or above the median abundance across all conditions. Color intensity reflects the magnitude of the deviation from the median (see scale at the bottom). (E–L) Differentially expressed genes in swimming control (SC) vs. water maze (WM)-trained animals at 1, 6, and 24 h after training.
When gene expression levels in swimming control animals were compared with water maze-trained animals 1, 6, or 24 h after training, 140 genes (11%) were found to be differentially expressed and were operationally defined as “memory-related genes” (MRGs) (Fig. 1A). The majority of these MRGs (110 of 140), were also PARGs, i.e., influenced by physical activity. Among MRGs, 91 genes were down-regulated (Fig. 1 B, E, G, and I) in the hippocampus of water maze-trained animals, whereas 55 genes were up-regulated (Fig. 1 C, F, H, and L).
A hierarchical clustering method was used to group memory related genes on the basis of similarity in their expression patterns (Fig. 1D). Genes represented by more than one probe set on the array, such as inducible nitric oxide synthase, inositol 1,4,5-trisphosphate receptor type 1, microtubule-associated protein 2, and Ca2+/calmodulin-dependent protein kinase II α, were clustered next to, or in, the immediate vicinity of each other, indicating that the effects of experimental noise or artifact are negligible. Although no information on the identity of the samples was used in the clustering, in some cases genes segregated according to their common biological functions. For example, genes encoding for membrane trafficking proteins, such as synaptotagmins 7 and 8, or syntaxin 2, 5, and 8, and most of the genes encoding for γ-aminobutyric acid (GABA) A and B type receptors were expressed concordantly. The most evident trait of the clustered data (Fig. 1D) was that MRGs showed entirely different temporal patterns of expression in swimming control vs. water maze-trained animals.
Although our data represented the average gene expression from two separate microarray analyses performed on pooled hippocampal RNA samples from naive, swimming control, and water maze-trained animals, there could be differences in gene expression between individual animals. To address this question and to confirm the reliability of the array data we selected 15 genes and quantitatively validated their differential expression in the hippocampal mRNA of individual animals by using real-time quantitative RT-PCR (see Table 2). Remarkably, the pattern of gene expression from sample to sample observed by microarrays closely paralleled the pattern observed by using real-time RT-PCR. The minimum and maximum correlation coefficients between the two profiles are 0.72 and 0.99, respectively (Table 2).
Fibroblast growth factor (FGF)-18 was the only MRG not influenced by physical activity that was increased 1, 6, and 24 h after water maze training (Fig. 2, which is published as supporting information on the PNAS web site, and Fig. 1 F, H, and L). To explore the effect of FGF-18 in spatial learning, we tested the effects of a single exogenous dose of FGF-18. Adult male rats were trained in a Morris water maze for two trials and then injected intracerebroventricular with 0.94 pmol of FGF-18 or vehicle. As shown in Table 1, animals treated with FGF-18 displayed significantly improved spatial learning behavior (P < 0.05) compared with vehicle-injected control animals. FGF-18 treatment induced a 49% reduction in the escape latency but no significant changes in motor activity.
Table 1.
Effects of a single exogenous administration of FGF-18 on water maze learning
| Treatment
|
Latency, sec | Distance, m | ||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 1 | Day 2 | |
| Control | 48.2 ± 16.1 | 37.1 ± 11.1 | 13.6 ± 4.3 | 10.4 ± 3.1 |
| FGF-18 | 46.6 ± 16.7 | 19.2 ± 6.3 | 15.2 ± 5.7 | 6.1 ± 2.0 |
, Day 1 vs. day 2, P < 0.05;
, control vs. FGF-18, P < 0.05.
Discussion
Our results show that both learning and physical activity have profound effects on hippocampal gene expression. Most of the MRGs, those differentially expressed between the swimming and spatial learning animal groups, were also affected during swimming alone, but with entirely different temporal patterns of expression as shown in the clustered data. Although learning and physical activity involve common groups of genes, the behavior of learning and memory can be distinguished from unique patterns of gene expression across time.
All of the MRGs identified have a recognized function and can be classified into six major groups based on their translated product (see Fig. 2): (i) cell signaling, (ii) synaptic proteins, (iii) cell–cell interaction and cytoskeletal proteins, (iv) apoptosis, (v) enzymes, and (vi) transcription or translation regulation. Some of these genes have been previously related to synaptic plasticity, memory, or cognitive disorders, whereas others provide a significant number of unique entry points that have not been recognized previously. The exact role and functional relationships of the genes and proteins implicated, however, are presumably those we cannot yet recognize. For this reason, in the following section we will discuss only some of the MRGs implicated by microarray analysis. As more time points, behavioral paradigms, and pathophysiological conditions are used for similar studies, and more complete high-density arrays become available, a more complete interpretive framework will emerge as to the key genes and pathways underlying learning and memory. To facilitate this exploration, the data generated in the present study and those produced in different behavioral (9) and pathophysiological (10) conditions are available online (Table 3 and www.brni-jhu.org/sebi/microarray-data).
Cell Signaling.
The group of genes involved in cell signaling is the largest and includes a subgroup of neuropeptides, growth factors, and their receptors. Among them is FGF-18, a member of the FGF family, which was shown to stimulate neurite outgrowth (12). Although the function of this peptide is still unknown, the other members of its family are important signaling molecules in several inductive and patterning processes, and act as brain organizer-derived signals during the formation of the early vertebrate nervous system. The expression of FGF-18 was induced by water maze training but not physical activity. This result, together with the ability of FGF-18 to enhance spatial memory when exogenously administered, is strong evidence in favor of its involvement in learning and memory.
Differential expression of interleukin-1β (IL-1β), interleukin 15 (IL-15), and interleukin-2 (IL-2) receptor α chain suggests a physiological role of brain cytokines in memory consolidation processes. Indeed, the reduction of IL-1β mRNA in water maze-trained animals is consistent with previous studies showing that central IL-1β administration and agents that induce central IL-1β activity impair the consolidation of memories that depend on the hippocampal formation (13).
Enhanced expression of corticotropin-releasing hormone in water-maze-trained animals is also in line with other evidence obtained in another learning paradigm (14).
The subgroup of G protein-coupled receptors includes two GABA B-type receptor splice variants, GABAB1d and GABAB2a. Functional GABAB receptors, whose function depends on dimerization of GABAB1 and GABAB2, are known to activate second messenger systems and modulate potassium and calcium channel activity, thereby controlling the presynaptic transmitter release and the postsynaptic silencing of excitatory neurotransmission (15). GABAB receptor agonists or antagonists are known to impair or facilitate, respectively, cognitive performance in the Morris water maze task as well as other kinds of learning (16). By reducing GABAB receptor signaling, the down-regulation of GABAB1d and GABAB2a 1 h after water maze training may exert a mnemonic effect similar to that produced by GABAB receptor antagonists.
Dopamine 1A and D4 receptors are down- and up-regulated, respectively, 1 h after water maze training. These receptors are coupled to different G proteins and their change in expression may allow for the modulation of a neuronal dopamine-mediated signal.
The opioid receptor-like receptor is decreased 1 h after water maze training. This receptor is a G protein-coupled receptor structurally related to the opioid receptors, whose endogenous ligand is the heptadecapeptide nociceptin, which has been implicated in sensory perception, memory process, and emotional behavior (17).
The adenosine receptor A1, which is negatively coupled to adenylate cyclase, decreased 1 h after water maze training. Adenosine is thought to exert a tonic inhibitory role on synaptic plasticity in the hippocampus (18). Its decrease, therefore, may exert a facilitory role during learning and memory.
The insulin receptor was increased in swimming control and decreased in water maze-trained rats, whereas the precursor of its endogenous ligand, insulin, was detectable only 24 h after water maze training. The fine balance of brain insulin and its receptor may regulate cognitive functions (19).
The subgroup of ligand-gated ion channels includes five GABAA receptor subunits which were all differentially expressed 1 h after water maze training. Four of them, α4, α5, β2, and γ2, were down-regulated, whereas one, the π subunit, was up-regulated. Changes in the expression of specific GABAA receptor subunits may affect the composition and pharmacology of GABAA receptor assemblies. These changes may also be relevant in consideration of the vast number of drugs such as anxiolytics, anticonvulsants, general anesthetics, barbiturates, ethanol, and neurosteroids, which are known to elicit at least some of their pharmacological effects through GABAA receptor subunits (20).
The expression of glutamate ionotropic receptors is dynamically regulated during spatial learning. N-methyl-d-aspartic acid receptor (NMDA-R) 1, which possesses all properties characteristic of the NMDA receptor–channel complex, is down-regulated 1 h after water maze training, whereas NMDA-R2A, which has regulatory activities, is up-regulated after 24 h. One l-α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptor α3 subunit is down-regulated 1 h after training. Two kainate receptors, GluR6 and GluR5–2, are up-regulated 6 and 24 h, respectively, after training. Plastic changes of different combinations of glutamate receptors might have profound effects on glutamate responsiveness (21).
The subgroup of ion channels includes several proteins that play a role in the maintenance of ionic homeostasis. Among these are 10 potassium (K+) channel subunits: 2 Shaker (Kcna5 and Kcna6), 2 Shab (Kcnb1 and Kcnb2), 1 Shal (Kcnd2) and 1 EAG-related (Kcnh5) voltage-dependent K+ channel subunits; 1 Ca2+-activated (Kcnn2) and 3 inwardly rectifying (Kcjn4, Kcjn11, and Kcjn16). Expression changes of different K+ channel subunits may alter the composition of the channel complexes and would affect cellular excitability (22). Although the exact contribution of each of the above subunits during spatial memory is unknown, 7 of 10 are down-regulated after water maze training and may produce increased excitability.
The subgroup of proteins involved in intracellular signaling includes several proteins involved in the intracellular homeostasis of calcium, sodium, and potassium ions. Among these is the frequenin homolog, also known as neuronal calcium sensor-1, which has recently been shown to regulate associative learning (23).
The subgroups of proteins involved in neurotransmitter transport includes GABA, glutamate, and serotonin transporters. The GABA and glutamate transporters are down-regulated 1, 6, or 24 h after water maze training, whereas the serotonin transporter is up-regulated after 1 h. Neurotransmitter uptake by nerve terminals and glial cells is crucial for providing a reservoir of transmitter or transmitter precursors and the termination of synaptic events (24). Changes in the expression of these transporters, therefore, may have profound effects on neurotransmission by controlling neurotransmitter levels at the synaptic cleft.
The subgroup of signaling enzymes includes a number of proteins previously implicated in learning and memory. After water maze training, a strong induction of the inducible form of nitric oxide synthase (iNOS) was observed. This enzyme produces nitric oxide (NO), a molecule involved in neurosynaptic transmission, and is induced in many pathological conditions. Although the role of NO in learning and memory is still unclear, some studies have reported that systemic NO inhibition had deleterious effects in water maze learning (25–27). The role of iNOS in the hippocampus, therefore, may go beyond its well established detrimental function in neurological disorders and could contribute to the mechanisms underlying learning and memory.
Two genes encoding enzymes involved in the mitogen-activated protein kinase (MAPK) signaling cascade, p38 MAPK and MAPK phosphatase, were found to be differentially expressed after water maze training. This signaling cascade has been previously implicated in the development of synaptic plasticity underlying learning and memory (28–30). However, there are three subfamilies of MAPKs that are activated by different upstream cascades and are involved in the regulation of distinct nuclear transcriptional factors (31). As suggested by the present observations and previous studies (32), long-term memory may involve different MAPKs and/or their MAPK phosphatases.
Differential expression of two Ca2+/calmodulin-dependent protein kinases, belonging to a class of signaling enzymes extensively implicated in memory formation and consolidation (33), was observed after water maze training.
Other proteins involved in signal transduction include Ania-3, a short form of the Homer family of proteins which bind to group I metabotropic glutamate receptors, inositol trisphosphate receptors, ryanodine receptors, and NMDA receptor-associated Shank proteins and have been implicated in synaptogenesis, signal transduction, receptor trafficking, and axon pathfinding (34). The long Homer forms are constitutively expressed and self-associate to function as adaptors to couple membrane receptors to intracellular pools of releasable Ca2+. The short Homer forms compete with the long Homer proteins for binding to signaling components, thus functioning as endogenous dominant-negative regulators of receptor-induced Ca2+ release from intracellular stores. Down-regulation of Ania-3 in water maze-trained animals may modulate the properties of the long Homer forms and be involved in activity-dependent alterations of synaptic structure and function.
Up-regulation of another signaling molecule, citron, was found 24 h after water maze training. Citron is a neuronal ρ-target molecule associated to the postsynaptic scaffold protein PSD-95, which plays an important role in the anchoring and clustering of neurotransmitter receptors at the synapses (35). The expression of citron may provide a crosstalk between the ρ signaling pathway, which has been implicated in the mechanisms of neuronal plasticity, and in neurotransmitter receptors such as the NMDA receptor.
Cell–Cell Interactions and Cytoskeletal Proteins.
The group of cell–cell interactions and cytoskeletal proteins includes a vast number of proteins whose change in expression may reflect the morphological adaptation of brain cells during formation of memory. Among them, for example, is δ-catenin, a component of the cell–cell adherens junctions expressed specifically in the nervous system. δ-catenin is down-regulated during neuronal migration and expressed in the apical dendrites of postmitotic neurons (36). Changes in δ-catenin expression, therefore, are considered to be fundamental for the establishment and maintenance of dendrites and synaptogenesis. δ-Catenin was originally discovered as an interactor with presenilin 1 (37), whose mutation causes early-onset familial Alzheimer's disease. In addition, hemizygosity of δ-catenin is associated with severe mental retardation in the cri-du-chat syndrome that is associated with severe mental retardation (38).
The hippocampal expression of several proteins involved in microtubule formation was reduced 1 h after water maze training. Among these are β-tubulin, neuraxin, and microtubule-associated protein 2 (MAP2) and 5. The reduced expression of MAP2, in particular, was confirmed in three redundant probe sets. Altered expression of MAP2, which is critical for dendritic stability (39), has been shown with contextual memory, long-term potentiation, aging, epilepsy, Alzheimer's disease, and Rett syndrome (40–45). We have recently found altered expression of MAP2 in a transgenic animal model of fragile X syndrome (10), which shows behavioral deficits in the Morris water maze (46). Expression of several others proteins involved cell–cell and cell–matrix interactions was found to be increased (intercellular adhesion molecule-1, C-CAM2a isoform) or more often decreased (neurexin 1, connexin 43, contactin 1, chondroitin sulfate proteoglycan 3, myelin-associated glycoprotein, and axonal glycoprotein). Cell adhesion molecules have already been implicated in synaptic plasticity, learning, and memory (47). Together, their changes may be critical in regulating cell–cell recognition and the establishment of mature dendritic relationships in the neuropil.
Apoptosis.
The group of proteins involved in apoptosis includes Bcl-2-related death gene product BOD-L, caspase 1 and 6, and DP5, which are all up-regulated after water maze training. In agreement with other studies (48), our data suggest that beyond their roles in cell death, apoptotic and anti-apoptotic cascades may play roles in synaptic plasticity.
Enzymes.
The group of enzymes includes two proteins involved in free radical metabolism, heme oxygenase 1 and superoxide dismutase 3, whose expression was reduced in the hippocampus of water maze-trained animals. Besides their role in oxidative stress, these enzymes may be implicated in other physiological roles such as learning and memory. Indeed, impaired spatial memory is found in mice overexpressing these two proteins (49, 50).
Transcription or Translation Regulation.
Among the group of differentially expressed genes involved in transcription or translation regulation is the up-regulated gene encoding for cyclin Ania-6a, whose splicing is dynamically controlled by different forms of neuronal stimulation (51), and Jun-B, which is induced after different memory tasks (52).
Synaptic Proteins.
The group of synaptic proteins includes a number of proteins that regulate membrane trafficking and fusion. They include synaptojanin 1, four members of the syntaxin family of proteins (syntaxin 2, 5, 8, and 12), five synaptotagmins (2, 4, 5, 7, and 8), and synaptosomal-associated protein-25. Differential expression of these proteins, which are involved in different steps of membrane trafficking and fusion (53), may regulate synaptic plasticity by affecting cellular functions such as secretion, endocytosis, and axonal growth.
The data presented here reveal distinct temporal gene expression profiles associated with learning and memory and demonstrate the utility of a cDNA microarray system as a means of dissecting the molecular basis of associative memory. It should be emphasized that the microarray provides estimates of changes in mRNA levels that cannot be correlated with the amount and function of the gene products. Translation and posttranslational modifications of many gene products and protein turnover have dramatic effects on function, and these cannot be inferred from expression analysis alone. Nevertheless, the approach used provides information on the gene expression changes that occur during learning and memory, and identify molecular targets and pathways whose modulation may allow new therapeutic approaches for improving cognition. As shown in previous studies, and in the present study for FGF-18, pharmacological or genetic modulation of some of these pathways can indeed be effective in facilitating learning and memory.
Supplementary Material
Abbreviations
LTM, long-term memory
STM, short-term memory
FGF, fibroblast growth factor
PARG, physical activity-related gene
MRG, memory-related gene
MAPK, mitogen-activated protein kinase
NMDA, N-methyl-d-aspartate
GABA, γ-aminobutyric acid
References
- 1.James W., (1890) Principles of Psychology (Holt, New York); reprinted (1999) (Thoemmes Press, Bristol, U.K.).
- 2.Davis H. P. & Squire, L. R. (1984) Psychol. Bull. 96, 518-559. [PubMed] [Google Scholar]
- 3.Stork O. & Welzl, H. (1999) Cell. Mol. Life Sci. 55, 575-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavallaro S., Meiri, N., Yi, C. L., Musco, S., Ma, W., Goldberg, J. & Alkon, D. L. (1997) Proc. Natl. Acad. Sci. USA 94, 9669-9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao W., Meiri, N., Xu, H., Cavallaro, S., Quattrone, A., Zhang, L. & Alkon, D. L. (2000) FASEB J. 14, 290-300. [DOI] [PubMed] [Google Scholar]
- 6.Alkon D. L., Nelson, T. J., Zhao, W. & Cavallaro, S. (1998) Trends Neurosci. 21, 529-537. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell K. T. & Alkon, D. L. (1999) Brain Res. 822, 114-125. [DOI] [PubMed] [Google Scholar]
- 8.Sun M. K., Nelson, T. J. & Alkon, D. L. (2000) Proc. Natl. Acad. Sci. USA 97, 12300-12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavallaro S., Schreurs, B. G., Zhao, W., D'Agata, V. & Alkon, D. L. (2001) Eur. J. Neurosci. 13, 1809-1815. [DOI] [PubMed] [Google Scholar]
- 10.D'Agata V., Warren, S. T., Zhao, W., Torre, E. R., Alkon, D. L. & Cavallaro, S. (2002) Neurobiol. Dis. 10, 211-218. [DOI] [PubMed] [Google Scholar]
- 11.Paxinos G. & Watson, C., (1998) The Rat Brain in Stereotaxic Coordinates (Academic, San Diego). [DOI] [PubMed]
- 12.Ohbayashi N., Hoshikawa, M., Kimura, S., Yamasaki, M., Fukui, S. & Itoh, N. (1998) J. Biol. Chem. 273, 18161-18164. [DOI] [PubMed] [Google Scholar]
- 13.Rachal Pugh C., Fleshner, M., Watkins, L. R., Maier, S. F. & Rudy, J. W. (2001) Neurosci. Biobehav. Rev. 25, 29-41. [DOI] [PubMed] [Google Scholar]
- 14.Lee E. H., Huang, A. M., Tsuei, K. S. & Lee, W. Y. (1996) Chin. J. Physiol. 39, 197-203. [PubMed] [Google Scholar]
- 15.Dutar P. & Nicoll, R. A. (1988) Nature 332, 156-158. [DOI] [PubMed] [Google Scholar]
- 16.Mott D. D. & Lewis, D. V. (1994) Int. Rev. Neurobiol. 36, 97-223. [DOI] [PubMed] [Google Scholar]
- 17.Calo G., Guerrini, R., Rizzi, A., Salvadori, S. & Regoli, D. (2000) Br. J. Pharmacol. 129, 1261-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Mendonca A. & Ribeiro, J. A. (1994) Neuroscience 62, 385-390. [DOI] [PubMed] [Google Scholar]
- 19.Park C. R. (2001) Neurosci. Biobehav. Rev. 25, 311-323. [DOI] [PubMed] [Google Scholar]
- 20.Smith T. A. (2001) Br. J. Biomed. Sci. 58, 111-121. [PubMed] [Google Scholar]
- 21.Madden D. R. (2002) Nat. Rev. Neurosci. 3, 91-101. [DOI] [PubMed] [Google Scholar]
- 22.Choe S. (2002) Nat. Rev. Neurosci. 3, 115-121. [DOI] [PubMed] [Google Scholar]
- 23.Gomez M., De Castro, E., Guarin, E., Sasakura, H., Kuhara, A., Mori, I., Bartfai, T., Bargmann, C. I. & Nef, P. (2001) Neuron 30, 241-248. [DOI] [PubMed] [Google Scholar]
- 24.Masson J., Sagne, C., Hamon, M. & El Mestikawy, S. (1999) Pharmacol. Rev. 51, 439-464. [PubMed] [Google Scholar]
- 25.Yamada K., Noda, Y., Nakayama, S., Komori, Y., Sugihara, H., Hasegawa, T. & Nabeshima, T. (1995) Br. J. Pharmacol. 115, 852-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman P. F., Atkins, C. M., Allen, M. T., Haley, J. E. & Steinmetz, J. E. (1992) NeuroReport 3, 567-570. [DOI] [PubMed] [Google Scholar]
- 27.Estall L. B., Grant, S. J. & Cicala, G. A. (1993) Pharmacol. Biochem. Behav. 46, 959-962. [DOI] [PubMed] [Google Scholar]
- 28.Kornhauser J. M. & Greenberg, M. E. (1997) Neuron 18, 839-842. [DOI] [PubMed] [Google Scholar]
- 29.Impey S., Obrietan, K. & Storm, D. R. (1999) Neuron 23, 11-14. [DOI] [PubMed] [Google Scholar]
- 30.Zhen X., Du, W., Romano, A. G., Friedman, E. & Harvey, J. A. (2001) J. Neurosci. 21, 5513-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis R. J. (1993) J. Biol. Chem. 268, 14533-14556. [Google Scholar]
- 32.Berman D. E., Hazvi, S., Rosenblum, K., Seger, R. & Dudai, Y. (1998) J. Neurosci. 18, 10037-10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayford M., Bach, M. E., Huang, Y. Y., Wang, L., Hawkins, R. D. & Kandel, E. R. (1996) Science 274, 1678-1683. [DOI] [PubMed] [Google Scholar]
- 34.Xiao B., Tu, J. C. & Worley, P. F. (2000) Curr. Opin. Neurobiol. 10, 370-374. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W., Vazquez, L., Apperson, M. & Kennedy, M. B. (1999) J. Neurosci. 19, 96-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho C., Zhou, J., Medina, M., Goto, T., Jacobson, M., Bhide, P. G. & Kosik, K. S. (2000) J. Comp. Neurol. 420, 261-276. [PubMed] [Google Scholar]
- 37.Zhou J., Liyanage, U., Medina, M., Ho, C., Simmons, A. D., Lovett, M. & Kosik, K. S. (1997) NeuroReport 8, 2085-2090. [DOI] [PubMed] [Google Scholar]
- 38.Medina M., Marinescu, R. C., Overhauser, J. & Kosik, K. S. (2000) Genomics 63, 157-164. [DOI] [PubMed] [Google Scholar]
- 39.Johnson G. V. & Jope, R. S. (1992) J. Neurosci. Res. 33, 505-512. [DOI] [PubMed] [Google Scholar]
- 40.Kosik K. S., Duffy, L. K., Dowling, M. M., Abraham, C., McCluskey, A. & Selkoe, D. J. (1984) Proc. Natl. Acad. Sci. USA 81, 7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leterrier J. F. & Eyer, J. (1992) J. Neurochem. 59, 1126-1137. [DOI] [PubMed] [Google Scholar]
- 42.Kaufmann W. E., Naidu, S. & Budden, S. (1995) Neuropediatrics 26, 109-113. [DOI] [PubMed] [Google Scholar]
- 43.Fukunaga K., Muller, D. & Miyamoto, E. (1996) Neurochem. Int. 28, 343-358. [DOI] [PubMed] [Google Scholar]
- 44.Yamanouchi H., Jay, V., Otsubo, H., Kaga, M., Becker, L. E. & Takashima, S. (1998) Acta Neuropathol. 95, 466-470. [DOI] [PubMed] [Google Scholar]
- 45.Woolf N. J., Zinnerman, M. D. & Johnson, G. V. (1999) Brain Res. 821, 241-249. [DOI] [PubMed] [Google Scholar]
- 46.The Dutch–Belgian Fragile X Consortium (1994) Cell 78, 23-33. [PubMed] [Google Scholar]
- 47.Wright J. W., Kramar, E. A., Meighan, S. E. & Harding, J. W. (2002) Peptides (Tarrytown, NY) 23, 221-246. [DOI] [PubMed] [Google Scholar]
- 48.Mattson M. P. & Duan, W. (1999) J. Neurosci. Res. 58, 152-166. [PubMed] [Google Scholar]
- 49.Gahtan E., Auerbach, J. M., Groner, Y. & Segal, M. (1998) Eur. J. Neurosci. 10, 538-544. [DOI] [PubMed] [Google Scholar]
- 50.Morgan D., Holcomb, L., Saad, I., Gordon, M. & Mahin Maines (1998) Brain Res. 808, 110-112. [DOI] [PubMed] [Google Scholar]
- 51.Berke J. D., Sgambato, V., Zhu, P. P., Lavoie, B., Vincent, M., Krause, M. & Hyman, S. E. (2001) Neuron 32, 277-287. [DOI] [PubMed] [Google Scholar]
- 52.Tischmeyer W. & Grimm, R. (1999) Cell. Mol. Life Sci. 55, 564-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jahn R. & Sudhof, T. C. (1999) Annu. Rev. Biochem. 68, 863-911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.