Abstract
The enzyme DHFR (dihydrofolate reductase) catalyses hydride transfer from NADPH to, and protonation of, dihydrofolate. The physical basis of the hydride transfer step catalysed by DHFR from Escherichia coli has been studied through the measurement of the temperature dependence of the reaction rates and the kinetic isotope effects. Single turnover experiments at pH 7.0 revealed a strong dependence of the reaction rates on temperature. The observed relatively large difference in the activation energies for hydrogen and deuterium transfer led to a temperature dependence of the primary kinetic isotope effects from 3.0±0.2 at 5 °C to 2.2±0.2 at 40 °C and an inverse ratio of the pre-exponential factors of 0.108±0.04. These results are consistent with theoretical models for hydrogen transfer that include contributions from quantum mechanical tunnelling coupled with protein motions that actively modulate the tunnelling distance. Previous work had suggested a coupling of a remote residue, Gly121, with the kinetic events at the active site. However, pre-steady-state experiments at pH 7.0 with the mutant G121V-DHFR, in which Gly121 was replaced with valine, revealed that the chemical mechanism of DHFR catalysis was robust to this replacement. The reduced catalytic efficiency of G121V-DHFR was mainly a consequence of the significantly reduced pre-exponential factors, indicating the requirement for significant molecular reorganization during G121V-DHFR catalysis. In contrast, steady-state measurements at pH 9.5, where hydride transfer is rate limiting, revealed temperature-independent kinetic isotope effects between 15 and 35 °C and a ratio of the pre-exponential factors above the semi-classical limit, suggesting a rigid active site configuration from which hydrogen tunnelling occurs. The mechanism by which hydrogen tunnelling in DHFR is coupled with the environment appears therefore to be sensitive to pH.
Keywords: dihydrofolate reductase, dynamics, hydrogen transfer, kinetics, protein motions, tunnelling
Abbreviations: DHFR, dihydrofolate reductase; DTT, dithiothreitol; H2F, 7,8-dihydrofolate; H4F, 5,6,7,8-tetrahydrofolate; KIE, kinetic isotope effect; NADPD, (4R)-[2H]NADPH; QM/MM, quantum mechanical and molecular mechanical
INTRODUCTION
It has long been accepted that electrons can be transferred in chemical reactions by quantum mechanical tunnelling with energies of activation deriving from heavy-atom motions [1]. However, only recently have several cases of hydrogen tunnelling been described where the probability of tunnelling was modulated by environmental dynamics and protein motions [2–15]. Several theoretical models that incorporate environmental motions have been developed to describe hydrogen transfer reactions [5,16–19]. The model proposed by Kuznetsov and Ulstrup [19] and developed further by Knapp and Klinman [5,13] for enzymatic transformations, which separates the faster e− and H co-ordinates from the environmental co-ordinate, distinguishes between passive motions that influence the probability of the formation of an active site configuration conducive to hydride transfer, and active protein motions that modulate the tunnelling distance and hence the reaction rate.
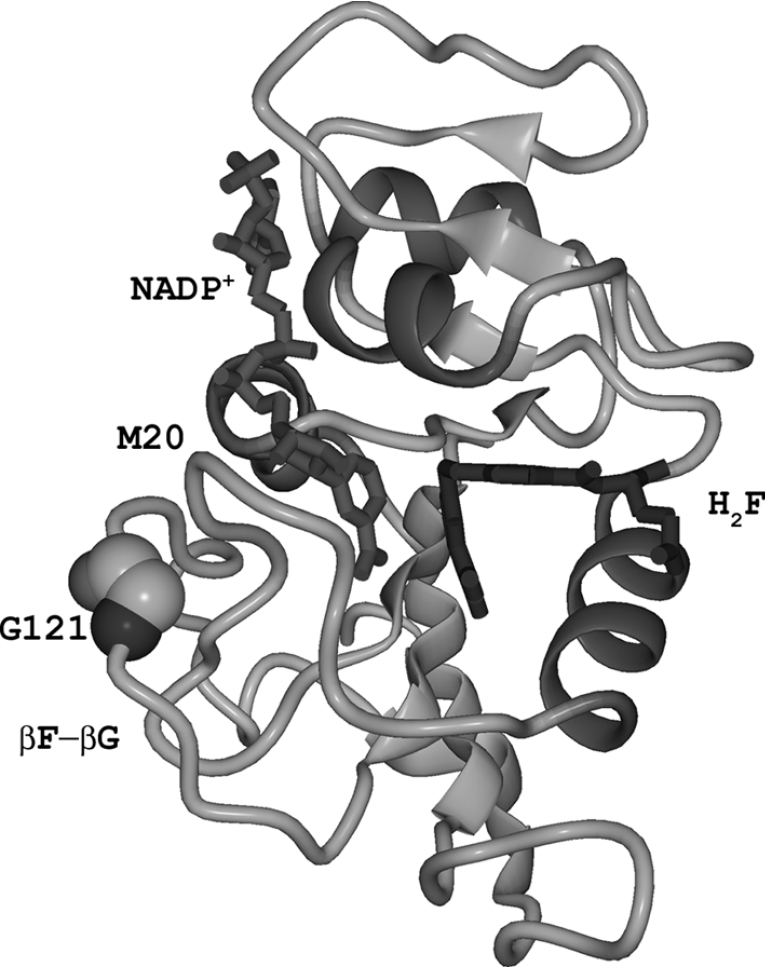
DHFR (dihydrofolate reductase) has long served as a paradigm for the study of issues relating to enzyme structure, dynamics and catalysis. DHFR catalyses the reduction of H2F (7,8-dihydrofolate) to H4F (5,6,7,8-tetrahydrofolate) with the concurrent oxidation of NADPH. DHFR is essential for maintaining intracellular levels of H4F, a cofactor involved in the biosynthesis of several amino acids, purines and thymidylate. DHFR from Escherichia coli is a monomeric enzyme comprising an eight-stranded β-sheet and four α-helices connected by flexible loop regions (Figure 1). The M20, βF-βG and βG-βH loops have been implicated in catalysis [20]. In the reactive ternary complex, when both H2F and NADPH are bound, the M20 loop adopts the closed conformation, which is stabilized through hydrogen bonds between residues in the M20 and the βF-βG loops [21]. The backbone atoms of the βF-βG loop display high dynamic mobility in NMR relaxation experiments, which has been interpreted to suggest a connection between the dynamic properties and the catalytic behaviour of DHFR [22–25].
Figure 1. Structure of DHFR from E. coli bound to NADP+ and H2F.
The PDB file 1RA2 [21] was used to generate the diagram. The βF-βG and the M20 loops as well as the position of the catalytically important loop residue Gly121 are indicated.
The complete kinetic scheme for DHFR from E. coli has been determined [26,27]. The reaction rate is strongly dependent on the pH of the solution. The chemical step, in which the pro-R hydrogen is transferred from C-4 of NADPH to C-6 of H2F and N-5 of the substrate is protonated, is characterized by a single pKa of approx. 6.5 with a rate variation of almost two orders of magnitude. This pKa can most likely be attributed to protonation of the substrate in the ternary enzyme complex [28]. At physiological pH, the overall rate of the conversion of H2F into H4F is limited by product release, while the chemical step is fast and essentially irreversible [26]. However, at pH values of 9.5 and above, hydrogen transfer is much slower and fully rate-determining.
Several computational studies using combined QM/MM (quantum mechanical and molecular mechanical) and free-energy perturbation approaches have shed some light on the mechanism of the hydride transfer step in DHFR catalysis [28–35]. QM/MM simulations and genomic sequence analysis identified a network of hydrogen bonds and van der Waals interactions in DHFR from E. coli between the surface βF-βG loop and the active site that may influence protein dynamics and promote catalysis [18,36]. Replacement of Gly121, a highly mobile residue located in the middle of the βF-βG loop over 19 Å (1 Å=0.1 nm) from the active site (Figure 1), with valine or leucine slowed the hydride transfer rate dramatically and weakened binding of NADPH [37]. Experimental and theoretical results suggested that this reduction in the rate of hydride transfer was due to significant differences in the stability of the tertiary structural elements surrounding the site of the mutation [22,33,38].
Two previous experimental studies suggested a coupling between protein dynamics and DHFR catalysis. Pre-steady-state measurements with DHFR from Thermotoga maritima indicated that protein fluctuations were coupled with hydride transfer which at low temperature displayed a significant contribution from tunnelling [8]. Steady-state experiments at the non-physiological pH value of 9, where hydride transfer is largely rate-determining for the E. coli enzyme, suggested a relatively rigid active site geometry and hydride transfer by extensive tunnelling, modulated by passive dynamics [2].
Here, we report measurements of the hydride transfer catalysed by DHFR from E. coli and the G121V-DHFR mutant, in which valine replaced the loop residue Gly121. While hydride transfer was slowed by almost two orders of magnitude in the mutant at physiological pH, hydrogen transfer was actively coupled with the environment for both proteins. The mechanism by which hydrogen transfer coupled with the environment was dependent on pH.
MATERIALS AND METHODS
Substrates and cofactors
H2F was prepared by dithionite reduction of folate (Sigma) as described previously [39]. NADPH was purchased from Sigma. NADPD ((4R)-[2H]NADPH) was prepared by reduction of NADP+ (Sigma) using the NADP+-dependent alcohol dehydrogenase from Thermoanaerobium brokii (Sigma) and purified by anion-exchange chromatography on Mono Q™ HR 5/5 (Amersham Biosciences). NADPH and NADPD concentrations were determined spectrophotometrically using a molar absorption coefficient (ϵ) at 339 nm of 6200 M−1·cm−1. Similarly, the concentration of H2F was measured assuming an ϵ of 28000 M−1·cm−1 at 282 nm for pH 7.4.
Protein purification
BL21(DE3) Star (Stratagene) cells were used for the production of DHFR from E. coli and its G121V mutant from a pET11c derived plasmid (Novagen) containing the respective coding regions essentially as described in [38]. Cells were grown in LB (Luria–Bertani) medium containing 0.27 mM ampicillin at 37 °C to an optical density at 600 nm of 0.6. Expression was induced by the addition of isopropyl β-thiogalactoside to a final concentration of 0.4 mM. Cells were harvested by centrifugation and resuspended in lysis buffer [25 mM potassium phosphate, pH 7.0, 0.5 M NaCl, 1 mM EDTA, 10%, v/v, glycerol and 1 mM DTT (dithiothreitol)]. The suspension was sonicated for 5 min on ice and centrifuged at 50000 g for 45 min. The supernatant was applied to a methotrexate column (10 ml; Sigma) pre-equilibrated with lysis buffer. The resin was extensively washed (∼300 ml) with the same buffer and elution of bound protein was achieved with 100 mM sodium borate (pH 8.3), 0.5 M NaCl, 10% glycerol and 3 mM folic acid. The eluted protein was dialysed extensively against 10 mM potassium phosphate (pH 7.0) containing 1 mM DTT. The dialysate was applied to a DEAE-Sepharose column (20 ml; GE Healthcare) previously equilibrated with 10 mM potassium phosphate (pH 7.0) containing 1 mM DTT and 5 mM EDTA. The bound protein was eluted with a gradient from 0 to 1 M NaCl in potassium phosphate (pH 7.0), 5 mM EDTA and 1 mM DTT and dialysed to remove salt. The protein concentrations were measured spectrophotometrically assuming an ϵ of 31100 M−1·cm−1 at 280 nm and by titration with methotrexate.
Pre-steady-state kinetic measurements
Pre-steady-state kinetic experiments were performed on an Applied Photophysics stopped-flow spectrophotometer essentially as described before [8]. Hydride transfer rates were measured following the fluorescence resonance energy transfer from the protein to reduced NADPH in potassium phosphate for optimal pH stability over the temperature range. The sample was excited at 292 nm and the emission was measured using an output filter with a cut-off at 400 nm. In a typical experiment for DHFR, the enzyme (40 μM, final concentrations) was preincubated with NADPH or NADPD (20 μM) in 100 mM potassium phosphate (pH 7.0) and 100 mM NaCl for at least 15 min to avoid hysteresis. The reaction was started by rapidly mixing with H2F (200 μM) in the same buffer. For G121V-DHFR, the enzyme (20 μM) was preincubated with NADPH or NADPD (10 μM) and the experiments were run as above. The hydride transfer rates did not change when the concentration of NADPH was reduced to 5 μM or doubled to 20 μM.
Steady-state kinetic measurements
All steady-state kinetic experiments were performed in MTEN buffer (50 mM MES buffer, 25 mM Tris, 25 mM ethanolamine and 100 mM NaCl) at pH 9.5. The pH was carefully adjusted at the experimental temperature with the relevant calibration buffers [borate (pH 10; Fisher Chemicals) and phosphate (pH 7; Fisher Chemicals)] at that temperature. DHFR (50 nM) was preincubated with NADPH/NADPD (50 μM) for 10 min at the experimental temperature prior to the addition of excess H2F (100 μM) to initiate the reaction. The steady-state rate was determined from the linear decrease in the absorbance of the reduced cofactor at 340 nm with time using a molar absorbance change for the DHFR reaction of 11800 M−1·cm−1 [40]. Each data point was measured at least six times.
RESULTS AND DISCUSSION
Temperature dependence of hydride transfer and KIEs (kinetic isotope effects) for DHFR catalysis
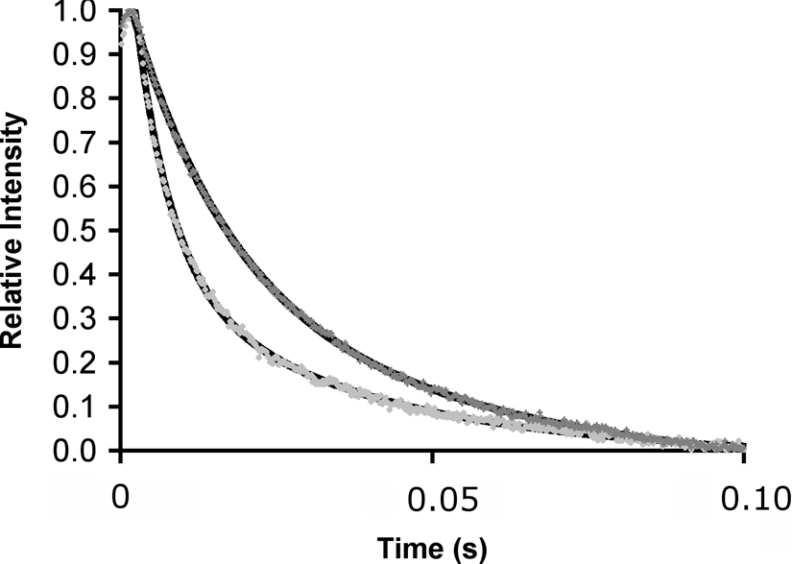
The rate of hydride transfer (kH) from NADPH to H2F was measured at pH 7 and 25 °C in single-turnover experiments by monitoring fluorescence resonance energy transfer from DHFR to the reduced cofactor in the presence of excess enzyme [26,37]. The decrease of the fluorescence intensity was best described by a double exponential (Figure 2). The rate constant of the slow step (∼6.5 s−1) was independent of the concentration of NADPH and did not depend on whether NADPH or NADPD was used as the cofactor. It most likely represented the slow release of cofactor and/or product. At 25 °C, the rapid decrease in fluorescence intensity preceding this slow step occurred with a first-order rate constant of 203.7±7.4 s−1 (see Supplementary Table 1 at http://www.BiochemJ.org/bj/394/bj3940259add.htm) in good agreement with previously published values of 220 s−1 [26,37] and 222.8±1.3 s−1 [8]. The slight variation is most likely a consequence of the different buffers used in these experiments. In the experiments reported here potassium phosphate was used because of its good pH stability over wide ranges of temperature. When NADPD was used as the cofactor, the rate constant was reduced to 75.1±5.4 s−1, resulting in a KIE of 2.7±0.2. This value was identical with the value of 2.8 that has been predicted computationally [31] and the published experimental value of 2.9±0.2 at ambient temperature [41].
Figure 2. Fluorescence energy transfer during DHFR catalysis.
Measurement of the rate of H- (light grey dots) and D- (dark grey) transfer catalysed by DHFR in a single turnover experiment measured by stopped-flow fluorescence resonance energy transfer from DHFR to the reduced cofactor at 25 °C. The relative intensity of fluorescence above 400 nm was measured after excitation at 292 nm. The fits to a double-exponential model for decreasing fluorescence intensity are also indicated. DHFR was preincubated with NADPH or NADPD and the reaction started through the addition of H2F. Final concentrations were: DHFR, 40 μM; reduced cofactor, 20 μM and H2F, 200 μM. The dead time of the experiments was approx. 0.002 s.
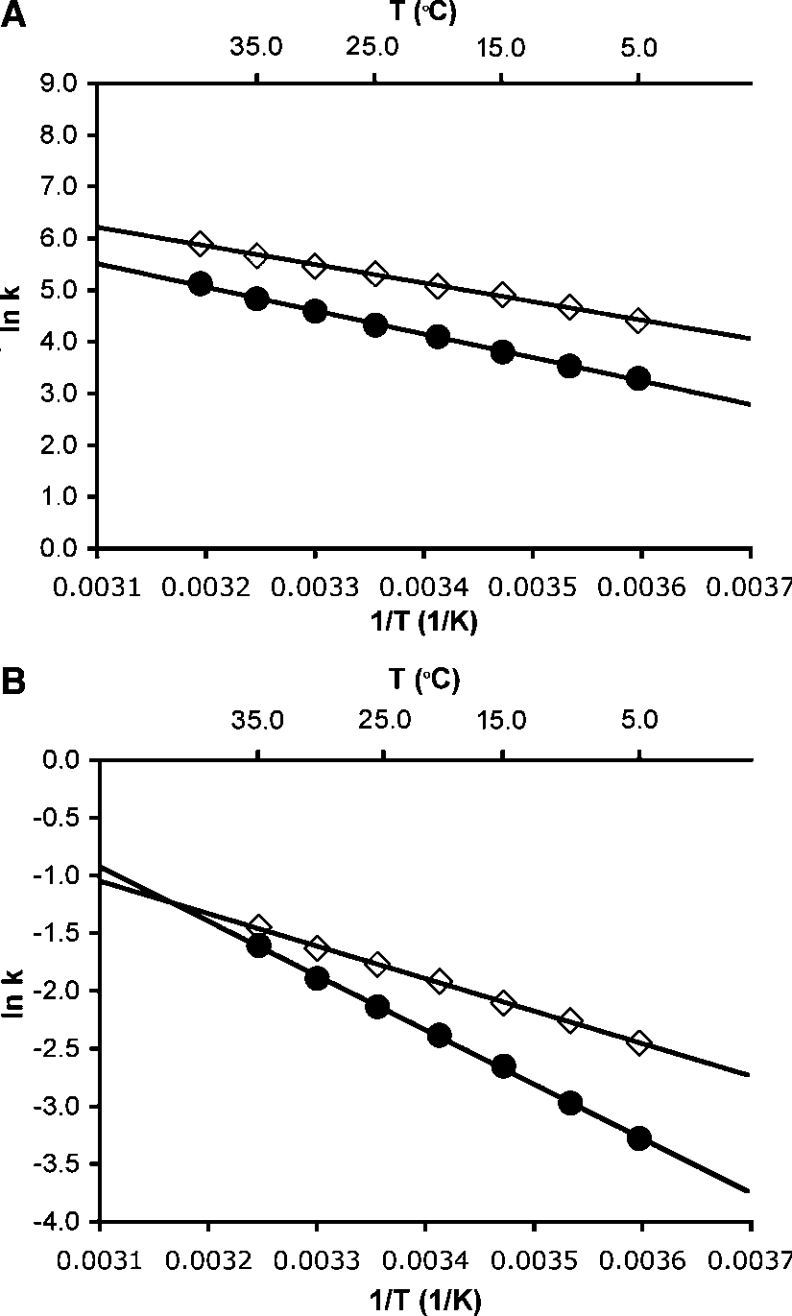
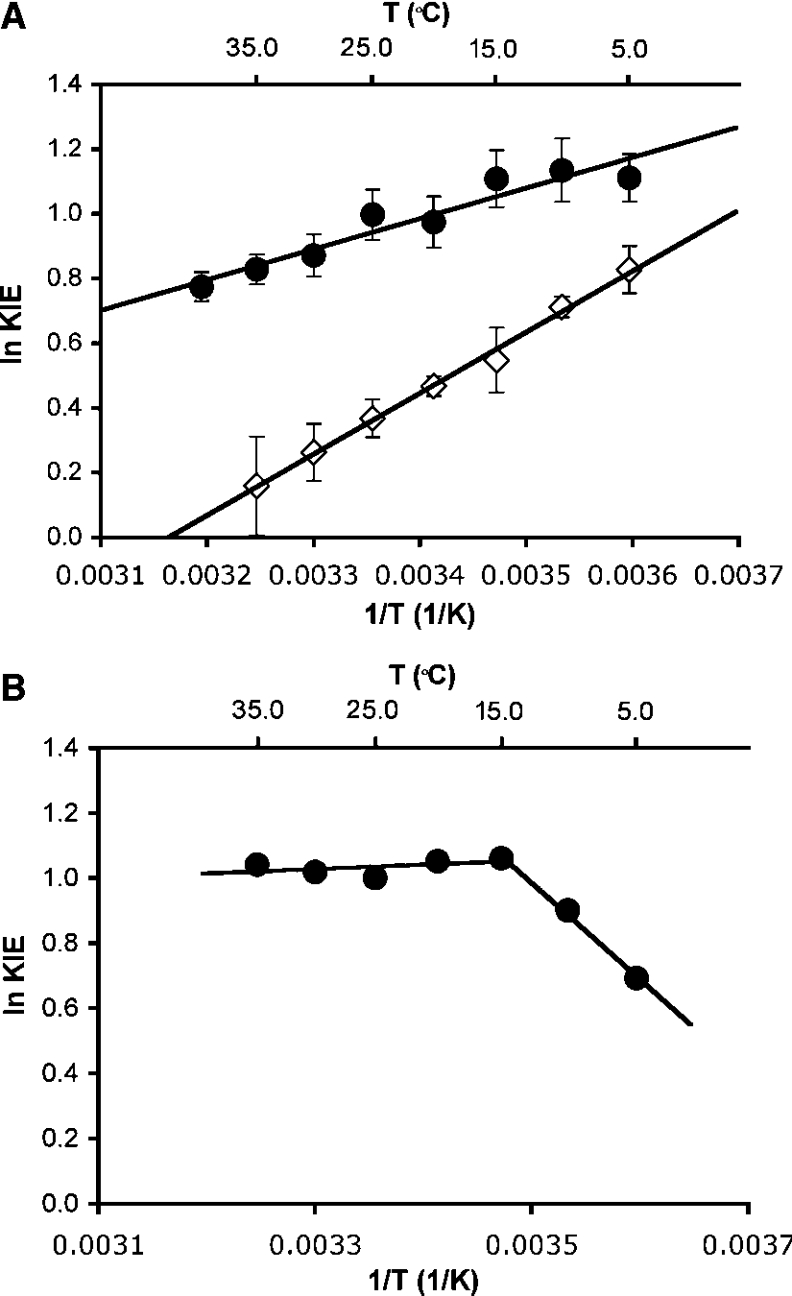
To evaluate whether hydride transfer during catalysis by DHFR was coupled to protein motions, the rate of the chemical step was measured at pH 7.0 as a function of the temperature between 5 and 40 °C for both 1H (H)- and 2H (D)-transfer. The temperature dependence of the rates was fitted to the empirical Arrhenius equation (ln k=ln A−EA/RT, where k is a first-order rate constant, A is the pre-exponential factor and EA is the experimental activation energy) to obtain the activation energy and the pre-exponential factor (Figure 3). The reaction rates showed a relatively strong dependence on the temperature, resulting in activation energies of EAH of 29.9±0.6 kJ·mol−1 and EAD of 37.7±0.7 kJ·mol−1, leading to an inverse pre-exponential factor AH/AD of 0.108±0.04 (AH=3.3±0.8×107 s−1; AD=3.07±0.81×108 s−1). The activation energy for H-transfer reported here was very similar to that measured previously in a different buffer but under identical conditions otherwise (28.2±0.9 kJ·mol−1) [8]. The relatively large difference in the activation energies for H- and D-transfer led to a temperature dependence of the primary KIEs (Figure 4). The KIE varied from 3.0±0.2 at 5 °C to 2.2±0.2 at 40 °C. The inverse ratio of the pre-exponential factors, which was well below the lower limit for the semi-classically calculated value of 0.71 [2,42], suggested a contribution from quantum mechanical tunnelling. Within a model of tunnelling through a rigid barrier, the temperature-dependent KIEs together with the observed values for AH/AD and EAH/D suggest a moderate amount of tunnelling [43]. Two recent computational studies of hydrogen transfer during catalysis by DHFR from E. coli suggested that the barrier for hydrogen transfer was significantly lowered through the inclusion of quantum mechanical effects [31,36]. Garcia-Viloca et al. [31] obtained an overall quantum mechanical correction of 13.4 kJ·mol−1 to the classical activation energy for hydrogen transfer. Most of this correction was attributed to quantization of nuclear vibrations, while only 2.9 kJ·mol−1 derived from hydrogen tunnelling. From the inverse ratio of the pre-exponential factors measured here, a previously proposed static model would predict moderate hydrogen tunnelling [43,44]. However, within the formalism of this model, elevated KIEs would be predicted in contrast with the rather small values measured here. The hydride transfer reaction catalysed by DHFR can therefore only be described adequately if protein motions coupled with hydrogen transfer are taken into account. Several theoretical approaches to hydrogen transfer models have been proposed that treat the hydrogen co-ordinates as fully quantum mechanical and incorporate various degrees of heavy atom motions to modulate the tunnelling barrier [16,17,45–47]. In one such model, a distinction was made between motions that actively modulate the tunnelling barrier and passive motions that merely help bring the substrates into reactive configurations (reviewed in [5,13]). This model predicts that when active dynamic motions are dominant and generate a temperature-dependent tunnelling distance, the KIEs become so temperature-dependent that the ratio of the pre-exponential factors is inverse. The experimentally observed inverse AH/AD and the temperature dependence of the KIEs suggest that at pH 7.0, DHFR actively modulates the tunnelling distance and hence the reaction rates [48]. Such active motions result in temperature-dependent KIEs and an inverse ratio of the pre-exponential factors, because the ideal tunnelling distance is different for the two isotopes. Such actively promoted tunnelling has been called rate-promoting vibrations [14,49,50], environmentally coupled tunnelling [13,43], and vibrationally enhanced ground state tunnelling [51]. The results reported here may also support the recent proposal of a correlation of changes in structural characteristics within DHFR from E. coli along the reaction pathway of hydrogen transfer through a network of hydrogen bonds and van der Waals interactions spanning the entire enzyme [18].
Figure 3. Arrhenius plots for H- and D-transfer during DHFR and G121V-DHFR catalysis at pH 7.0.
Temperature dependence of the H- (◇) and D-transfer (●) rate constants for the reactions catalysed by DHFR (A) and G121V-DHFR (B) is shown. Each data point is the average of at least six measurements; the error bars are obscured by the symbols. Fitting the average rate value for every temperature to the Arrhenius equation yields the following parameters: DHFR, EAH=29.9±0.6 kJ·mol−1, AH=3.3±0.8×107 s−1 and EAD=37.7±0.6 kJ·mol−1, AD=3.07±0.80×108 s−1; G121V-DHFR, EAH=23.3±0.3 kJ·mol−1, AH=2.2±0.2×103 s−1 and EAD=39.0±0.6 kJ·mol−1, AD=8.1±2.0×105 s−1.
Figure 4. Temperature dependence of the KIEs for H- and D-transfer from NADPH/NADPD to H2F.
(A) Results from pre-steady-state measurements at pH 7.0 for catalysis by DHFR (●) and G121V-DHFR (◇). The best fit through the data yielded ΔEAH/D of 7.83±0.98 kJ·mol−1 and AH/AD of 0.109±0.043 for DHFR and ΔEAH/D of 15.83±0.44 kJ·mol−1 and AH/AD of 0.0024±0.0004 for G121V-DHFR. (B) Steady-state measurements for DHFR catalysis at pH 9.0 and 5.0. The best fit through the data between 15 and 35 °C is indicated and yields ΔEAH/D of 1.11±0.79 kJ·mol−1 and AH/AD of 1.81±0.19.
Steady-state kinetics of DHFR at pH 9.5
In a previous study, the temperature dependence of the deuterium and tritium isotope effects at pH 9.0, which was measured under steady-state conditions, also revealed environmentally coupled hydrogen transfer [2]. However, in contrast with the results presented here, for H-transfer at pH 7.0, the KIEs were independent of temperature. The rates for hydride transfer in DHFR catalysis have been shown to decrease steeply with increasing pH [26]. However, the rate of product release, which is rate-determining at physiological pH, is pH-independent; hence hydride transfer becomes rate limiting at elevated pH and, at pH 9.5 and above, the steady-state rate reflects the rate of the chemical step only [2,26]. Since at pH 9.0 the chemical step is not fully rate-determining, hydride transfer rates were measured under steady-state conditions at pH 9.5 to exclude kinetic complexity at pH 9.0 as a basis for the observed difference in the temperature dependence of transfer at elevated and physiological pH. We have therefore determined the temperature dependence of the steady-state turnover rates for the reduction of H2F with NADPH and NADPD during DHFR catalysis at pH 9.5 [26] from the decrease in the absorbance at 340 nm due to the oxidation of the cofactor. At 25 °C, the rates for H- and D-transfer were 0.927±0.012 and 0.341±0.008 s−1, resulting in a KIE of 2.72±0.1 (see Supplementary Table 2 at http://www.BiochemJ.org/bj/394/bj3940259addd.htm). The KIE measured here was essentially identical to that calculated in a previous study [KIE (H/D)=2.92±0.076] from the measured H/T and D/T KIEs at pH 9.0 [2].
Within the temperature range of 15 and 35 °C the KIEs remained constant within the error of the experiment (Figure 4), while below 15 °C the KIEs decreased sharply. Fitting the KIEs between 15 and 35 °C to the Arrhenius equation resulted in a very small difference in the activation energy for H- and D-transfer (ΔEAH/D=1.11±0.79 kJ·mol−1) and an isotope effect for the pre-exponential factor AH/AD of 1.81±0.19, larger than the upper semi-classical limit of 1.41 [42]. A similar temperature dependence and enhanced KIE for the pre-exponential factors had been observed previously for the H/T and D/T KIEs at pH 9.0 [2]. A larger value for AH/AD of 4.0±1.5 has been reported previously in steady-state experiments at pH 9.0 [2]. While an AH/AD ratio outside the semi-classical range has widely been accepted as an indication for quantum mechanical tunnelling, a recent computational study resulted in a ratio of pre-exponential factors of 1.9 [52], identical within error with the results reported here. Interestingly, a larger value was obtained when the tunnelling contributions were omitted.
The temperature-independent KIEs at elevated pH were in sharp contrast with the strong temperature dependence of the KIEs at pH 7 and suggested a relatively rigid active site configuration from which hydrogen transfer occurs with a contribution from environmentally coupled hydrogen tunnelling. Within the model linking catalysis and dynamics proposed by Klinman and co-workers [43], the elevated value for AH/AD together with the temperature-independent KIEs suggested contributions from passive dynamics that do not actively modulate the tunnelling distance. This is in sharp contrast with the behaviour of DHFR at physiological pH, where hydrogen transfer is facilitated by active modulation of the tunnelling barrier. It is interesting to point out in this context that a recent computational study of DHFR catalysis revealed a small but significant temperature variation of the KIEs as a consequence of two competing temperature effects [53]. It is perhaps not surprising that in such a system the temperature dependence varies with reaction conditions.
DHFR has evolved an active site structure that is organized to support hydrogen tunnelling and to use active dynamics to promote hydrogen transfer at physiological pH. Relatively small increases in the pH of the solution, however, lead to a stiffening of the active site that no longer allows DHFR to actively promote hydrogen transfer, resulting in a reduction of the reaction rate by more than one order of magnitude. Interestingly, at pH 9.5, DHFR displays a temperature dependence of the reaction rates similar to that of the structurally more rigid DHFR from the thermophilic bacterium T. maritima [8]. Both enzymes appear to rely on passive motions only to generate active site configurations conducive to hydrogen transfer. In addition, the hydride transfer rates for the thermophilic DHFR at pH 7 and for the mesophilc DHFR at pH 9.5 were similar in the experimentally accessible temperature range [8,39]. These observations may suggest an evolutionary pattern in which catalysis progressed from a relatively rigid active site structure of DHFR from the ancient thermophile T. maritima to a more flexible and kinetically more efficient structure in E. coli that actively promotes hydrogen transfer at physiological pH.
Temperature dependence of hydride transfer and KIEs for catalysis by G121V-DHFR
Although residue 121 in the βF-βG loop of DHFR is on the exterior of the protein and approx. 19 Å from the centre of the enzyme, experimental studies have shown that mutations of Gly121 can reduce the rate of hydride transfer by up to 163-fold [20,37]. Together with NMR and computational studies, these results have been interpreted as evidence for the existence of a network of coupled dynamic motions that include residues in exterior loops of DHFR and promote hydride transfer [18]. It has been postulated that mutation of Gly121 disrupts this network of coupled motions leading to the observed reduction in the reaction rate and altered dynamic properties of the enzyme [52]. The rate of the hydride transfer from NADPH to H2F catalysed by G121V-DHFR was measured at pH 7.0 in pre-steady-state experiments. The decrease in the fluorescence intensity was best described by a double exponential. A first event was observed that occurred with a rate constant of approx. 3.5 s−1 at 25 °C and did not show a KIE when NADPD replaced NADPH. This process had previously been interpreted as a conformational change associated with a rearrangement of the reduced nicotinamide ring into the enzyme's active site [20]. Surprisingly, this fast isotopically insensitive step had not been observed under similar conditions in a previous study of G121V-DHFR catalysis, perhaps due to the different buffers used in the two studies [37]. This initial step was followed by a slow decrease in the fluorescence intensity, the rate of which did not change when the concentration of NADPH was varied. The slow step was therefore most likely due to the oxidation of the cofactor that occurred with a first-order rate constant of 0.171±0.004 s−1 at 25 °C (see Supplementary Table 3 at http://www.BiochemJ.org/bj/394/bj3940259add.htm). This rate constant is somewhat lower than that published previously [37], most likely as a consequence of the different reaction conditions. In the present study, potassium phosphate, which shows little variation of pH with changing temperature, was used rather than the amine-based MTEN buffer. At 25 °C, a KIE of 1.450±0.042 was measured when NADPD was used instead of NADPH.
Similar to the wild-type enzyme, a relatively strong temperature dependence of the hydride transfer rates and the KIEs was observed between 5 and 35 °C (Figures 3 and 4). The rates increased almost 3-fold in the temperature range leading to activation energies of EAH of 23.3±0.3 and EAD of 39.0±0.6 kJ·mol−1 for H- and D-transfer respectively. The activation energy for H-transfer catalysed by G121V-DHFR was smaller than that measured for the wild-type, indicating that the reduced catalytic efficiency of the mutant was a consequence of a smaller pre-exponential factor (AH=2.2±0.2×103 s−1; AD=8.1±2.0×105 s−1). The difference in activation energies (ΔEA=EAH−EAD) of −15.7 kJ·mol−1 was twice that observed for wild-type DHFR, leading to more strongly inverse pre-exponential factors AH/AD of 0.0025±0.0007. These data are consistent with a model in which the reaction occurred with a significant contribution from quantum mechanical tunnelling coupled with active dynamic motions.
The results presented here indicate that, while replacing residue 121 in DHFR with valine leads to a significant reduction in the rate of hydrogen transfer, the general mechanism by which the reaction is coupled with the environment is unaltered, at least in a qualitative sense. Molecular dynamics, CD and fluorescence experiments had suggested that the reduced catalytic efficiency of the mutant may arise from structural effects on the overall fold of the protein [38]. NMR experiments indicated that in G121VDHFR the closed conformation, in which hydrogen transfer occurs in the wild-type, was destabilized [22]. Geometric constraints imposed by the bulky isopropyl group in G121V-DHFR may be transmitted to the active site to produce a lower number of active site configurations favourable for hydride transfer. In agreement with this explanation, a computational study by Thorpe and Brooks [33] suggested that H2F and NADPH populate preferentially a region of configuration space of DHFR that is conducive to the reaction, while substrate and cofactor become trapped in unproductive configurations in G121V-DHFR. The results presented here indicate, however, that once NADPH and H2F are bound in the reactive configuration, both mutant and wild-type actively promote hydride transfer.
The reduced catalytic efficiency of G121V-DHFR was mainly a consequence of the significantly reduced pre-exponential factors. The Eyring interpretation of the temperature dependence of the reaction rates relates activation energy and pre-exponential factor to activation enthalpy and entropy, Δη‡ and ΔS‡. The activation enthalpies and entropies for hydride transfer by DHFR and G121V-DHFR were obtained by fitting the temperature dependence of the reaction rates to the Eyring equation [k/T=kB/h exp (ΔS‡/R) exp(−ΔH‡/RT), where k is a first-order constant, kB the Boltzmann constant and h the Planck constant; R is the universal gas constant]. While the activation enthalpy favoured hydride transfer by G121V-DHFR by 6.3±0.2 kJ·mol−1, the activation entropy strongly favoured the reaction catalysed by the wild-type enzyme. An increase in the activation entropy (−TΔS‡) for the mutant indicated that a significantly higher molecular reorganization was required to reach the transition state in G121V-DHFR (TΔS‡G121V=−56.4±0.8 kJ·mol−1) than in the wild type enzyme (TΔS‡WT=−32.9±0.8 kJ·mol−1).
In summary, our results suggest that the mechanism by which hydrogen transfer in DHFR is coupled with protein fluctuations, depends on the pH of the environment. Under physiological conditions, DHFR increases the reaction rate through optimization of the coupling of environmental dynamics with quantum mechanical tunnelling. Only relatively small changes in pH lead to a stiffening of the active site and a loss of active promotion of hydrogen transfer. On the other hand, the mechanism is relatively robust towards mutation of residue 121, the identity of which is known to be critical for efficient hydride transfer. Like in the wild-type enzyme, the tunnelling distance is modulated by active dynamics in the kinetically compromised mutant G121V-DHFR. The reduced hydride transfer rates of this mutant are therefore not the consequence of altered dynamics, but most likely of geometric constraints [33,38] that lead to a higher entropic cost for the formation of active site conformations conducive to the reaction.
Online data
Acknowledgments
This work was supported by the BBSRC (Biotechnology and Biological Sciences Research Council) and Cardiff University.
References
- 1.Marcus R. A., Sutin N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- 2.Sikorski R. S., Wang L., Markham K. A., Rajagopalan P. T. R., Benkovic S. J., Kohen A. Tunneling and coupled motion in the Escherichia coli dihydrofolate reductase catalysis. J. Am. Chem. Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- 3.Basran J., Patel S., Sutcliffe M. J., Scrutton N. S. Importance of barrier shape in enzyme-catalyzed reactions: vibrationally assisted hydrogen tunneling in tryptophan tryptophylquinone-dependent amine dehydrogenases. J. Biol. Chem. 2001;276:6234-6242. doi: 10.1074/jbc.M008141200. [DOI] [PubMed] [Google Scholar]
- 4.Francisco W. A., Knapp M. J., Blackburn N. J., Klinman J. P. Hydrogen tunneling in peptidylglycine α-hydroxylating monooxygenase. J. Am. Chem. Soc. 2002;124:8194–8195. doi: 10.1021/ja025758s. [DOI] [PubMed] [Google Scholar]
- 5.Knapp M. J., Klinman J. P. Environmentally coupled hydrogen tunneling: linking catalysis to dynamics. Eur. J. Biochem. 2002;269:3113–3121. doi: 10.1046/j.1432-1033.2002.03022.x. [DOI] [PubMed] [Google Scholar]
- 6.Kohen A., Klinman J. P. Hydrogen tunneling in biology. Chem. Biol. 1999;6:R191–R198. doi: 10.1016/s1074-5521(99)80058-1. [DOI] [PubMed] [Google Scholar]
- 7.Kohen A., Klinman J. P. Protein flexibility correlates with degree of hydrogen tunneling in thermophilic and mesophilic alcohol dehydrogenases. J. Am. Chem. Soc. 2000;122:10738–10739. [Google Scholar]
- 8.Maglia G., Allemann R. K. Evidence for environmentally coupled hydrogen tunneling during dihydrofolate reductase catalysis. J. Am. Chem. Soc. 2003;125:13372–13373. doi: 10.1021/ja035692g. [DOI] [PubMed] [Google Scholar]
- 9.Rickert K. W., Klinman J. P. Nature of hydrogen transfer in soybean lipoxygenase 1: separation of primary and secondary isotope effects. Biochemistry. 1999;38:12218–12228. doi: 10.1021/bi990834y. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal N., Hong B. Y., Mihai C., Kohen A. Vibrationally enhanced hydrogen tunneling in the Escherichia coli thymidylate synthase catalyzed reaction. Biochemistry. 2004;43:1998–2006. doi: 10.1021/bi036124g. [DOI] [PubMed] [Google Scholar]
- 11.Masgrau L., Basran J., Hothi P., Sutcliffe M. J., Scrutton N. S. Hydrogen tunneling in quinoproteins. Arch. Biochem. Biophys. 2004;428:41–51. doi: 10.1016/j.abb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Doll K. M., Bender B. R., Finke R. G. The first experimental test of the hypothesis that enzymes have evolved to enhance hydrogen tunneling. J. Am. Chem. Soc. 2003;125:10877–10884. doi: 10.1021/ja030120h. [DOI] [PubMed] [Google Scholar]
- 13.Klinman J. P. Dynamic barriers and tunneling. New views of hydrogen transfer in enzyme reactions. Pure Appl. Chem. 2003;75:601–608. [Google Scholar]
- 14.Antoniou D., Caratzoulas S., Kalyanaraman C., Mincer J. S., Schwartz S. D. Barrier passage and protein dynamics in enzymatically catalyzed reactions. Eur. J. Biochem. 2002;269:3103–3112. doi: 10.1046/j.1432-1033.2002.03021.x. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Viloca M., Gao J., Karplus M., Truhlar D. G. How enzymes work: analysis by modern rate theory and computer simulations. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 16.Borgis D., Hynes J. T. Curve crossing formulation for proton transfer reactions in solution. J. Phys. Chem. 1996;100:1118–1128. [Google Scholar]
- 17.Bruno W. J., Bialek W. Vibrationally enhanced tunneling as a mechanism for enzymatic hydrogen transfer. Biophys. J. 1992;63:689–699. doi: 10.1016/S0006-3495(92)81654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal P. K., Billeter S. R., Rajagopalan P. T. R., Benkovic S. J., Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuznetsov A. M., Ulstrup J. Proton and hydrogen atom tunnelling in hydrolytic and redox enzyme catalysis. Can. J. Chem. 1999;77:1085–1096. [Google Scholar]
- 20.Rajagopalan P. T. R., Lutz S., Benkovic S. J. Coupling interactions of distal residues enhance dihydrofolate reductase catalysis: mutational effects on hydride transfer rates. Biochemistry. 2002;41:12618–12628. doi: 10.1021/bi026369d. [DOI] [PubMed] [Google Scholar]
- 21.Sawaya M. R., Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 22.Venkitakrishnan R. P., Zaborowski E., McElheny D., Benkovic S. J., Dyson H. J., Wright P. E. Conformational changes in the active site loops of dihydrofolate reductase during the catalytic cycle. Biochemistry. 2004;43:16046–16055. doi: 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- 23.Osborne M. J., Schnell J., Benkovic S. J., Dyson H. J., Wright P. E. Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 24.Falzone C. J., Wright P. E., Benkovic S. J. Dynamics of a flexible loop in dihydrofolate-reductase from Escherichia coli and its implication for catalysis. Biochemistry. 1994;33:439–442. doi: 10.1021/bi00168a007. [DOI] [PubMed] [Google Scholar]
- 25.McElheny D., Schnell J. R., Lansing J. C., Dyson H. J., Wright P. E. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fierke C. A., Johnson K. A., Benkovic S. J. Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry. 1987;26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 27.Miller G. P., Benkovic S. J. Stretching exercises: flexibility in dihydrofolate reductase catalysis. Chem. Biol. 1998;5:R105–R113. doi: 10.1016/s1074-5521(98)90616-0. [DOI] [PubMed] [Google Scholar]
- 28.Cummins P. L., Gready J. E. Energetically most likely substrate and active-site protonation sites and pathways in the catalytic mechanism of dihydrofolate reductase. J. Am. Chem. Soc. 2001;123:3418–3428. doi: 10.1021/ja0038474. [DOI] [PubMed] [Google Scholar]
- 29.Castillo R., Andres J., Moliner V. Catalytic mechanism of dihydrofolate reductase enzyme: a combined quantum-mechanical/molecular-mechanical characterization of transition state structure for the hydride transfer step. J. Am. Chem. Soc. 1999;121:12140–12147. [Google Scholar]
- 30.Garcia-Viloca M., Truhlar D. G., Gao J. Importance of substrate and cofactor polarization in the active site of dihydrofolate reductase. J. Mol. Biol. 2003;327:549–560. doi: 10.1016/s0022-2836(03)00123-2. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Viloca M., Truhlar D. G., Gao J. Reaction-path energetics and kinetics of the hydride transfer reaction catalyzed by dihydrofolate reductase. Biochemistry. 2003;42:13558–13575. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 32.Thorpe I. F., Brooks C. L. Barriers to hydride transfer in wild type and mutant dihydrofolate reductase from E. coli. J. Phys. Chem. B. 2003;107:14042–14051. [Google Scholar]
- 33.Thorpe I. F., Brooks C. L. The coupling of structural fluctuations to hydride transfer in dihydrofolate reductase. Proteins Struct. Funct. Bioinform. 2004;57:444–457. doi: 10.1002/prot.20219. [DOI] [PubMed] [Google Scholar]
- 34.Rod T. H., Radkiewicz J. L., Brooks C. L. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rod T. H., Brooks C. L. How dihydrofolate reductase facilitates protonation of dihydrofolate. J. Am. Chem. Soc. 2003;125:8718–8719. doi: 10.1021/ja035272r. [DOI] [PubMed] [Google Scholar]
- 36.Agarwal P. K., Billeter S. R., Hammes-Schiffer S. Nuclear quantum effects and enzyme dynamics in dihydrofolate reductase catalysis. J. Phys. Chem. B. 2002;106:3283–3293. [Google Scholar]
- 37.Cameron C. E., Benkovic S. J. Evidence for a functional role of the dynamics of glycine-121 of Escherichia coli dihydrofolate reductase obtained from kinetic analysis of a site-directed mutant. Biochemistry. 1997;36:15792–15800. doi: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- 38.Swanwick R. S., Shrimpton P. J., Allemann R. K. Pivotal role of Gly121 in dihydrofolate reductase from Escherichia coli: the altered structure of a mutant enzyme may form the basis of its diminished catalytic performance. Biochemistry. 2004;43:4119–4127. doi: 10.1021/bi036164k. [DOI] [PubMed] [Google Scholar]
- 39.Maglia G., Javed M. H., Allemann R. K. Hydride transfer during catalysis by dihydrofolate reductase from Thermotoga maritima. Biochem. J. 2003;374:529–535. doi: 10.1042/BJ20030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone S. R., Morrison J. F. Kinetic mechanism of the reaction catalyzed by dihydrofolate reductase from Escherichia coli. Biochemistry. 1982;21:3757–3765. doi: 10.1021/bi00259a006. [DOI] [PubMed] [Google Scholar]
- 41.Miller G. P., Benkovic S. J. Strength of an interloop hydrogen bond determines the kinetic pathway in catalysis by Escherichia coli dihydrofolate reductase. Biochemistry. 1998;37:6336–6342. doi: 10.1021/bi973065w. [DOI] [PubMed] [Google Scholar]
- 42.Bell R. P. London: Chapman and Hall; 1980. The Tunnel Effect in Chemistry; pp. 88–105. [Google Scholar]
- 43.Knapp M. J., Rickert K., Klinman J. P. Temperature-dependent isotope effects in soybean lipoxygenase-1: correlating hydrogen tunneling with protein dynamics. J. Am. Chem. Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 44.Jonsson T., Glickman M. H., Sun S. J., Klinman J. P. Experimental evidence for extensive tunneling of hydrogen in the lipoxygenase reaction: implications for enzyme catalysis. J. Am. Chem. Soc. 1996;118:10319–10320. [Google Scholar]
- 45.Borgis D., Hynes J. T. Dynamic theory of proton tunneling transfer rates in solution: general formulation. Chem. Phys. 1993;170:315–346. [Google Scholar]
- 46.Antoniou D., Schwartz S. D. Activated chemistry in the presence of a strongly symmetrically coupled vibration. J. Chem. Phys. 1998;108:3620–3625. [Google Scholar]
- 47.Antoniou D., Schwartz S. D. Internal enzyme motions as a source of catalytic activity: rate-promoting vibrations and hydrogen tunneling. J. Phys. Chem. B. 2001;105:5553–5558. [Google Scholar]
- 48.Knapp M. J., Klinman J. P. Kinetic studies of oxygen reactivity in soybean lipoxygenase-1. Biochemistry. 2003;42:11466–11475. doi: 10.1021/bi0300884. [DOI] [PubMed] [Google Scholar]
- 49.Mincer J. S., Schwartz S. D. A computational method to identify residues important in creating a protein promoting vibration in enzymes. J. Phys. Chem. B. 2003;107:366–371. [Google Scholar]
- 50.Caratzoulas S., Mincer J. S., Schwartz S. D. Identification of a protein-promoting vibration in the reaction catalyzed by horse liver alcohol dehydrogenase. J. Am. Chem. Soc. 2002;124:3270–3276. doi: 10.1021/ja017146y. [DOI] [PubMed] [Google Scholar]
- 51.Sutcliffe M. J., Scrutton N. S. A new conceptual framework for enzyme catalysis: hydrogen tunneling coupled to enzyme dynamics in flavoprotein and quinoprotein enzymes. Eur. J. Biochem. 2002;269:3096–3102. doi: 10.1046/j.1432-1033.2002.03020.x. [DOI] [PubMed] [Google Scholar]
- 52.Watney J. B., Agarwal P. K., Hammes-Schiffer S. Effect of mutation on enzyme motion in dihydrofolate reductase. J. Am. Chem. Soc. 2003;125:3745–3750. doi: 10.1021/ja028487u. [DOI] [PubMed] [Google Scholar]
- 53.Pu J. Z., Ma S. H., Gao J. L., Truhlar D. G. Small temperature dependence of the kinetic isotope effect for the hydride transfer reaction catalyzed by Escherichia coli dihydrofolate reductase. J. Phys. Chem. B. 2005;109:8551–8556. doi: 10.1021/jp051184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.