Abstract
Tristetraprolin (TTP) is a zinc-finger protein that binds to AREs (AU-rich elements) within certain mRNAs and causes destabilization of those mRNAs. Mice deficient in TTP develop a profound inflammatory syndrome with erosive arthritis, autoimmunity and myeloid hyperplasia. Previous studies showed that TTP is phosphorylated extensively in intact cells. However, limited information is available about the identities of these phosphorylation sites. We investigated the phosphorylation sites in human TTP from transfected HEK-293 cells by MS and site-directed mutagenesis. A number of phosphorylation sites including Ser66, Ser88, Thr92, Ser169, Ser186, Ser197, Ser218, Ser228, Ser276 and Ser296 were identified by MS analyses using MALDI (matrix-assisted laser-desorption–ionization)-MS, MALDI-tandem MS, LC (liquid chromatography)–tandem MS and multidimensional protein identification technology. Mutations of Ser197, Ser218 and Ser228 to alanine in the human protein significantly increased TTP's gel mobility (likely to be stoichiometric), whereas mutations at the other sites had little effect on its gel mobility. Dephosphorylation and in vivo labelling studies showed that mutant proteins containing multiple mutations were still phosphorylated, and all were able to bind to RNA probes containing AREs. Confocal microscopy showed a similar cytosolic localization of TTP among the various proteins. Ser197, Ser218 and Ser228 are predicted by motif scanning to be potential sites for protein kinase A, glycogen synthase kinase-3 and extracellular-signal-regulated kinase 1 (both Ser218 and Ser228) respectively. The present study has identified multiple phosphorylation sites in the anti-inflammatory protein TTP in mammalian cells and should provide the molecular basis for further studies on the function and regulation of TTP in controlling pro-inflammatory cytokines.
Keywords: cytokine, inflammation, mass spectrometry (MS), phosphorylation site, site-directed mutagenesis, tristetraprolin
Abbreviations: ARE, AU-rich element; CIAP, calf intestine alkaline phosphatase; CMV, cytomegalovirus; EMSA, electrophoretic mobility-shift assay; ERK2, extracellular-signal-regulated kinase 2 [p42 MAPK (mitogen-activated protein kinase)]; FCS, fetal-calf serum; ESI, electrospray ionization; GM-CSF, granulocyte–macrophage colony-stimulating factor; HA, haemagglutinin; hTTP, human TPP (tristetraprolin); IMAC, immobilized metal ion affinity chromatography; JNK, c-Jun N-terminal kinase; KO, knockout; LC, liquid chromatography; LPS, lipopolysaccharide; MALDI-MS, matrix-assisted laser-desorption–ionization MS; MAPK, mitogen-activated protein kinase; MBP, maltose-binding protein; MK2, MAPK-activated protein kinase 2; MS/MS, tandem MS; mTTP, mouse TPP; MudPIT, multidimensional protein identification technology; m/z, mass-to-charge ratio; Ni-NTA, Ni2+-nitrilotriacetate; TFA, trifluoroacetic acid; TNFα, tumour necrosis factor-α; TZF, tandem zinc finger; WT, wild-type
INTRODUCTION
TTP (tristetraprolin) is the best-understood member of a small family of tandem CCCH zinc-finger proteins. Similar tandem CCCH zinc-finger sequences have been found in many species, ranging from humans to yeasts and plants. In mammals, three members of the specific TTP family of tandem CCCH zinc-finger proteins have been characterized: TTP (also known as ZFP36, TIS11, G0S24 and NUP475), ZFP36L1 (also known as TIS11b, cMG1, ERF1, BRF1 and Berg36) and ZFP36L2 (also known as TIS11d, ERF2 and BRF2) [1]. Gene KO (knockout) studies have shown that TTP deficiency causes a profound inflammatory syndrome with erosive arthritis, autoimmunity and myeloid hyperplasia [2]. This is believed to be due almost entirely to excessive production of TNFα (tumour necrosis factor-α) and GM-CSF (granulocyte–macrophage colony-stimulating factor), both of whose mRNAs are direct targets of TTP. The excessive cytokine production is due, at least in part, to the stabilization of these mRNAs in cells from the KO mice [2–4]. For these reasons, TTP can be thought of as an anti-inflammatory or arthritis-suppressor protein [5].
The cDNAs encoding TTP were originally cloned by three groups on the basis of its very rapid and dramatic transcriptional induction in fibroblasts in response to insulin, phorbol esters and serum [6–8]. Recently we have shown that TTP protein is a very-low-abundance cytosolic protein whose levels are also dramatically induced by LPS (lipopolysaccharide) and FCS (fetal-calf serum) [9]. The protein is relatively stable once induced, in contrast with its labile mRNA [9]. In addition, TTP in normal tissues and in stimulated cells exhibits a larger molecular mass on SDS/polyacrylamide gels than the predicted size [9], probably due to extensive phosphorylation of TTP in intact cells and tissues. This conclusion is supported by the fact that dephosphorylation of TTP from transfected HEK-293 cells and LPS-stimulated mouse RAW 264.7 cells results in a marked decrease in apparent TTP size on SDS gels, resulting in an apparent size close to that of TTP expressed in, and purified from, Escherichia coli under denaturing conditions [9–11].
Several groups have studied TTP phosphorylation. We and others have shown that TTP could be phosphorylated in intact cells and/or in cell-free systems by ERK2 {extracellular-signal-regulated kinase 2 [p42 MAPK (mitogen-activated protein kinase)]} [10,12,13], p38 MAPK [10–12,14], JNK (c-Jun N-terminal kinase) [12] and MAPK-activated protein kinase 2 (MAPKAPK2 or MK2) [15–18]. Several investigations have studied the effect of phosphorylation on TTP function(s). For example, Ser220 of mTTP (mouse TTP) has been shown to be phosphorylated in vivo and in vitro [13], but not to affect protein nuclear-to-cytoplasmic shuttling [19]. Phosphorylation of Ser178 of mTTP increases its binding to 14-3-3 proteins and provides one of the mechanisms targeting TTP in the cytoplasm [20]. Similarly, dephosphorylation of mTTP by CIAP (calf intestinal alkaline phosphatase) prevents its binding to 14-3-3 proteins [20]. hTTP (human TTP) expressed in HEK-293 cells and then dephosphorylated by CIAP is able to bind more tightly to an ARE (AU-rich element) probe than phosphorylated hTTP [11]. Finally, recombinant TTP from E. coli binds to a TNFα ARE probe with higher affinity than TTP purified from mammalian cells [10].
In the present study we have identified several phosphorylation sites in hTTP from transfected HEK-293 cells, including several that cause electrophoretic shifts in protein migration, implying stoichiometric phosphorylation [21,22]. We have utilized a number of approaches, including in vivo labelling, site-directed mutagenesis, peptide mapping, Edman sequencing and MS. We report the identification of a number of high-probability phosphorylation sites (i.e., those identified with multiple phosphopeptides and/or by more than one method), including Ser66, Ser88, Thr92, Ser169, Ser186, Ser197, Ser218, Ser228, Ser276 and Ser296, as well as a large number of other sites with lower probability [i.e., those only identified by MudPIT (multidimensional protein identification technology)]. When Ser197, Ser218 and Ser228 were mutated to alanine residues, major changes in the electrophoretic mobility of hTTP were observed. These mobility changes are consistent with these three sites in hTTP being involved in stoichiometric phosphorylation. Each is part of a proline-directed protein kinase site that is completely conserved in all known mammalian TTP orthologues. All of the proteins tested were able to bind to the TNFα ARE, promote degradation of the bound mRNA, and were predominantly localized in the cytosol of transfected human cells. Ser197, Ser218 and Ser228 are predicted by motif scanning to be potential sites for protein kinase A (Ser197), glycogen synthase kinase-3 (Ser218) and ERK1 (both Ser218 and Ser228) respectively. Identification of the anti-inflammatory protein TTP as a hyperphosphorylated protein in mammalian cells should help further studies on the function and regulation of TTP in controlling the biosynthesis of pro-inflammatory cytokines such as TNFα.
EXPERIMENTAL
Protein expression plasmids
The construction of the WT (wild-type) pHis-hTTP expression plasmid [CMV.(his)6N.hTTP] was described previously [11] (CMV is cytomegalovirus). This plasmid contains DNA sequence encoding six consecutive histidine residues [(his)6] that were inserted between the sequences for the initiator methionine residue and the second aspartate residue of full-length hTTP (GenBank® accession no. NP_003398). The plasmid was then used as a template for the construction of 40 site-directed mutant plasmids using the primer-overlapping site-directed mutagenesis technique [10,23] and by recombination of fragments from the existing constructs. These mutant sites were selected on the basis of sequence comparisons among TTP proteins from all available mammalian species, including full-length amino acid sequences of TTP from human, mouse, rat, cow and sheep, partial amino acid sequences deduced from expressed sequence tags and genomic sequences from chimpanzee, dog, horse and pig. Ten plasmids contained single site mutations: S88A, S90A, S93A, S186A, S197A, S214A, S218A, S228A, S296A, T271A; eight plasmids contained two site mutations: S214A/S218A, S214A/S228A, S218A/S228A, S197A/S214A, S197A/S218A, S197A/S228A, S197A/S296A and S93A/S197A; five plasmids contained three site mutations: S88A/S90A/S93A, S197A/S214A/S218A, S197A/S214A/S228A, S197A/S218A/S228A and S214A/S218A/S228A; three plasmids contained four site mutations: S214A/S218A/S228A/S296A, S88A/S90A/S93A/S197A and S197A/S214A/S218A/S228A; five plasmids contained five site mutations: S88A/S90A/S93A/S214A/S218A, S88A/S90A/S93A/S214A/S228A, S88A/S90A/S93A/S218A/S228A, S197A/S214A/S218A/S228A/S296A and S88A/S197A/S214A/S218A/S228A; four plasmids contained six site mutations: S88A/S90A/S93A/S214A/S218A/S228A, S88A/S197A/S214A/S218A/S228A/S296A, S88A/S186A/S197A/S214A/S218A/S228A and S88A/S197A/S214A/S218A/S228A/T271A; four plasmids contained seven site mutations: S88A/S186A/S197A/S214A/S218A/S228A/S296A, S88A/S197A/S214A/S218A/S228A/S296A/T271A, S88A/S90A/S93A/S214A/S218A/S228A/S296A and S88A/S90A/S93A/S197A/S214A/S218A/S228A; and one plasmid contained eight site mutations: S88A/S90A/S93A/S197A/S214A/S218A/S228A/S296A. Unless other-wise specified, all amino acid numbers refer to hTTP (RefSeq NP_003398).
Plasmid pHis-hTTP-Flag was constructed to include DNA sequences encoding a FLAG peptide (DYKDDDDK) at the C-ter-minus of the full-length His-hTTP protein as described previously [24]. Plasmid pHA-hTTP was the plasmid (CMV.hTTP.tag) encoding the full-length hTTP with the influenza-virus haemagglutinin epitope tag at the C-terminus [25]. This plasmid was used for the construction of the following plasmids encoding truncated hTTP protein fragments: hTTP (Full: S228A), hTTP (97–173), hTTP (1–173), hTTP (1–173 with deletion of residues 64–82), hTTP (97–326 S228A), hTTP (67–173), hTTP (67–206), hTTP (67–230), hTTP (97–206) and hTTP (97–230). Plasmid pMBP-hTTP was used for the expression and purification of hTTP as an MBP (maltose-binding protein) fusion protein or as a non-fusion hTTP as described previously [12]. All insert sequences were confirmed by dRhodamine Terminator Cycle Sequencing (PerkinElmer Life Sciences, Gaithersburg, MD, U.S.A.).
Protein expression and purification
HEK-293 cells were transfected with plasmids using the calcium phosphate precipitation method described previously [10,26]. The transfected cells were lysed in lysis buffer (10 mM Hepes, pH 7.6, 3 mM MgCl2, 40 mM KCl, 0.5% (v/v) Nonidet P40, 8 μg/ml leupeptin and 0.5 mM PMSF) as described previously [10]. The cell lysate was centrifuged at 1000 g for 5 min and the supernatant was further centrifuged at 10000 g for 10 min. Histidine-tagged hTTP from the 10000 g supernatant was bound to Ni-NTA (Ni2+-nitrilotriacetate) beads (Qiagen, Madison, WI, U.S.A.) and eluted with elution buffers (100, 200 or 250 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl and 0.05% Tween 20, pH 8.0) as described previously [10]. Glycerol (20%, v/v) was added to the eluted protein samples. The samples were then frozen in liquid nitrogen and stored at −70 °C. The non-fusion hTTP was purified from MBP–hTTP fusion protein following PreScission protease (Amersham Biosciences, Uppsala, Sweden) digestion of the fusion protein expressed in, and purified from, E. coli as described previously [12].
Protein concentration determination, SDS/PAGE and immunoblotting
Protein concentrations were determined with the Bio-Rad Dye assay kit with modifications as described previously [10]. SDS/PAGE, using the Tris/glycine gel buffer system and immunoblotting, followed described procedures [12]. The primary antibody was an anti-MBP-hTTP serum raised in New Zealand White rabbits against the purified MBP–hTTP fusion protein [10]. The secondary antibodies were affinity-purified goat anti-rabbit IgG (heavy+light) horseradish peroxidase conjugate with human IgG absorbed (Bio-Rad Laboratories).
In vivo labelling, Ni-NTA purification, immunoprecipitation and dephosphorylation
Proteins in HEK-293 cells were labelled with [32P]orthophosphate following a previously published procedure [10] and were lysed directly on the Petri dishes in lysis buffer (50 mM NaH2PO4, 250 mM NaCl, 50 mM NaF, 10 mM imidazole, 1 mM PMSF, 1 μg/ml leupeptin and 0.5% Nonidet P40). Histidine-tagged hTTP was purified by Ni-NTA beads as described previously [10] or immunoprecipitated by the anti-MBP–hTTP serum following a similar procedure, using 20 μl of the antiserum per 100 μl of the 10000 g supernatant [10]. The immunoprecipitated pellet was washed three times and suspended in 20 μl of Ni-NTA wash buffer (20 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl and 0.05% Tween 20, pH 8.0) before being analysed by SDS/PAGE. The dephosphorylation procedure was similar to that described previously [10].
Immunocytochemistry and confocal microscopy
HEK-293 cells were grown overnight on glass coverslips in 24-well tissue-culture plates (Becton Dickinson, Lincoln Park, NJ, U.S.A.). The cells were transfected with pHis-hTTP (50 ng of DNA/1 ml per well) and incubated overnight as described above. After another 24 h incubation the cells were processed for immunocytochemistry as described previously [10]. The primary antiserum was the anti-MBP–hTTP serum (1:5000 dilution), and goat anti-rabbit Alexa Fluor® 488 (Molecular Probes, Eugene, OR, U.S.A.) was used at a 1:1000 dilution as the secondary antibody. The slides were examined and imaged with an LSM510 UV confocal microscope (Zeiss, Thornwood, NY, U.S.A.).
RNA ARE-binding activity assay
EMSAs (electrophoretic mobility-shift assays) were performed to evaluate the ARE-binding activity of TTP purified from HEK-293 cells following previously reported procedures [3,10]. The RNA probe (100000–200000 c.p.m./reaction) was prepared from [α-32P]UTP and T7 RNA polymerase using the RiboProbe In Vitro Transcription System (Promega Corp., Madison, WI, U.S.A.). The binding probe was made using a mouse TNFα mRNA ARE region (nucleotides 1281–1350 of GenBank® accession no. X02611) as the template. The TTP–probe complexes and free probes were detected by autoradiography or phosphorimaging (Molecular Dynamics, Sunnyvale, CA, U.S.A.).
HPLC separation, peptide mapping and Edman sequencing
WT and mutant hTTP proteins were purified from HEK-293 cells with Ni-NTA beads as described above. The eluted proteins were digested at 37 °C for 24 h with tosylphenylalanylchloromethane (‘TPCK’)-treated trypsin (Sigma, St. Louis, MO, U.S.A.) as described previously [10]. The peptides were separated using a C18 column (Amersham Biosciences) by reversed-phase HPLC with a linear gradient from buffer A [0.065% TFA (trifluoroacetic acid) in 2% acetonitrile] to 100% of buffer B (0.05% TFA in 100% acetonitrile) over 90 min and a flow rate of 0.5 ml/min. The fractions were collected in 96-well plates with Frac-950 (Amersham Biosciences) and the radioactivity of every fraction was counted using scintillation counters [Beckman or MicroBeta (PerkinElmer, Gaithersburg, MD, U.S.A.)]. The N-terminal sequences of peptides were determined by Edman sequencing with an N-terminal protein sequencer (Procise model 49X cLC Protein Sequencing System; Applied Biosystems, Foster City, CA, U.S.A.) and analysed with ABI 610A 2.1 data-analysis software and FASTA programs [12,27].
In-gel digestion and MALDI-MS (matrix-assisted laser-desorption–ionization MS)
TTP purified from HEK-293 cells by Ni-NTA affinity beads was separated by SDS/10%-(w/v)-PAGE. After the gel had been stained with Commassie Blue or silver reagent, the protein band was excised from the gel and subjected to automatic tryptic digestion using an Investigator™ Progest Protein Digestion Station (Genomic Solutions, Ann Arbor, MI, U.S.A.) and sequencing-grade modified trypsin (Promega). The tryptic peptide mixtures were freeze-dried and reconstituted in 5 μl of an acetonitrile/water (1:1, v/v) with 0.1% formic acid. The tryptic peptide solution (0.5 μl) was mixed with 0.5 μl of MALDI matrix consisting of a saturated solution of α-cyano-4-hydroxycinnamic acid in ethanol/water/formic acid (45:45:10, by vol.). MALDI analyses were performed as described in [28] using a Voyager Super DE STR (Applied Biosystems, Framingham, MA, U.S.A.) delayed-extraction time-of-flight mass spectrometer equipped with a nitrogen laser (337 nm) to desorb and ionize the samples. A close external calibration, using two points to bracket the mass range of interest, was used. MALDI analyses were performed at least twice, and the mass accuracy of the instrument with external calibration was greater than 0.01% in the reflector mode.
LC/MS/MS (liquid chromatography–tandem MS)
TTP purified from HEK-293 cells by Ni-NTA affinity beads was further purified by HPLC using an Agilent (Palo Alto, CA, U.S.A.) HP Series 1100 HPLC system. TTP (250 μl) was diluted to 600 μl with water, loaded on to a Zorbax C4 reversed-phase column (25 cm×4.6 mm; Amersham Biosciences, Piscataway, NJ, U.S.A.), and eluted at a flow rate of 1 ml/min using a linear water/acetonitrile/0.5% TFA gradient of 5% acetonitrile for 10 min, then 5–95% acetonitrile for 30 min. Fractions were collected at 1 min intervals, freeze-dried, resuspended in water, and analysed by MALDI-MS. The TTP-containing fractions were then pooled and stored at −80 °C until further analysis. The HPLC-purified TTP was further suspended in 10 mM ammonium bicarbonate buffer, pH 8, and subjected to tryptic digestion at 37 °C overnight using 2 μg of porcine trypsin (Promega). The resulting tryptic peptides were subjected to LC/MS/MS analysis using a Q-Tof (quadrupole time-of-flight) Ultima Global (Waters/Micromass, Milford, MA, U.S.A.) hybrid tandem mass spectrometer. This instrument is equipped with a nanoflow electrospray source and consists of a quadrupole mass filter and an orthogonal acceleration time-of-flight mass spectrometer. For the MS/MS experiments, a parent ion is selected with the first mass analyser and transmitted into a collision cell where fragmentation is induced by collision with argon atoms. The resulting fragment ions were detected with the second mass analyser. In this type of experiment, only ions resulting from fragmentation of the selected parent ion are observed.
Briefly, hTTP peptide mixtures (8 μl) were separated using a Hypersil C18 (‘pepmap’) column (15 cm×75 μm internal diameter; LC Packings, San Francisco, CA, U.S.A.) and an Agilent Cap1100 HPLC system consisting of a micro autosampler and binary pumps for delivering the gradients. The peptides were eluted at a flow rate of ≈300 nl/min with a linear gradient of 5–75% acetonitrile+0.1% formic acid over 35 min, and the purified peptides were subjected to tandem MS analysis. During the LC/MS experiments, either manual selection of the parent ion or automated data-dependent acquisition software was employed for the tandem-MS acquisitions. The collision energy used for these experiments was set according to the charge state and the m/z (mass-to-charge ratio) of the precursor, as determined from a charge state recognition algorithm. Data analysis was accomplished with a MassLynx data system, MaxEnt deconvolution software and ProteinLynx software supplied by the manufacturer. LC–tandem MS analyses were performed in duplicate and the mass accuracy was greater than 0.01%.
IMAC [immobilized metal ion (Fe3+) affinity chromatography]–MALDI-tandem MS
Phosphorylation sites in hTTP from HEK-293 cells were also determined using MALDI-tandem MS following enrichment of phosphopeptides using IMAC) [29]. Following trypsin digestion, TTP peptide mixtures were loaded on to the IMAC column in 100 mM acetic acid and incubated at 37 °C with slow rotation for 30 min. To remove the unbound peptides, the column was washed with 3×30 μl of water, followed by 3×30 μl of 100 mM acetic acid. The resin with bound phosphopeptides was analyzed directly by MALDI-MS and MALDI-tandem MS. MALDI-tandem MS analyses were performed with either a Micromass Q-Tof Ultima Global (Waters/Micromass, Milford, MA, U.S.A.) hybrid tandem mass spectrometer equipped with a MALDI source or a Voyager Proteomics 4700 (Applied Biosystems, Framingham, MA, U.S.A.) instrument. Both instruments are equipped with a nitrogen laser (337 nm) to desorb and ionize the samples and utilize time-of-flight mass spectrometers as the mass analyser. For MALDI-tandem MS, the solutions (0.5 μl) and/or IMAC resin were mixed with 0.5 μl MALDI matrix consisting of either 5 mg/ml α-cyano-4-hydroxycinnamic acid in acetonitrile/water (50:50, v/v) (containing 0.1% formic acid) or a 3:1 dilution of a saturated solution of α-cyano-4-hydroxycinnamic acid in acetonitrile/water (50:50, v/v) containing 0.1% formic acid. For the tandem MS experiments, a parent ion was selected manually with MS-1 (the first mass analyser) and transmitted into a collision cell, where fragmentation was induced. The resulting fragment ions were detected with the second mass analyser. In this type of experiment, only ions resulting from fragmentation of the selected parent ion are observed. All analyses were performed twice, and the mass accuracy with external calibration was better than 0.01% in the reflector mode.
MudPIT
Plasmid pHis-hTTP-Flag was used to transfect HEK-293 cells. The supernatant was prepared from the lysate of the transfected cells by centrifugation at 100000 g for 45 min, supplemented with 15% (v/v) glycerol, and stored at −70 °C as described in [24]. TTP was purified from the 100000 g supernatant using Ni-NTA affinity beads, and was eluted with 250 mM imidazole in the elution buffer as described above. The eluted protein was precipitated with 1 vol. of chloroform and 4 vol. of methanol. The protein pellet was washed with 3 vol. of methanol, suspended in 10 mM ammonium bicarbonate at pH 8.5, and dried using a Speed-Vac apparatus. Purified TTP was digested separately with three different proteases, including modified trypsin (Roche Diagnostics), subtilisin (Boehringer Mannheim), and elastase (Boehringer Mannheim) according to the triple-digest protocol [30]. The three digestion mixtures were pooled together, separated by a triphasic microcapillary column containing a 7 cm length of 5-μm-particle-diameter Polaris C18-A material (Metachem, Ventura, CA, U.S.A.), 6 cm of 5-μm-particle-size Partisphere strong cation exchanger (Whatman) and 3 cm of 5-μm-particle size hydrophilic-interaction-chromatography material (PolyLC, Columbia, MD, U.S.A.) and analysed by tandem MS as described in [31]. Tandem-MS spectra were initially analysed by a software algorithm 2TO3 to identify spectra containing a prominent 98 Da (−H3PO4) neutral loss (or 49 loss from a +2 charge state or 33 loss from a +3 charge state) from the precursor. The resulting tandem-MS spectra were searched using SEQUEST against the protein database to identify the peptide. The resulting SEQUEST output files were filtered by DTASelect for modifications. The spectra from three datasets containing the prominent 98 Da neutral loss were further analysed for unique tandem-MS fragmentation patterns of phosphopeptides as previously used [30,31] using SEQUEST-PHOS, a modified SEQUEST program to model the neutral loss (i.e., −33, −49, −98) in the theoretical fragment-ion spectrum (J. D. Venable and J. R. Yates III, unpublished work). In this manner the masses of phosphorylated peptides were identified and matched against predicted proteolytic peptides. Specific sites of phosphorylation were assigned on the basis of biological predictions.
RESULTS
Phosphorylation of TTP in intact cells
HEK-293 cells were labelled with [32P]orthophosphate following transfection with pHis-hTTP. TTP was eluted from Ni-NTA affinity beads with 100, 200 and 250 mM imidazole solutions and was detected by autoradiography (Figure 1A) and by immunoblotting with anti-MBP-hTTP antibodies (results not shown). The autoradiograph showed that the purified TTP was highly phosphorylated in HEK-293 cells and that 32P-labelled TTP was essentially the only labelled phosphoprotein recovered in the eluted fraction (Figure 1A). For MS analyses, TTP was partially purified as described above without [32P]orthophosphate labelling. Silver staining indicated that TTP was the major protein in the imidazole elution (Figure 1B) and was identified by immunoblotting using anti-MBP-hTTP serum (Figure 1C) and by MALDI-MS analysis following tryptic digestion (results not shown).
Figure 1. Phosphorylation of TTP in intact cells and purification of TTP for MS analyses.
(A) Phosphorylation of TTP in intact HEK-293 cells. Lanes 1, 2, 3 and 4 represent [32P]orthophosphate-labelled proteins eluted with 100, 200, and 250 mM imidazole buffer and the residual protein bound to the Ni-NTA beads respectively. (B, C) Purification of TTP for MS analysis. TTP was purified with Ni-NTA beads from transfected HEK-293 cells, separated by SDS/PAGE and detected by silver staining (B) or by immunoblotting with the anti-MBP-hTTP antibodies (C).
Identification of phosphopeptides of TTP by MALDI-MS
To identify phosphopeptides of TTP expressed in intact human cells, the TTP band on SDS/PAGE was excised from the gel, digested with trypsin and analysed by MALDI-MS (Figure 2). The major peptide ions corresponding to tryptic peptides of TTP are indicated (Figure 2A). In addition, these data suggested the presence of several monophosphorylated peptides (T21 and T1) as well as multiple phosphorylation sites in the T26 and T20-21 or T21-22 peptides (tryptic peptides T20-21 and T21-22 from hTTP have identical molecular mass and composition, but not sequence). The MALDI spectrum in the mass range of the ions corresponding to tryptic peptides T26 (amino acids 271–314), T21 (amino acids 196–242) and T20-21/T21-22 (amino acids 195–242/196–243) was expanded and is shown in Figure 2(B). From these data, ions corresponding in mass to the addition of one, two and three phosphate groups could be determined by the addition of 80 Da per phosphate group to the ions corresponding in mass to non-phosphorylated tryptic peptides. Generally speaking, MALDI-MS is not a very quantitative technique. Moreover, the addition of negatively charged modifications on a peptide (i.e. phosphorylation) would reduce its sensitivity under positive-ion conditions in the mass spectrometer. Because very little of the non-phosphorylated T21 and T20-21/T21-22 peptides (<8%) relative to the phosphorylated T21 and T20-21/T21-22 peptides (normalized to 100%) were observed in the MALDI spectrum, this suggests, however, that, stoichiometrically, at least one phosphorylation site in tryptic peptides T21 and T21-21/T21-22 was normally present.
Figure 2. Identification of phosphopeptides of hTTP by MALDI-MS.
(A) Linear MALDI-MS analysis of the in-gel tryptic digest of TTP purified from HEK-293 cells. The major ions corresponding in mass to tryptic peptides derived from His-hTTP are indicated. Some of the observed peptide ions correspond to phosphorylated tryptic peptides as indicated (e.g. +P on T26). The ion labelled with an asterisk (*) corresponds to a background ion. (B) Expanded MALDI spectrum in the mass range of the ions corresponding to tryptic peptides T26 (amino acids 271–314), T21 (amino acids 196–242) and T20-21/T21-22 (amino acids 195–242/196–243). These data show ions corresponding to the addition of multiple phosphate groups (+P, +2P and +3P) to the ions corresponding in mass to non-phosphorylated tryptic peptides T26, T21 and T20-21/T21-22.
Identification of phosphorylation sites in TTP by tandem MS
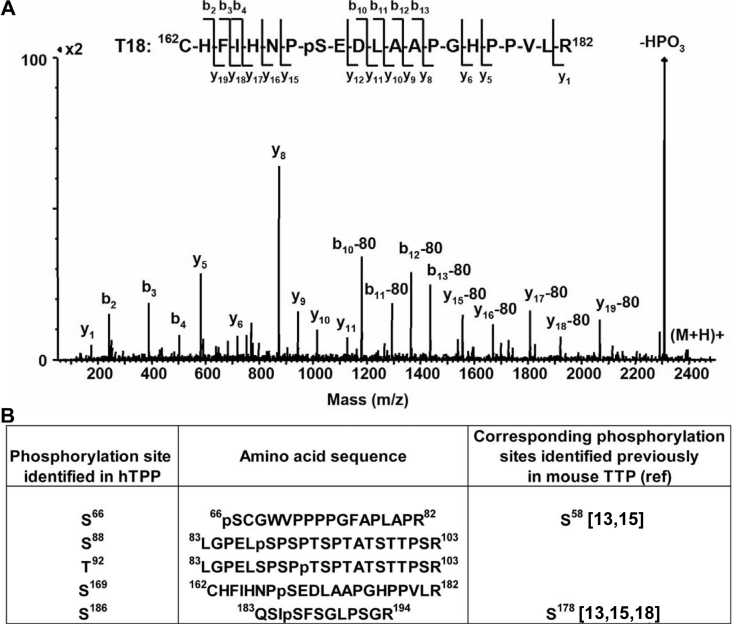
The protein purified from HEK-293 cells by Ni-NTA beads was further purified by HPLC to remove imidazole before digestion. Only a minor HPLC peak was identified by MALDI-MS as containing TTP. The HPLC-purified TTP was digested with trypsin and the tryptic peptides were analysed by LC–tandem MS. A representative deconvoluted LC–ESI (electrospray ionization)-tandem MS spectrum of a monophosphorylated tryptic peptide of TTP is shown in Figure 3(A). The MaxEnt deconvolution software can be applied to any low-mass multiply charged spectrum to resolve the multiply charged peaks on to a singly charged axis. The resulting tandem-MS spectrum obtained by transformation of all ions to the single-charge state for the triply charged ion of m/z 796.4, which corresponds in mass to monophosphorylated tryptic peptide T18 (amino acids 162–182), shows an abundant ion which corresponds in mass to the loss of HPO3 from the molecular ion. The observation of this fragment ion indicates the presence of a phosphate group on this peptide. In addition, a series of b and y ions, as well as the loss of HPO3 from the b and y ions, were observed. These structurally informative fragment ions allowed the assignment of the site of phosphorylation in this peptide to Ser169 of TTP. In addition to LC–tandem MS, MALDI-tandem MS and IMAC–MALDI-tandem MS were used to identify phosphorylation sites at Ser66 in peptide T4, Ser88 and Thr92 in peptide T5, Ser169 in peptide T18, and Ser186 in peptide T19 of TTP. Figure 3(B) summarizes the phosphorylation sites in TTP identified by these tandem MS analyses. The tandem-MS spectrum of the doubly charged ion of m/z 1082.5 (results not shown), which corresponds in mass to tryptic peptide five (L83GPELSPSPTSPTATSTTPSR103) plus one phosphate group, showed several fragment ions which correlated in mass to a phosphate group at either Ser88 or Thr92 in hTTP. To confirm the site of phosphorylation in this peptide, synthetic peptides containing a monophosphorylation site at either Ser6 (LGPELpSPSPTSPTATSTTPSR; ‘p’ indicates ‘phospho’) or at Thr10 (LGPELSPSPpTSPTATSTTPSR) were purchased and analysed by LC–tandem MS. From these data, it was determined that this peptide in TTP exists as a mixture of two phosphorylated T5 tryptic peptides, one with a phosphate located at Ser88 and one with a phosphate located at Thr92 (results not shown). In addition, the phosphorylation site at Ser88 was confirmed by Edman sequencing using a phosphopeptide purified by HPLC following tryptic digestion of in vivo-labelled hTTP from HEK-293 cells (results not shown). Tandem MS analyses of peptides from gel-based digests did not identify phosphorylation sites in peptides at the C-terminus of TTP, probably due to unfavourable sizes of the peptides for the tandem MS analyses [15].
Figure 3. Identification of phosphorylation sites at Ser66, Ser88, Thr92, Ser169 and Ser186 in hTTP by LC–tandem MS and MALDI-tandem MS.
(A) The deconvoluted LC–ESI-tandem MS spectrum of the triply charged ion of m/z 796.4, which corresponds in mass to monophosphorylated tryptic peptide T18 (amino acids 162–182), shows the loss of HPO3 from the molecular ion. In addition, a series of b and y ions, as well as the loss of HPO3 from the b and y ions, were observed, which allow the assignment of the site of phosphorylation in this peptide to Ser169 of TTP. (B) Summary of the phosphorylation sites in hTTP confirmed by LC–tandem MS and/or MALDI-tandem MS.
Characterization of phosphorylation sites in TTP by MudPIT
Concurrently, TTP was purified from transfected HEK-293 cells with Ni-NTA resin, concentrated by the chloroform/methanol precipitation method and digested with modified trypsin, subtilisin and elastase. Sequence coverage was approx. 96–97% using this triple-digestion protocol. The digested peptides were then pooled and analysed with LC–MS–MudPIT. Using this approach, a number of phosphorylated peptides were identified on the basis of the observation of loss of H3PO4 from the protonated molecule. The following sites were assigned as phosphorylated in TTP from HEK-293 cells: Ser197 (five unique phosphopeptides), Ser228 (four unique phosphopeptides), Ser276 (four unique phosphopeptides) and Ser296 (four unique phosphopeptides) (Table 1). In addition, numerous other phosphopeptides, including the previously found sites at Ser66, Ser88, Thr92, Ser169 and Ser186 (Figure 3), were assigned with lower degrees of confidence by this method [Table 1 and Supplementary Table 1 (http://www.BiochemJ.org/bj/394/bj3940285add.htm) for a complete list]. The multiple unique phosphopeptides recovered by the MudPIT analyses indicated that Ser197, Ser228, Ser276 and Ser296 were probably the major phosphorylation sites of TTP. These results are consistent with the MALDI-MS data (Figure 2), where up to three sites of phosphorylation were observed for tryptic peptide T20-21/T21-22 (amino acids 195–242/196–243) and up to two sites of phosphorylation were observed for tryptic peptide T26 (amino acids 271–314).
Table 1. Identification of phosphorylation sites in hTTP by MudPIT.
The bold letters in the peptide sequence alignment represent the phosphorylation sites of hTTP identified in the present study. A complete list of the data is provided online as Supplementary Table 1 (http://www.BiochemJ.org/bj/394/bj3940285add.htm).
Identification of the major phosphorylation sites affecting TTP's electrophoretic mobility
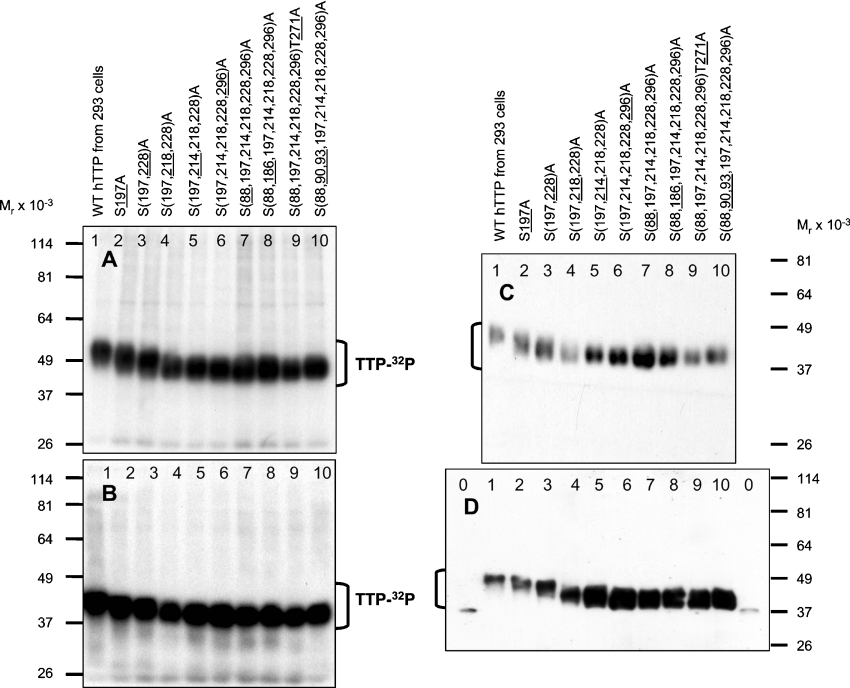
Previous studies showed that dephosphorylation of TTP from transfected HEK-293 cells markedly increased its electrophoretic mobility on SDS/PAGE [9–11,13]. We used the retardation of TTP's electrophoretic mobility as an indicator of probable stoichiometric phosphorylation in intact cells [21,22]. Under this paradigm, mutation of the specific site to alanine should abrogate the retarded mobility on SDS/polyacrylamide gels. Western blotting with anti-MBP-hTTP antibodies showed that TTP purified under denaturing conditions after expression in E. coli migrated as a single sharp band with an apparent Mr of 38000 (Figure 4A, lane 1). In contrast, WT TTP migrated as a broad band or series of bands at an Mr of approx. 40000–50000 (Figure 4A, lane 2). The S197A mutant had a major effect on the electrophoretic mobility of TTP, as shown by the fact that the gel mobility of TTP with the S197A mutation was much higher than that of TTP with non-mutated Ser197 (Figure 4A; compare lane 4 with lane 9, or lane 5 with lane 6). Triple mutations at Ser214, Ser218 and Ser228 also dramatically affected the electrophoretic mobility of the protein (Figure 4A; compare lane 2 with lane 9). Finally, the S88A, S90A, S93A and S296A mutations did not significantly affect the electrophoretic mobility of the protein (Figure 4A; compare lane 7 with lane 8, and lane 3 with lane 4).
Figure 4. Effect of mutation in TTP on its electrophoretic mobility.
Proteins in the 10000 g supernatants from transfected HEK-293 cells were used for immunoblotting (A), Ni-NTA purification (B) or dephosphorylation (C). (A) Immunoblotting. Proteins in the 10000 g supernatants from cells transfected with WT pHis-hTTP (lane 2) and seven mutants (lanes 3–9) were probed with anti-MBP-hTTP antibodies. Non-fusion TTP purified from E. coli was used as a size standard (lane 1). (B) Silver staining. WT (lane 5) and mutant TTP (lanes 1–4 and lanes 6–9) were purified from transfected HEK-293 cells with 100 mM imidazole elution and separated by SDS/10%-(w/v)-PAGE, followed by silver staining of the gel. (C) Immunoblotting. 10000 g supernatants from transfected HEK-293 cells were treated with (+) or without (−) CIAP as indicated. Mutant proteins (lane pairs 3–7) were then separated by SDS/10%-PAGE and probed with anti-MBP-hTTP antibodies. Non-fusion TTP purified from E. coli (lane 1) and WT TTP from HEK-293 cells (lane 2) was used as size standards for estimating the extent of the dephosphorylation reactions. Note that S197A/S214A/S218A/S228A/S296A is shown as S(197,214,218,228,296)A (etc.).
Silver staining of SDS/PAGE gels of TTP purified from HEK-293 cells showed that TTP with many of these same alanine mutations resulted in the transformation of the WT protein bands into almost a single band when the S197A mutation was present in every mutant (Figure 4B). Since significant amounts of most of these mutant proteins still migrated more slowly than TTP purified from E. coli (Figure 4A), we subjected the mutant proteins to ‘global’ dephosphorylation with CIAP. The results showed that CIAP treatment of all of these mutant proteins increased their migration (Figure 4C), suggesting the presence of additional phosphorylation sites in these mutant proteins in intact cells. We next investigated the effects of individual alanine mutations at Ser88, Ser90, Ser93, Ser186, Ser197, Ser214, Ser218, Ser228, Thr271 and S296 on the electrophoretic mobility of TTP proteins using pairwise comparisons (Figure 5). Protein samples were paired in a way that the samples in each pair have only one mutation difference to maximize the effect of direct comparison in each pair of the proteins. Immunoblotting showed that protein with S197A (Figure 5A), S218A (Figure 5B) or S228A (Figure 5C) mutation exhibited higher electrophoretic mobility on SDS/PAGE than that without any of the three mutations in each pair (e.g., lane 20 versus lane 21 in Figure 5A for S197A; lane 4 versus lane 5 in Figure 5B for S218A; lane 4 versus lane 5 in Figure 5C for S228A). These data suggested that Ser197, Ser218 and Ser228 are three of the major sites that significantly affect the electrophoretic mobility of TTP. These data are compatible with previous data from our laboratory showing that the S220A mutation in mTTP (corresponding to the S228A mutation in human) profoundly affected electrophoretic mobility of the protein when expressed in NIH 3T3 cells [13]. We also analysed individual mutations at Ser88, Ser90, Ser93, Ser186, Ser214, Ser296 and Thr271 with similar pairwise comparisons, and much less or no significant effects on gel mobility of the proteins were observed (results not shown).
Figure 5. Pairwise comparison of WT TTP with TTP with serine-to-alanine mutations at Ser197, Ser218 and Ser228.
Proteins in the 10000 g supernatants of transfected HEK-293 cells were separated by SDS/10%-PAGE, and TTP was detected by immunoblotting with anti-MBP-hTTP antibodies. The underlined numbers 197, 218, and 228 above the gel lanes represent the sites of the three serine residues mutated to alanine residues in addition to the mutations of TTP in the preceding lane in the pairwise comparison respectively. (A) Pairwise comparison of TTP proteins with or without the Ser197 mutation. (B) Pairwise comparison of TTP proteins with or without the Ser218 mutation. (C) Pairwise comparison of TTP proteins with or without the Ser228 mutation. Note: S197A/S214A is shown as S(197,214)A (etc.).
Evidence for TTP as a hyperphosphorylated protein by in vivo labelling of truncated TTP fragments
Previous studies have suggested that mutations at Ser52 and/or Ser178 in mTTP (corresponding to Ser60 and Ser186 in the human sequence) do not significantly affect the electrophoretic mobility of mouse TTP expressed in NIH 3T3 cells [15,20]. To try to identify additional phosphorylation sites in TTP, we transfected the same number of HEK-293 cells with the same amount of plasmids for the truncated TTP fragments and labelled the cells with the same amount of [32P]orthophosphate. Autoradiographic and immunoblotting results indicated that the different kinds of truncated TTP fragments were differentially labelled (Figure 6, left panel) and were expressed at different levels in HEK-293 cells (Figure 6, right panel). Comparison of the resulting autoradiographs and corresponding immunoblots indicated that additional sites were likely to be present in the N- and C-terminal regions of TTP (Figure 6). On the basis of the relative amount of labelling in Figure 6 (left panel) and the relative amount of TTP protein/peptides in Figure 6 (right panel), these results indicated that: (1) TTP with the S228A mutation was phosphorylated in HEK-293 cells (lanes 1 in Figures 6A and 6B respectively and thereafter); (2) the central region of TTP containing the tandem zinc-finger domain was not or was only weakly phosphorylated in HEK-293 cells (lanes 2); (3) the N-terminal 67 amino acids were highly phosphorylated, whereas residues between 67 and 173 were only slightly phosphorylated (compare lanes 3 with lanes 6); (4) certain amino acid residue(s) between 206 and 230 were likely to be significantly phosphorylated in HEK-293 cells (compare lanes 6 with lanes 7, and lanes 6 with lanes 8); and (5) there was no evidence for a significant decrease in phosphorylation when comparing residues 1–173 with a similar construct in which residues 64–82 had been deleted (compare lanes 3 with lanes 4).
Figure 6. 32P-labelling of TTP fragments in intact cells.
Human HEK-293 cells were transfected with ten plasmids encoding HA–hTTP with the S228A mutation, or with truncated fragments, as indicated. The cells were incubated with [32P]orthophosphate and HA-hTTP proteins and peptides were immunoprecipitated from HEK-293 cell extracts using anti-HA serum, then separated by SDS/PAGE. The gel was then either exposed to X-ray film (left panel) or used for Western blotting (right panel). The positions of the immunoreactive bands in the right panel are indicated on the left panel by an asterisk. Lane 1 contained full-length TTP with the S228A mutation; lanes 2, 3 and 6–10 contained the indicated fragments of WT TTP. Lane 4 contained the N-terminal fragment 1–173, with deletion of residues 64–82. Lane 5 contained the C-terminal residues 97–326 with the S228A mutation.
Evidence for TTP as a hyperphosphorylated protein by in vivo labelling of TTP mutant proteins
To further investigate TTP phosphorylation in intact cells, we labelled the same number of HEK-293 cells with the same amount of [32P]orthophosphate following transfection with the same amount of the WT or nine serine/threonine-to-alanine mutant plasmids. The WT and the mutant TTP proteins were purified from the 10000 g supernatant using Ni-NTA affinity beads (Figure 7A) or by immunoprecipitation with the anti-MBP-hTTP antibodies (Figure 7B). The autoradiographs showed that the proteins appeared to be labelled to similar extents, despite their extensive mutations (Figures 7A and 7B). Some of this can be accounted for by the increased levels of expression of some of the mutant proteins, as shown by immunoblotting in Figure 7(C). It was also possible that mutations of TTP at the major phosphorylation sites might expose other minor sites to more extensive phosphorylation than in the WT protein, because similar HPLC profiles from phosphopeptide mapping were observed among the various proteins following complete digestion with trypsin (results not shown). A parallel transfection experiment was done with the same plasmids, and TTP in the 10000 g supernatant and TTP from E. coli were detected by immunoblotting with the same antibodies (Figure 7D). In both cases (Figures 7C and 7D), the TTP protein with a single mutation at Ser197 migrated more quickly than the WT protein (lanes 1 versus lane 2). The TTP protein with double mutations at the Ser197 and Ser228 sites resulted in even faster migration than the mutated protein at Ser197 alone (lanes 2 compared with lanes 3). Furthermore, TTP protein with triple mutations at Ser197, Ser218 and Ser228 sites migrated more rapidly than the protein with the double mutations (compare lanes 3 with lanes 4). There was no significant increase in migration using TTP proteins with additional mutant sites, including Ser296 (lanes 6), Ser88 (lanes 7), Ser186 (lanes 8), Thr271 (lanes 9) or Ser90-plus-Ser93 (lanes 10). However, the WT protein, as well as all of the nine mutant TTP proteins tested in this experiment, migrated more slowly than TTP purified from E. coli (Figure 7D). These cell-labelling results supported previous findings that mutations at Ser197, Ser218 and Ser228 were three of the major sites affecting the electrophoretic mobility of TTP (Figures 4 and 5), but that even the most extensively mutated proteins were still phosphorylated (Figure 4C).
Figure 7. 32P-labelling of the WT and mutant TTP proteins in intact cells.
WT and nine mutant TTP proteins were labelled with [32P]orthophosphate in HEK-293 cells (A–C). The labelled proteins were either purified with Ni-NTA affinity beads and eluted with imidazole buffer (A, C) or immunoprecipitated with anti-MBP-hTTP antibodies (B). A parallel experiment without 32P was performed, and 10000 g supernatants from the HEK-293 cells were used for immunoblotting with anti-MBP-hTTP antibodies (D). The protein identities are shown at the top of each gel lane. Lane ‘0’ on each side of the immunoblot (D) contained the non-fusion TTP purified from E. coli. (A) and (B) are autoradiographs and panels (C) and (D) are immunoblots. The underlined numbers above the gel lanes represent the sites of serine/threonine residues mutated to alanine in addition to the mutations of TTP in the preceding lane. Note: S197A/S228A is shown as S(197,228)A (etc.).
Effect of serine mutations on ARE-binding and de-adenylation activity of TTP
The relationship between the serine/threonine mutations and the TNFα mRNA ARE-binding activity and RNA degradation was investigated using RNA-gel-shift and in vitro-de-adenylation assays of TTP purified from HEK-293 cells using Ni-NTA beads and eluted with 100 mM imidazole. Preliminary data suggest that proteins containing alanine mutations at one or more of the electrophoretic-mobility-altering phosphorylation sites can still bind ARE-containing RNA probes and promote their de-adenylation, but more quantitative comparisons of these mutants with the WT protein will require considerable further experimentation (results not shown). These results suggest that all of the mutant proteins tested possessed the essential structures for ARE-binding under these assay conditions.
Effect of serine mutations on subcellular localization of TTP
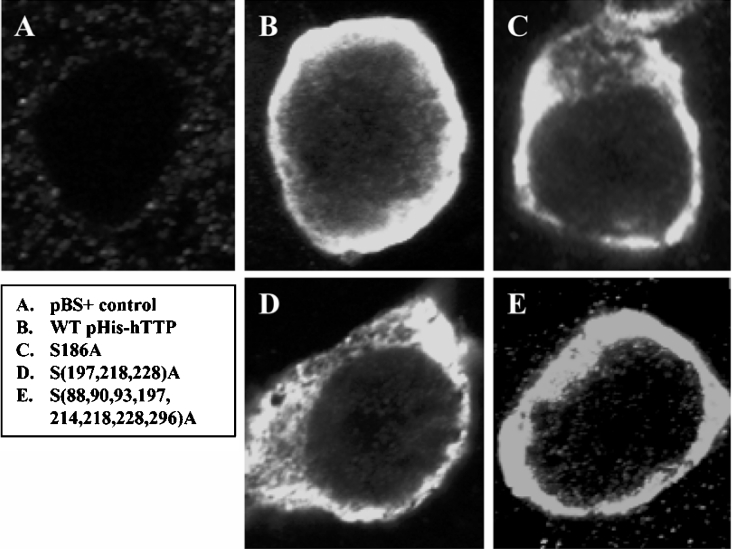
HEK-293 cells were transfected with pBS+ and pHis-hTTP, encoding the WT TTP protein and mutant TTP proteins with S186A, S197A/S218A/S228A or S88A/S90A/S93A/S186A/S214A/S218A/S228A/S296A mutations. Following immunostaining with anti-MBP-hTTP antibodies, confocal microscopy showed that most of the immunofluorescence was detected in the cytosol of the HEK-293 cells transfected with both WT and mutant plasmids and that there was no significant difference among the proteins tested (Figure 8).
Figure 8. Effect of serine mutations on the cellular localization of TTP.
HEK-293 cells were transfected with pBS+ control plasmid (A) and pHis-hTTP plasmids encoding WT TTP (B) and mutant TTP with S186A (C), S197A/S218A/S228A [shown as S(197,218,228)A] or S88A/S90A/S93A/S197A/S214A/S218A/S228A/S296A [shown as S(88,90,93,197,214,218,228,296)A] mutations. The cells were stained with the anti-MBP-hTTP serum and labelled with goat anti-rabbit–Alexa Fluor 488. Immunofluorescence was recorded by confocal microscopy.
DISCUSSION
It has been known for many years that TTP is highly phosphorylated in intact cells and that this modification can affect the electrophoretic mobility of the protein [13]. The primary objective of the present study was to identify as many phosphorylation sites as possible, with a secondary objective to identify the major phosphorylation sites that affect electrophoretic mobility, using hTTP from transfected HEK-293 cells. In the present study, LC–tandem MS and MALDI-tandem MS identified phosphorylation sites at Ser66, Ser88, Thr92, Ser169, and Ser186 in various tryptic peptides of hTTP, and direct protein sequencing confirmed a phosphorylation site at Ser88. MudPIT analysis identified phosphopeptides with numerous phosphorylation sites in hTTP; these phosphopeptides included the five sites listed above as well as other high-probability sites (those detected in multiple unique phosphopeptides) at Ser197, Ser218, Ser228, Ser276 and Ser296 and numerous lower probability sites. Finally, site-directed mutagenesis showed that alanine mutations of TTP at Ser197, Ser218 and Ser228 increased the electrophoretic mobility of the mutant proteins on SDS/PAGE with a Tris/glycine gel buffer system, an indication of stoichiometric changes in phosphorylation [21,22,32]. We suggest that Ser66, Ser88, Thr92, Ser169, Ser186, Ser197, Ser218, Ser228, Ser276 and Ser296 are the major sites of phosphorylation in TTP from transfected HEK-293 cells for the following reasons: (1) phosphopeptides containing Ser66, Ser88, Thr92, Ser169 and Ser186 observed by MudPIT were confirmed by LC–tandem MS, MALDI-tandem MS or protein sequencing; (2) Ser197, Ser218 and Ser228 strongly affected the electrophoretic mobility of TTP; (3) more than four copies of phosphopeptides containing Ser197, Ser228, Ser276 and Ser296 were identified from triply digested TTP using MudPIT; and (4) 32P-labelling studies showed that the truncated TTP peptides containing Ser218 and Ser228 were highly phosphorylated in intact cells. In addition, the MALDI-MS analysis of the in-gel tryptic digest of hTTP showed ions corresponding in mass to tryptic peptide T20-21/T21-22 (amino acids 195–242/196–243) plus the addition of one, two and three phosphate groups. Other potential sites (with fewer unique phosphopeptides observed by MudPIT) may represent lower-stoichiometry sites of phosphorylation in TTP. All of the sites reported here are conserved in TTP from various mammalian species [Table 2 and Supplementary Table 2 (http://www.BiochemJ.org/bj/394/bj3940285add.htm) for a complete sequence alignment]. In addition, previous studies have demonstrated that Ser58, Ser176, Ser178, Ser220, Thr250 and Ser264 of mTTP are phosphorylated in vivo and/or in vitro [15]; these sites correspond to Ser66, Ser184, Ser186, Ser228, Thr257 and Thr271 of hTTP. In the present study we only investigated the phosphorylation sites in hTTP expressed in HEK-293 cells. It is possible that the major phosphorylation sites, as well as the total number of phosphorylation sites, may be different in other cell lines and in normal cells and tissues under various physiological conditions.
Table 2. Alignment of amino acid sequences of mammalian TTP and the phosphorylation sites identified in the present and in other studies.
The amino acid sequences used in the alignment include human (NP_003398), mouse (NP_035886), rat (P47973), bovine (P53781), sheep (AY462109), pig (CB288050, CB286240), horse (CD536573, and CD536523), chimpanzee (CR555169) and dog (AAEX01054372.1 and AAEX01054371.1). The sequences were aligned with the PILEUP program from GCG (Genetics Computer Group). The three repeats of tetraproline residues are underlined. The bold residues within the sequence alignment are the phosphorylation sites identified in the present and other studies and the CCCH residues in the tandem zinc-finger-binding motifs. The conserved serine, threonine, and tyrosine residues, which correspond to the phosphorylation sites in hTTP identified in the present study, are indicated at the top of the sequence alignment. Those corresponding to the previously identified sites in mTTP are indicated at the bottom of the sequence alignment. A total of 40 site-directed mutants were used in the present study and were described in the Experimental section. A complete sequence alignment is provided online as Supplementary Table 2 (http://www.BiochemJ.org/bj/394/bj3940285add.htm).
We attempted to correlate specific phosphorylation sites of TTP with effects on its electrophoretic mobility. Previous studies have demonstrated that the Mr of TTP, as determined by SDS/PAGE, is dramatically increased upon phosphorylation induced by mitogens and cytokines in intact cells. For example, hTTP has a predicted Mr of 34086, and the protein purified from E. coli migrated as a single band at an Mr of 36000 [12]. However, TTP expressed in HEK-293 cells migrated as multiple bands of Mr 40000–50000. Similarly, mTTP has a calculated Mr of 33613, yet migrated as multiple bands of Mr 40000–50000 in LPS-stimulated RAW 264.7 cells [9]. This change in SDS/PAGE migration can be attributed to phosphorylation, since the larger bands were transformed into a single band of lower apparent Mr on dephosphorylation with alkaline phosphatase [9–11,14,16]. No such change was seen when TTP was treated with PNGase F (peptide:N-glycosidase F), an enzyme used to remove N-linked glucans from proteins [10]. Here we demonstrated, by site-directed mutagenesis and in vivo labelling studies, that Ser197, Ser218 and Ser228 are three of the major sites of phosphorylation that significantly affect hTTP's gel mobility. However, it appears that phosphorylation at additional sites can contribute to its gel-mobility changes, since the gel mobility of the mutant protein could be further increased following CIAP dephosphorylation.
Our results not only confirm several phosphorylation sites reported previously in mTTP, but also provide evidence for a number of new phosphorylation sites (Table 3). Previous studies suggested that Ser52, Ser178 and Ser220 of mTTP (corresponding to Ser60, Ser186 and Ser228 of hTTP) are the major phosphorylation sites of mTTP (Table 3) [15,18,20]. We did not find Ser60 as a major site in the present study, perhaps due to poor coverage of the N-terminus; however, we confirmed Ser186 and Ser228 as major sites, and in addition identified Ser66, Ser88, Thr92, Ser169, Ser197, Ser218, Ser276 and Ser296 as high-probability sites in hTTP. We found that S197A, S218A and S228A all affected TTP's electrophoretic mobility on SDS gels, whereas the S220A of mTTP (corresponding to S228A in hTTP) was previously shown to have this effect [13]. Our data showing that sites at Thr257 and Ser271 of hTTP were also phosphorylated in intact cells confirm data from cell-free assays showing that Thr250 and Ser264 of mTTP could be phosphorylated by MK2 in vitro [15]. Some probable phosphorylation sites described here have not been reported previously, including the high-probability sites at Ser88, Thr92, Ser169, Ser197, Ser218, Ser276 and Ser296 and the lower-probability sites at Ser12, Ser21, Ser41, Ser43, Ser46, Ser48, Ser90, Ser93, Thr95, Thr106, Thr111, Tyr158, Ser160, Ser188, Thr196, Ser207, Ser210, Ser217, Ser230, Ser233, Thr238, Ser252, Ser273, Ser279, Tyr284 and Ser294 (Table 3). Although previous studies showed that Ser52, Ser105 and Ser316 of mTTP were phosphorylated in intact cells [15], we did not note phosphorylation of the equivalent sites at Ser60, Ser113 or Ser323 of hTTP. These differences might be due to the differences in cell cultures and/or analytical methods used by the two groups.
Table 3. Comparison of the phosphorylation sites of TTP reported in the present and in previous studies.
Phosphorylation sites of hTTP at Ser66, Ser88, Thr92, Ser69, Ser186, Ser197, Ser218, Ser228, Ser276 and Ser296 were confirmed by more than one analysis in the present study. Phosphorylation sites of hTTP at Ser66, Ser184, Ser186, Ser228, Thr257 and Thr271 correspond to the reported phosphorylation sites of mTTP at Ser58, Ser176, Ser178, Ser220, Thr250 and Ser264. All of the identified sites are conserved in mammalian TTP orthologues (see Table 2). For brevity the one-letter code for amino acids is used in the body of the Table: S, serine; T, threonine; Y, tyrosine.
| Points of comparison | The present study (hTTP) | Previous studies (mTTP) | Reference(s) |
|---|---|---|---|
| Electrophoretic mobility-shift | S197, S218, S228 | S220 | [13] |
| Major sites | S66, S88, T92, S169, S186, S197, S218, S228, S276, S296 | S52, S178, S220 | [13,15] |
| Confirmed sites | S66, S184, S186, S228, T257, T271 | S58, S176, S178, S220, T250, S264 | [13,15,18] |
| Confirmation | T257, S271 confirmed as in vivo sites | T250, S264 reported as in vitro sites | [15] |
| Unconfirmed sites | Unconfirmed in this study | S52, S105, S316 reported as in vivo sites | [15] |
| New sites | S12, S21, S41, S43, S46, S48, S88, S90, T92, S93, T95, T106, T111, Y158, S160, S169, S188, T196, S197, S207, S210, S217, S218, S230, S233, T238, S252, S273, S276, S279, Y284, S294, S296 | Not reported previously |
One of the major tasks of future research will be to investigate the functional consequence(s) of phosphorylation. We previously showed that TTP expressed in HEK-293 cells and then dephosphorylated by CIAP is able to bind more tightly to an ARE probe than native phosphorylated TTP [11]. However, there was no apparent effect of TTP phosphorylation by p42/ERK2, p38 or JNK MAPKs [12], or by MK2 [15], on its mRNA-binding activity. TTP purified after expression in E. coli exhibited about a 2-fold greater affinity for the TNFα mRNA ARE than the protein purified from transfected HEK-293 cells [10]. In the present study, all of the TTP mutants examined exhibited ARE-binding activity using EMSAs, and several of the mutants exhibited activity in the in vitro-de-adenylation assay; however, without considerable further experimentation, we cannot exclude quantitative differences in these activities between the normal protein and the mutants. Confocal microscopy following immunostaining showed that the subcellular localization of TTP was not apparently altered by the S186A, the S197A/S218A/S228A or the S88A/S90A/S93A/S186A/S214A/S218A/S228A/S296A mutations. Additional studies will be needed to determine the precise relationship between the status of phosphorylation at individual sites and ARE-binding activity, as well as effects of TTP phosphorylation on RNA de-adenylation and degradation and its own subcellular localization and stability.
The pattern of LPS-stimulated TTP induction in RAW 264.7 macrophages consists of a gradual increase over 1–3 h, accompanied by an apparent increase in phosphorylation [9,14,16]. This pattern of TTP biosynthesis and phosphorylation occurring in parallel is different from a more typical protein phosphorylation pattern, in which previously synthesized protein is phosphorylated within a few minutes by an activated protein kinase. This unusual pattern raises interesting questions about the types of protein kinases involved, their regulation and their effects on the protein. In all, 12 of the phosphorylation sites reported here are potential sites for proline-directed protein kinases, including Ser41, Ser46, Ser88, Ser90, Ser93, Ser197, Ser218, Ser228, Thr238, Thr257, Thr271 and Ser296 (Table 2). Motif scanning (http://scansite.mit.edu) [33,34] suggests that hTTP is a potential substrate for a variety of protein kinases, including: (1) ERK1 sites at Ser41 (PWSLSP), Ser88 (PELSP), Ser218 (PSSSP) and Ser228 (PLSP) in hTTP, in agreement with the consensus sequence motif [P(1–4)X(S/T)P] as described in [35]; (2) p38 MAPK sites at Ser93 (PTSP) and Thr238 (PGTP) in hTTP, in agreement with the consensus sequence motif [PX(S/T)P] as described in [36]; (3) a Cdc2 (cell division control 2) kinase site at Thr238 (GTPLAR) in hTTP, in agreement with the consensus sequence motif [X(S/T)PX(R/K)] as described in [37]; (4) a glycogen synthase kinase-3 site at Ser218 (SPSSS) in hTTP, in agreement with the consensus sequence motif [(S/T)XXX(S/T)] as described in [38]; (5) protein kinase A sites at Ser197 (RTSP) and Thr257 (RATP) in hTTP, in agreement with the consensus sequence motif [R(1–2)X(S/T)X] as described in [37]; (6) a protein kinase Cμ site at Ser66 (LVEGRS) in hTTP, in agreement with the consensus sequence motif [LVXXXS] as described in [39]; and (7) a protein kinase C site at Ser252 (PSCR) in hTTP, in agreement with the consensus sequence motif [X(S/T)X(R/K)] as described in [37]. Of these, Ser66, Ser88, Ser90, Ser93, Ser197, Ser218, Ser228, Ser252, Thr271 and Ser296 are conserved in all mammalian orthologues of TTP identified so far, along with their corresponding surrounding sequence motifs [Table 2 and Supplementary Table 2 (http://www.BiochemJ.org/bj/394/bj3940285add.htm)]. Understanding the importance of these potential phosphorylation reactions will require further studies using a variety of techniques to confirm the types of protein kinases and their target sites in TTP.
There are two TTP-related proteins in human cells, namely ZFP36L1 (also known as TIS11b, cMG1, ERF1, BRF1 and Berg36) and ZFP36L2 (also known as TIS11d, ERF2 and BRF2) [1]. A third related protein was recently identified that seems to be expressed only in rodents as a placenta-specific protein [40]. Recent studies have shown that mice deficient in ZFP36L1 develop chorioallantoic fusion defects and embryonic lethality [41], and mice with decreased levels of an N-terminally truncated form of ZFP36L2 exhibit female infertility and disrupted early embryonic development [42]. Some of the phosphorylation sites reported here are located in relatively conserved sequence blocks among all three proteins of the TTP family, including Thr106, Tyr158, Ser184, Ser186, Ser217, Ser218, Ser273, Ser276, Tyr284, Ser294 and Ser296. The phosphorylation site reported in mTTP at Ser316 [15], corresponding to Ser323 in hTTP, is localized at the C-terminus and is conserved in all of the sequences in the TTP family. On the other hand, some of the major phosphorylation sites identified in hTTP (Ser66, Ser88, Thr92, Ser169, Ser186, Ser197 and Ser228) and mTTP (Ser52, Ser178 and Ser220) appear to be specific to TTP. It will be informative to identify all of the phosphorylation sites in the other family members, as well as the remaining sites in TTP, and to determine the roles that these phosphorylation events play in the regulation of mRNA metabolism by these proteins.
In summary, the present study reports the observation of many novel phosphorylation sites in hTTP from transfected HEK-293 cells, as well as a subset of these sites that affect its electrophoretic mobility on SDS/PAGE. On the basis of direct identification, and the preservation in orthologues from mammalian species of the relevant serine/threonine and their surrounding motifs, the high-probability in vivo sites in hTTP are Ser66, Ser88, Thr92, Ser169, Ser186, Ser197, Ser218, Ser228, Ser276 and Ser296. Of these high-probability sites, those most affecting electrophoretic mobility (i.e., those likely to be stoichiometric) are Ser197, Ser218 and Ser228. Ser197, Ser218 and Ser228 are predicted by motif-scanning programs to be potential sites for protein kinase A, glycogen synthase kinase-3 and ERK1 (both Ser218 and Ser228) respectively. All mutant proteins bound with roughly similar affinities to the same TNFα mRNA ARE probe and all were predominantly localized in the cytosol of the transfected human cells. The effects of these phosphorylation events on TTP function and/or stability require further investigation. Our identification of multiple phosphorylation sites in TTP from mammalian cells should provide the molecular basis for further studies on the regulation of TTP in its control of pro-inflammatory cytokine biosynthesis.
Online data
Acknowledgments
This work was supported in part by NIH (National Institutes of Health) Intramural Research Training Award TAXP003505 and by the Intramural Research Program of the NIH, NIEHS (National Institute of Environmental Health Sciences). A preliminary report of this study was presented at the Human Proteomics Organization Third World Congress in Beijing, People's Republic of China, on 25–27 October 2004. We thank Dr Wi S. Lai for some plasmids and transfection extracts, Ms Rui Lin for technical assistance, Ms Jane S. Tuttle for advice on mammalian cell culture and Dr Robert M. Petrovich and Dr Mohamed Trebak for helpful comments on the manuscript.
References
- 1.Blackshear P. J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 2.Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F., Blackshear P. J. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 3.Carballo E., Lai W. S., Blackshear P. J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 4.Carballo E., Lai W. S., Blackshear P. J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 5.Phillips K., Kedersha N., Shen L., Blackshear P. J., Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor α, cyclooxygenase 2, and inflammatory arthritis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuBois R. N., McLane M. W., Ryder K., Lau L. F., Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J. Biol. Chem. 1990;265:19185–19191. [PubMed] [Google Scholar]
- 7.Lai W. S., Stumpo D. J., Blackshear P. J. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 8.Varnum B. C., Lim R. W., Sukhatme V. P., Herschman H. R. Nucleotide sequence of a cDNA encoding TIS11, a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4:119–120. [PubMed] [Google Scholar]
- 9.Cao H., Tuttle J. S., Blackshear P. J. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J. Biol. Chem. 2004;279:21489–21499. doi: 10.1074/jbc.M400900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao H. Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications. Biochemistry. 2004;43:13724–13738. doi: 10.1021/bi049014y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carballo E., Cao H., Lai W. S., Kennington E. A., Campbell D., Blackshear P. J. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao H., Dzineku F., Blackshear P. J. Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases. Arch. Biochem. Biophys. 2003;412:106–120. doi: 10.1016/s0003-9861(03)00012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor G. A., Thompson M. J., Lai W. S., Blackshear P. J. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J. Biol. Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- 14.Zhu W., Brauchle M. A., Di Padova F., Gram H., New L., Ono K., Downey J. S., Han J. Gene suppression by tristetraprolin and release by the p38 pathway. Am. J. Physiol. 2001;281:499–508. doi: 10.1152/ajplung.2001.281.2.L499. [DOI] [PubMed] [Google Scholar]
- 15.Chrestensen C. A., Schroeder M. J., Shabanowitz J., Hunt D. F., Pelo J. W., Worthington M. T., Sturgill T. W. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 16.Mahtani K. R., Brook M., Dean J. L., Sully G., Saklatvala J., Clark A. R. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ming X. F., Stoecklin G., Lu M., Looser R., Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor G. A., Thompson M. J., Lai W. S., Blackshear P. J. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol. Endocrinol. 1996;10:140–146. doi: 10.1210/mend.10.2.8825554. [DOI] [PubMed] [Google Scholar]
- 20.Johnson B. A., Stehn J. R., Yaffe M. B., Blackwell T. K. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 21.Rangel-Aldao R., Kupiec J. W., Rosen O. M. Resolution of the phosphorylated and dephosphorylated cAMP-binding proteins of bovine cardiac muscle by affinity labeling and two-dimensional electrophoresis. J. Biol. Chem. 1979;254:2499–508. [PubMed] [Google Scholar]
- 22.Rodriguez P., Bhogal M. S., Colyer J. Stoichiometric phosphorylation of cardiac ryanodine receptor on serine 2809 by calmodulin-dependent kinase II and protein kinase A. J. Biol. Chem. 2003;278:38593–38600. doi: 10.1074/jbc.C301180200. [DOI] [PubMed] [Google Scholar]
- 23.Lai W. S., Kennington E. A., Blackshear P. J. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 24.Lai W. S., Kennington E. A., Blackshear P. J. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai W. S., Blackshear P. J. Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin- mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly(A) tail. J. Biol. Chem. 2001;276:23144–23154. doi: 10.1074/jbc.M100680200. [DOI] [PubMed] [Google Scholar]
- 27.Wooldridge A. A., MacDonald J. A., Erdodi F., Ma C., Borman M. A., Hartshorne D. J., Haystead T. A. Smooth muscle phosphatase is regulated in vivo by exclusion of phosphorylation of threonine 696 of MYPT1 by phosphorylation of serine 695 in response to cyclic nucleotides. J. Biol. Chem. 2004;279:34496–34504. doi: 10.1074/jbc.M405957200. [DOI] [PubMed] [Google Scholar]
- 28.Deterding L. J., Prasad R., Mullen G. P., Wilson S. H., Tomer K. B. Mapping of the 5′-2-deoxyribose-5-phosphate lyase active site in DNA polymerase β by mass spectrometry. J. Biol. Chem. 2000;275:10463–10471. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- 29.Zhou W., Merrick B. A., Khaledi M. G., Tomer K. B. Detection and sequencing of phosphopeptides affinity bound to immobilized metal ion beads by matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2000;11:273–282. doi: 10.1016/s1044-0305(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 30.MacCoss M. J., McDonald W. H., Saraf A., Sadygov R., Clark J. M., Tasto J. J., Gould K. L., Wolters D., Washburn M., Weiss A., et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venable J. D., Yates J. R., 3rd Impact of ion trap tandem mass spectra variability on the identification of peptides. Anal. Chem. 2004;76:2928–2937. doi: 10.1021/ac0348219. [DOI] [PubMed] [Google Scholar]
- 32.Hofmann F., Beavo J. A., Bechtel P. J., Krebs E. G. Comparison of adenosine 3′:5′-monophosphate-dependent protein kinases from rabbit skeletal and bovine heart muscle. J. Biol. Chem. 1975;250:7795–7801. [PubMed] [Google Scholar]
- 33.Obenauer J. C., Cantley L. C., Yaffe M. B. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids. Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yaffe M. B., Leparc G. G., Lai J., Obata T., Volinia S., Cantley L. C. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 35.Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 36.Hawkins J., Zheng S., Frantz B., LoGrasso P. p38 MAPK substrate specificity differs greatly for protein and peptide substrates. Arch. Biochem. Biophys. 2000;382:310–313. doi: 10.1006/abbi.2000.2005. [DOI] [PubMed] [Google Scholar]
- 37.Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 38.Frame S., Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikawa K., Toker A., Johannes F.-J., Songyang Z., Cantley L. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 40.Blackshear P. J., Phillips R. S., Ghosh S., Ramos S. V., Richfield E. K., Lai W. S. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the tristetraprolin family of CCCH tandem zinc finger proteins. Biol. Reprod. 2005;73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 41.Stumpo D. J., Byrd N. A., Phillips R. S., Ghosh S., Maronpot R. R., Castranio T., Meyers E. N., Mishina Y., Blackshear P. J. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Mol. Cell. Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos S. B., Stumpo D. J., Kennington E. A., Phillips R. S., Bock C. B., Ribeiro-Neto F., Blackshear P. J. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.