Abstract
Shewanella oneidensis contains four genes that encode proteins that have high sequence identity with yeast OYE (Old Yellow Enzyme, an NADPH oxidoreductase), the well-studied archetype of the OYE protein family. The present paper describes the first comparative study of OYEs that are present in a single bacterial species, performed to gain insight into their biochemical properties and physiological importance. The four proteins [named SYE1–SYE4 (Shewanella Yellow Enzyme 1–4)] were expressed as glutathione S-transferase fusion proteins in Escherichia coli. The yield of SYE2, however, was too low for further characterization, even after expression attempts in S. oneidensis. The SYE1, SYE3 and SYE4 proteins were found to have characteristics similar to those of other OYE family members. They were identified as flavoproteins that catalyse the reduction of different α,β-unsaturated carbonyl compounds and form charge transfer complexes with a range of phenolic compounds. Whereas the properties of SYE1 and SYE3 were very similar, those of SYE4 were clearly different in terms of ligand binding, catalytic efficiency and substrate specificity. Also, the activity of SYE4 was found to be NADPH-dependent, whereas SYE1 and SYE3 had a preference for NADH. It has been suggested that yeast OYE protects the actin cytoskeleton from oxidative stress. There are indications that bacterial OYEs are also involved in the oxidative stress response, but their exact role is unclear. Induction studies in S. oneidensis revealed that yeast and bacterial OYEs may share a common physiological role, i.e. the protection of cellular components against oxidative damage. As only SYE4 was induced under oxidative stress conditions, however, a functional divergence between bacterial OYEs is likely to exist.
Keywords: acrolein, flavoprotein, NADPH oxidoreductase, Old Yellow Enzyme (OYE), oxidative stress response, Shewanella oneidensis
Abbreviations: CHP, cumene hydroperoxide; CT, charge transfer; GST, glutathione S-transferase; GTN, glycerol trinitrate; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; (Le)OPR, (Lycopersicon esculentum) 12-oxophytodienoate reductase; MR, morphinone reductase; NEM, N-ethylmaleimide; NG, nitroglycerin; OYE, Old Yellow Enzyme; PETN, pentaerythritol tetranitrate; SELDI, surface-enhanced laser-desorption–ionization; SYE, Shewanella Yellow Enzyme; t-BOOH, t-butylhydroperoxide; TNT, 2,4,6,-trinitrotoluene; VIS, visible
INTRODUCTION
The OYE (Old Yellow Enzyme, an NADPH oxidoreductase) family constitutes a subfamily of the Class I flavin-dependent α,β-barrel oxidoreductases [1]. OYE from Saccharomyces carlsbergensis was the first flavin-containing enzyme to be identified [2] and is the archetype of this rapidly growing family, of which new members are now primarily being discovered through genome sequencing projects. Some members of the OYE family are related quite distantly to OYE, including the oestrogen-binding protein of Candida albicans [3], the bile-acid-inducible flavoenzymes BaiH and BaiC [4] and trimethylamine dehydrogenase [5]. More closely related enzymes have been characterized in other yeasts, Gram-positive and Gram-negative bacteria, monocotyledonous and dicotyledonous plants, Caenorhabditis elegans and Trypanosoma cruzi [6–16]. Bacterial homologues that have been characterized so far include PETN (pentaerythritol tetranitrate) reductase [17], GTN (glycerol trinitrate) reductase [18], MR (morphinone reductase) [19], 2-cyclohexenone reductase [20], the xenobiotic reductases A and B from Pseudomonas sp. [21] and NEM (N-ethylmaleimide) reductase [22]. More recently, the first OYE homologue from a Gram-positive bacterium, YqjM from Bacillus subtilis, was studied [23].
With only one accessible active site containing the non-covalently bound cofactor FMN, the enzyme acts through a Bi Bi Ping Pong mechanism in which the coenzyme NAD(P)H and the substrate use the same binding site. The physiological reductant of OYE is assumed to be NADPH. Only GTN reductase and MR show a strong preference for NADH [18,19]. Oxidation by molecular oxygen is not considered to be a physiological reaction because of the low rate and the production of H2O2 and O2−. In general, quinones and α,β-unsaturated aldehydes and ketones can act as oxidants in which the olefinic bond is reduced [6]. OYE can also catalyse the slow aromatization of cyclic enones, such as cyclohexenone [24].
Oxidized members of the OYE family have the ability to form CT (charge transfer) complexes with a variety of phenolic ligands, which are held parallel to the si face of the flavin by π overlap. This results in long-wavelength optical transitions of between 500 and 800 nm and in reciprocal perturbations of the oxidized flavin absorption spectrum (350–500 nm region). The broad long-wavelength absorbance arises from a CT interaction between the phenolate (electron donor) and oxidized FMN (electron acceptor) [25], the result of the transfer of the flavin–phenolate complex from its ground state to an excited state upon irradiation by light of the appropriate energy. The energy of the transition and the Hammett para-constant of the p-substituted phenolic compounds have been shown to be positively correlated [25,26], indicating that the phenolate is the electron donor.
The physiological substrate of OYE is still unknown despite the fact that several members of the OYE family have been studied extensively at the molecular level and that crystallographic structures of six members of the OYE family are available, namely trimethylamine dehydrogenase [27], Sacch. carlsbergensis OYE [28], PETN reductase [29], OPR (12-oxophytodienoate reductase) [30], MR [31] and YqjM [32]. It has been suggested that yeast OYE is involved in the oxidative stress response, as OYE was found to protect the actin cytoskeleton from oxidative stress [33]. Furthermore, it was shown previously that the expression of YqjM is up-regulated substantially in B. subtilis upon the induction of oxidative stress conditions [23].
Shewanella oneidensis is an important model organism in bio-remediation studies because it is characterized by unique respiratory capabilities, such as the possibility to reduce heavy metals [34]. The coding sequences of four OYE homologues, which we designated SYE1 (Shewanella Yellow Enzyme 1) to SYE4, can be identified in this organism by BLAST analyses. In the present paper, we report the first detailed and comparative study of the different OYE homologues that are present in a single bacterial species. The differences in biochemical characteristics between the SYEs are discussed and are compared with those of other OYE family members. Induction studies were performed to gain an insight into the physiological role of the different proteins. The results are discussed with regard to the probable divergence in physiological function of bacterial OYE family members.
EXPERIMENTAL
Materials and reagents
Q Sepharose FF, High Load 16/60 Superdex™ 200 prep grade, glutathione–Sepharose 4 FF and nitrocellulose membrane were purchased from Amersham Biosciences; PVDF membrane was from Applied Biosystems; maleic acid, thrombin, progesterone, 1,4-androstadiene-3,17-dione, CHP (cumene hydroperoxide), t-BOOH (t-butylhydroperoxide), paraquat, FAD and FMN were from Sigma–Aldrich; NG (nitroglycerin), NADH and NADPH were from Merck KGaA; 2-cyclohexen-1-one, acrolein and all p-substituted phenols were from Acros Organics; and NEM, picric acid and fumaric acid were from Fluka. Oligonucleotide sequencing and PCR primers were synthesized at Sigma–Genosys.
Database searches and sequence alignments
Database searches were performed with the BLAST server from the NCBI (National Center for Biotechnology Information) using the blastp and tblastn options, including all non-redundant GenBank® CDS (coding sequence) translations and the RefSeq Proteins, PDB, SwissProt, PIR (Protein Information Resource) and PRF (Protein Research Foundation) databases. The NCBI bl2seq tool [35] was used to obtain the percentage identity and percentage similarity between pairwise-aligned sequences. Amino acid sequences were aligned with the ClustalW program [36a] using default parameters.
Bacterial strains and plasmids
S. oneidensis MR-1 [BCCM™ (Belgian Coordinated Collections of Microorganisms)/LMG (Laboratorium voor Microbiologie, Universiteit Gent); strain number LMG 19005] was used for the preparation of genomic DNA, for SYE induction analysis and for overexpression studies; Escherichia coli MC1061 [BCCM™/LMBP (Laboratory of Molecular Biology Plasmid) collection; accession number LMBP 472] and XL1-Blue (Stratagene) were used for cloning purposes; BL21(DE3) (Novagen), Top10 (Invitrogen) and LMG194 (Invitrogen) strains were used for protein expression. S. oneidensis and E. coli were routinely grown in LB (Luria–Bertani) medium under constant shaking at 28 °C or 37 °C respectively, unless indicated otherwise. The pGEX-4T-2 vector was purchased from Amersham Biosciences, the pBAD-TOPO vector was from Invitrogen and pACYCDuet-1 was from Novagen.
Cloning and production of the SYE proteins
The different sye genes were amplified by PCR and cloned in the pGEX-4T-2 vector. All four pGEX constructs, upon induction of the tac promoter with IPTG (isopropyl β-D-thiogalactoside), result in the production of a fusion protein between GST (glutathione S-transferase) and one of the SYE proteins. The correct sequence of the obtained constructs was confirmed by direct sequencing of the plasmid DNA, using Dye Terminator Cycle Sequencing (Applied Biosystems). Two additional constructs were made to allow co-expression of the sye1 and sye2 genes. First, a genomic fragment containing both sye1 and sye2 was cloned downstream of the arabinose promoter of the pBAD-TOPO expression vector (construct pBADSYE1-2). Secondly, the sye1 and sye2 genes were each cloned after one of the two T7 promoters in the pACYCDuet-1 vector (pACYCSYE1-2 construct).
Each of the four pGEX constructs was transformed into E. coli BL21(DE3). Optimal expression was reached at 28 °C. Induction was performed with 0.5 mM IPTG at an A600 of 0.6, after which the cells were incubated for another 15 h. After centrifugation at 3313 g for 15 min at 4 °C, the bacterial pellet was washed with 50 mM Tris/HCl, pH 8.0, resuspended in the same buffer and kept frozen at −80 °C until further use. pGEX-SYE2 was also transformed into S. oneidensis by electroporation, and expression was analysed under similar conditions. pBADSYE1-2 was transformed into E. coli strains Top10 and LMG194. Expression was assayed at 37 °C using L-arabinose concentrations ranging from 0.00002 to 0.2%. pACYCSYE1-2 was transformed in E. coli BL21(DE3), grown at 28 °C and induced with 0.5 mM IPTG.
Purification of the recombinant GST-tagged SYE proteins
Frozen cells were thawed on ice, ruptured by sonication using a Branson Sonifier, and centrifuged at 20800 g for 30 min at 4 °C. Appropriate volumes of supernatant were loaded on to a 10 ml Q Sepharose FF column equilibrated with 50 mM Tris/HCl, pH 8.0. Using a step gradient of the same buffer containing 1 M NaCl, the different GST fusion proteins eluted between 200 and 300 mM NaCl. The fractions containing GST–SYE1, GST–SYE3 or GST–SYE4 were pooled, dialysed against PBS and loaded on to an 8 ml glutathione–Sepharose 4 FF column. The GST–SYE proteins were eluted in 50 mM Tris/HCl, pH 8.0, supplemented with 10 mM reduced glutathione and then loaded on to a High Load 16/60 Superdex™ 200 prep grade gel filtration column to remove the final contaminants.
Removal of the GST-tag and purification of the untagged SYE proteins
After the two initial purification steps (see above), thrombin was added in a 1 unit/1 mg ratio in order to enzymatically cleave the GST-tag from the different fusion proteins. An incubation time of 8 h at room temperature (22 °C) was sufficient to cleave GST–SYE1 and GST–SYE4 completely. The untagged SYE1 and SYE4 proteins were separated from GST using an 8 ml glutathione–Sepharose 4 FF column. The flow-through was concentrated and applied on to a High Load 16/60 Superdex™ 200 prep grade gel filtration column to obtain pure protein. GST–SYE3 was digested only partially, even after an increased incubation time of 24 h. SYE3 was therefore separated from GST and the remaining GST–SYE3 using glutathione–Sepharose affinity chromatography. As we also observed complex formation between SYE3 and GST–SYE3, a final gel filtration step was needed to dissociate the complexes using PBS containing 1 M NaCl, 10% (v/v) glycerol and 5 mM dithiothreitol.
Analytical methods
SDS/PAGE (15% gels) was performed using the method of Laemmli [36b]. Before loading, samples were heated for 5 min at 95 °C in loading buffer containing 12% (w/v) 2-mercaptoethanol as a reducing agent. Proteins were visualized by staining the gels with Coomassie Brilliant Blue R-250 (Fluka). Final purity was determined by silver-staining. For N-terminal sequencing, proteins were blotted on to a PVDF membrane in protein transfer buffer [3.03 g of Tris/HCl (pH 8.0), 14.4 g of glycine and 200 ml of methanol in 1 litre] during 3 h at 50 V using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). The identity of the different recombinant SYE proteins was confirmed by N-terminal amino acid sequence analysis and peptide fingerprint mapping. N-terminal sequencing was performed on a 476A pulsed liquid sequencer (Applied Biosystems) connected to an online phenylthiohydantoin-amino-acid analyser. Protein fragments were generated with trypsin and their mass was determined on an electro-spray Q-TOF mass spectrometer (Micromass, Wythenshawe, UK). The peptides were identified using the Mascot search engine (http://www.matrixscience.com). Protein concentration was determined spectrophotometrically using molar absorption coefficients ϵ458=11500 M−1·cm−1, ϵ459=10900 M−1·cm−1 and ϵ462=11800 M−1·cm−1 for SYE1, SYE3 and SYE4 respectively. For each protein, the absorption coefficient of enzyme-bound FMN was determined by comparing the flavin absorbance of the pure protein with the absorbance of the free flavin released upon denaturation in 2% SDS.
Analysis of the flavin prosthetic group
The identity of the flavin prosthetic group was determined by TLC, as described by Marshall et al. [37]. Briefly, purified SYE proteins were boiled in PBS for 5 min and the denatured protein was removed by centrifugation at 20800 g for 10 min. Supernatant samples were loaded on to 5 cm×10 cm Silica gel 60 F254 plates (Merck KGaA) next to standards of FAD and FMN. The plates were developed in 2% (w/v) Na2HPO4, air-dried and examined under UV light for areas of quenched fluorescence. The corresponding RF values were determined.
Ligand-binding titrations
The six different GST–SYE fusion proteins were titrated up to saturation with increasing concentrations of a variety of p-substituted phenolic ligands in 0.1 M Tris/HCl, pH 8.0. Absorption spectra were recorded between 300 and 800 nm using an Uvikon 943 double-beam UV/VIS (visible) spectrophotometer (Kontron Instruments). The spectral changes at the absorbance maximum of the resulting long-wavelength transitions were plotted against the ligand concentration. The dissociation constants for the protein–ligand interaction were determined using the software SigmaPlot version 8.0 (Jandel Scientific). The same software was used to perform linear regression analysis on the plots between the Hammett para-constants of the ligands and the wave numbers of the absorbance maxima of the CT transitions.
Photoreduction and steady-state kinetic analyses
Photoreduction experiments were performed under anaerobic conditions, by irradiation with a slide projector and using 5 mM EDTA as electron donor. Absorption spectra were recorded at regular time intervals.
All kinetic parameters were determined under strict anaerobic conditions and all assays were performed in triplicate. Reaction mixtures consisting of 200 mM sodium phosphate buffer, pH 7.3, 60–100 μM NAD(P)H and various concentrations of substrate were prepared within a glove box (Coy Laboratories, Grass Lake, MI, U.S.A.) and transferred to sealable quartz cuvettes (Hellma). The concentrations of NADH and NADPH were determined spectrophotometrically using a molar absorption coefficient of 6220 M−1·cm−1 at 340 nm. Enzyme solutions were prepared in sealed flasks and made anaerobic by bubbling with N2 gas. Reactions were initiated by the addition of enzyme to the reaction mixtures using a Hamilton needle. Enzyme concentrations were varied depending on the observed reaction rate with each substrate. SYE activity was assayed by following the oxidation of NAD(P)H using an Uvikon 943 spectrophotometer. Apparent steady-state kinetic constants were determined by least-squares fitting procedures to the standard Michaelis–Menten equation using SigmaPlot software.
Western blot analyses
Purified recombinant SYE1 protein was used to generate polyclonal antiserum in rabbits (Eurogentec). Cross-reactivity of the SYE1 antiserum (dilution of 1:1000) was evaluated with the purified SYE1, SYE3 and SYE4 proteins and with crude extracts of S. oneidensis and the different overexpression cultures. For induction experiments, full grown S. oneidensis cultures were diluted (1:100) in LB medium and grown for 3 h at 28 °C. A selection of substrates and peroxides was added to the cultures, and samples were taken at regular intervals between 0 and 4 h after induction. For each sample, equal concentrations of crude extracts were separated by SDS/PAGE (15% gels) and transferred on to a nitrocellulose membrane by electroblotting. Detection was performed using a goat anti-rabbit peroxidase conjugate (Sigma, dilution 1:25000) in combination with a chemiluminescent system (Roche Molecular Biochemicals). Pure SYE4 (20 ng) was also loaded on to the gels as an internal molecular-mass marker and as a standard for the estimation of changes in protein expression.
SELDI (surface-enhanced laser-desorption–ionization) ProteinChip analysis
PS20 ProteinChip arrays (Ciphergen Biosystems, Fremont, CA, U.S.A.) were used to verify the molecular mass of the different recombinant SYE proteins, as well as to confirm the cross-reactivity of the anti-SYE1 antibody with the different SYE proteins. Before use, the serum was loaded on to a Protein G affinity column, and the purified antibodies were dialysed against PBS. Following this, 10 ng of the serum was added to each spot of the PS20 chip and incubated for 1 h at room temperature in a humidity chamber, and the remaining pre-activated sites were blocked for 30 min with 0.5 M Tris/HCl, pH 8.0, according to the manufacturer's instructions. The antibody solution was removed and the whole array was washed twice with PBS containing 0.1% (v/v) Triton X-100 (wash buffer) for 15 min and three times with PBS for 5 min, on a rotary shaker. Wash buffer (5 μl) containing 0.1 M NaCl (binding buffer) or 5 μl of binding buffer spiked with 50 ng of pure SYE1, SYE3 or SYE4 was added to the spots containing covalently bound antibody, and incubation was continued for 4 h at room temperature in a humidity chamber. The array was then washed three times with binding buffer and once in PBS. After a final rinse with water, two additions of 0.5 μl of the energy-absorbing molecule sinapinic acid were added. The spots were air-dried and mass analysis was performed using the SELDI ProteinChip system (PBS II series, Ciphergen Biosystems).
RESULTS AND DISCUSSION
The OYE homologues of S. oneidensis
We were able to identify four OYE homologues during a BLAST search of the S. oneidensis genome using the amino acid sequence of Sacch. carlsbergensis OYE isoform 1 (OYE1; NCBI accession number Q02899) as a probe. For our convenience, we designated the four proteins SYE1–SYE4, consistent with the nomenclature used for Kluyveromyces lactis (KYE1) [7] and Hansenula polymorpha (HYE1, HYE2 and HYE3) [14]. SYE1 (NCBI accession number AAN55488) and SYE4 (NCBI accession number AAN56390) are both annotated as FMN-binding oxidoreductases [34] showing 38% identity (57% similarity) and 36% identity (55% similarity) with OYE1 respectively. SYE2 (NCBI accession number AAN55487) and SYE3 (NCBI accession number AAN57126) are both described as putative NEM reductases that share 32% identity (51% similarity) and 36% identity (54% similarity) with OYE1 respectively. The sye2 gene is located 8 nt downstream of sye1, whereas sye3 and sye4 are each positioned in different genomic regions. Since a gene encoding a transcriptional regulator from the LysR family can be found upstream from sye1, it is possible that sye1 and sye2 belong to an operon under the control of the lysR promoter. Analysis of the DNA sequences flanking the sye3 and sye4 genes revealed the presence of open reading frames with deduced amino acid sequences similar to transcriptional regulators of the LysR and TetR family respectively. These regulators usually control the expression of proteins that are involved in the stress response.
Database comparisons and sequence alignments
The deduced amino acid sequences of the four SYEs were compared with sequence information available in the nr database using the NCBI blastp and tblastn programs. The four proteins show high sequence similarities to a wide variety of members belonging to the OYE family of α,β-barrel flavoprotein oxidoreductases. The highest percentages of identity and similarity were found with, from SYE1 to SYE4, Vibrio parahaemolyticus NEM reductase 2 (NCBI accession number BAC62116; 79% identity and 88% similarity), Photobacterium profundum SS9 NEM reductase (NCBI accession number YP_133310; 71% identity and 85% similarity), Vibrio cholerae NEM reductase (NCBI accession number NP_233377; 72% identity and 82% similarity) and P. profundum SS9 NADH-dependent flavin oxidoreductase (YP_133033; 78% identity and 87% similarity).
The amino acid sequence alignment in Figure 1 reveals that the active-site residues and the functionally important residues involved in the binding of the FMN group, which are strictly conserved in the known OYE homologues, are also conserved in the four SYEs, meaning that all requirements are fulfilled for a functional FMN-binding site. His187, His/Asn190 and Tyr192 {numbering according to LeOPR [Lycopersicon esculentum (tomato) OPR], see Figure 1} are the active-site residues, of which Asn190 is conserved in yeast OYE but often substituted by histidine in other members of the OYE family, and also in SYE2. However, a comparison of the crystal structures from LeOPR and Sacch. carlsbergensis OYE revealed that the Nδ1 of His190 in OPR occupies the same position in the active site as the corresponding asparagine Nδ1 in OYE [30]. This suggests that the electronic effects on the substrate carbonyl oxygen are similar.
Figure 1. Sequence alignment of members of the OYE family.
So_SYE1, S. oneidensis SYE1 (NCBI accession number AAN55488); So_SYE2, S. oneidensis SYE2 (NCBI accession number AAN55487); So_SYE3, S. oneidensis SYE3 (NCBI accession number AAN57126); So_SYE4, S. oneidensis SYE4 (NCBI accession number AAN56390); Le_OPR (NCBI accession number CAB43506); Sc_OYE, Saccharomyces cerevisiae OYE isoform 1 (NCBI accession number A39495); and Bs_YqjM, B. subtilis OYE homologue YqjM (NCBI accession number BAA12619); *, active site residues in OPR; +, #, amino acid residues from OPR that form side-chain and main-chain hydrogen bonds respectively with FMN. In the consensus sequence, bold capital letters are residues that are conserved in all sequences, capital letters are residues that are conserved in all but one sequence and lower case letters are residues that are conserved in all but two sequences; c (underlined) represents conserved substitutions. Data for structural elements shown for OPR are taken from [30]. The α-helices and β-strands that form the eight-stranded α,β barrel are indicated as α1–α8 and β1–β8 respectively. The extra-barrel helices are indicated as αA and αB, and extra-barrel strands are shown as βA–βF.
In general, the residues that make side-chain interactions with FMN are strictly conserved within the OYE family, whereas the residues that contact FMN by their main chain are not, although their Cα positions are structurally well conserved, which is essential to establish the FMN hydrogen-bonding network. This general rule also applies to the SYEs in which residues Thr37, Gln110 and Arg239 (LeOPR numbering) that contact FMN by their side chain are strictly conserved. Differences can be seen in the residues Pro35, Ala68, Gly308, Gly309, Tyr310, Gly330, Arg331 and Leu332 that make interactions through the peptide backbone with FMN. Pro35, Gly308 and Gly330 are conserved in the four sequences. Ala68 is replaced by glycine in a conservative substitution in SYE1 and SYE2, and Tyr310 and Arg331 in SYE2 are changed into leucine and threonine respectively. More variation is observed in the SYE residues corresponding to Gly309 (Asn326 in OYE) and Leu332 (Phe350 in OYE), but both are also converted into non-conserved residues within most other OYE family members.
Functional overexpression and purification of the recombinant SYEs
The four sye genes were inserted in the pGEX-4T-2 vector downstream of the GST coding sequence and were transformed into E. coli BL21(DE3). Cultures were grown initially at 37 °C, but each fusion protein was mainly present either as colourless apoprotein or in inclusion bodies. The incubation temperature was therefore lowered, as well as the IPTG concentration: growth at 28 °C and addition of 0.5 mM IPTG yielded large amounts of soluble GST–SYE1, GST–SYE3 and GST–SYE4. Despite testing several overexpression conditions, GST–SYE2 remained predominantly present in inclusion bodies. This was still the case for the expression experiments in S. oneidensis. Why the expression level of soluble SYE2 was low is unclear. In OYE1, it has been shown that mutation of the active-site residue Asn194 into histidine results in a low yield of purifiable protein, possibly by interference of the bulkier histidine residue with binding of the FMN group [38]. All SYE proteins have an asparagine residue at the corresponding position, except for SYE2, which contains a histidine residue. Also, Arg331, which makes two interactions with the FMN group, is replaced by a threonine residue in SYE2. Another explanation for the low yield might be that, as yeast OYE is known to form functional homo- and hetero-dimers and the sye2 gene is located only 8 nt downstream of the sye1 gene, SYE2 and SYE1 form a heterodimer in vivo. Coexpression of the two genes with the pBADSYE1-2 and pACYCSYE1-2 constructs was tested in different E. coli strains using increasing concentrations of the inducer L-arabinose (0.00002–0.2%) or 0.5 mM IPTG respectively. No visible induction bands were detected on Coomassie-Blue-stained polyacrylamide gels, although Western blot analysis showed very low levels of expression in the cultures induced with 0.002–0.2% L-arabinose.
The GST–SYE1, GST–SYE3 and GST–SYE4 fusion proteins were purified in three subsequent chromatographic steps for further analysis. The yield of all three proteins was at least 50 mg/l. The GST moiety was cleaved off enzymatically, and the processed proteins were purified further by affinity chromatography and gel filtration. The elution volume of the different SYE proteins on a Superdex™ 200 column corresponded to a molecular mass of approx. 80 kDa, meaning that the native proteins form dimers in solution.
UV/VIS spectral analysis and identification of the flavin prosthetic group
Spectroscopic characterization of the pure native proteins revealed absorbance spectra that were typical of a flavin-containing enzyme with maxima at 374 nm and 458 nm, 371 nm and 459 nm, and 381 nm and 462 nm for SYE1, SYE3 and SYE4 respectively. The spectral properties of SYE4 resemble those of LeOPR [12] and OYE1 [39], while the spectra of SYE1 and SYE3 are more comparable with the spectrum of the YqjM protein [23]. Boiling the different enzymes resulted in a colourless pellet of denatured protein and a bright yellow supernatant, which suggests that the cofactor is bound non-covalently, as in the other known OYE homologues. The SYE4 protein, however, had to be boiled twice as long to remove the prosthetic group completely, suggesting a much tighter binding compared with SYE1 and SYE3. For all three proteins, the released cofactor was identified as FMN by TLC analysis, as it co-migrated with the FMN standard, resulting in comparable RF values.
Binding of p-substituted phenols and the formation of CT complexes
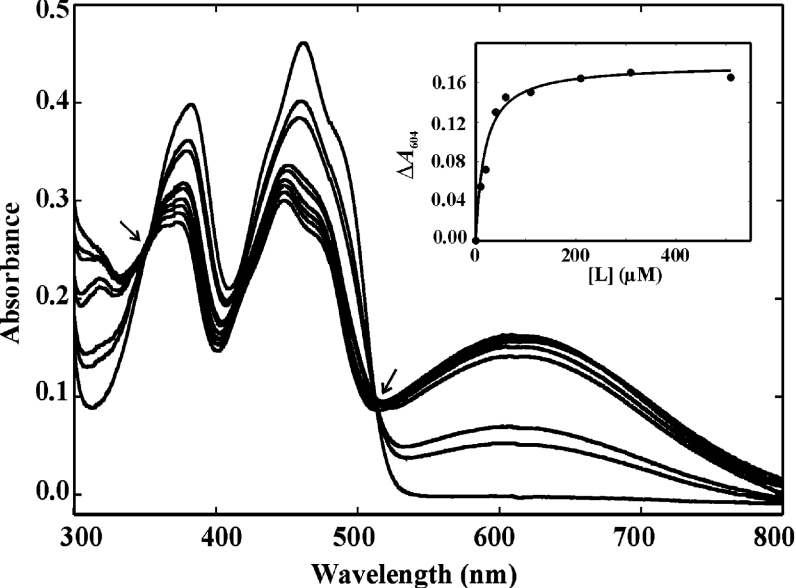
A distinguishing feature of OYE family members is their ability to form spectroscopically distinct CT complexes upon titration with compounds that have a phenolic hydroxy group [12,23,25]. All six GST–SYEs were therefore titrated with a series of nine phenolic compounds to compare the characteristics of the anticipated long-wavelength transitions. As an example, the typical spectrum obtained by titration of SYE4 with p-chlorophenol is shown in Figure 2. In most cases, an increase in ligand concentration resulted in spectral changes that are typical for the formation of a CT complex, i.e. the appearance of new long-wavelength optical transitions in the absorbance spectrum. A comparison of the absorption maximum for each of the formed CT complexes and their dissociation constants (Kd) is shown in Table 1. In general, the Kd values for the untagged proteins were lower than for the tagged GST–SYE proteins, whereas their long-wavelength absorbance maxima were rather similar. This was also the case for the GST-tagged and untagged LeOPR protein [12]. Although the dissociation constants of the SYE proteins were higher than those reported for OYE1, they were generally lower than those of LeOPR [12] and YqjM [23]. This difference was most significant for the Kd values of the GST–SYE4 CT complexes, with ligands having a pKa of 9.34 or more. The correlation between the pKa of the phenol and the dissociation constant of the CT complex was most apparent in the SYE4 and GST–SYE4 proteins: the higher the pKa of the ligand, the higher the wavelength maximum of the complex (the less energy required for the CT transition) and the higher the dissociation constant. As was also observed for the LeOPR CT complexes [12], there was a hypsochromic shift of the absorbance maximum of each of the SYE CT complexes compared with those of OYE1 [25]. This indicates that the redox potential of the enzyme-bound FMN is more positive and/or the redox potential of the phenolic ligand is more negative in the OYE1 CT complexes compared with the L. esculentum and bacterial counterparts [12,23].
Figure 2. Spectral changes in SYE4 upon titration with p-chlorophenol.
Absorbance spectra were recorded between 300 and 800 nm in 0.1 M Tris/HCl, pH 8, at 22 °C. SYE4 (40 μM) was titrated with 0, 10, 20, 40, 60, 110, 210, 310 and 510 μM of p-chlorophenol. The long-wavelength transition that appeared with increasing ligand concentration has an absorbance maximum that is centred at 604 nm. The isosbestic points at 354 and 512 nm are indicated by arrows. The inset shows the SYE4–ligand complex formation (ΔA604) as a function of ligand concentration ([L]). The data points were fitted to the hyperbolic equation ΔA=ΔAmax/1+(Kd/[L]) from which the dissociation constant Kd was determined using SigmaPlot.
Table 1. Comparison of binding of p-substituted phenols by the different GST–SYE proteins.
The SYE proteins were titrated with the indicated phenolic ligands for which the phenolic pKa of the free ligand (according to [39]) is given. The dissociation constants (μM) Kd±S.D. derived from the titration data are given, and the absorbance maximum of each of the long-wavelength CT complexes, CTmax (nm), is shown in parentheses. *, no detection of long-wavelength optical transitions.
| Ligand | pKa | SYE1 | GST–SYE1 | SYE3 | GST–SYE3 | SYE4 | GST–SYE4 |
|---|---|---|---|---|---|---|---|
| Pentafluorophenol | 5.40 | 19.23±4.23 (550) | * | 32.11±12.22 (560) | * | 1.99±0.26 (550) | 0.95±0.21 (560) |
| p-Nitrophenol | 7.05 | 2.38±0.47 (550) | 73.59±28.70 (550) | 9.71±3.75 (555) | 76.76±16.55 (550) | 2.09±0.17 (550) | 5.14±1.41 (550) |
| p-Hydroxybenzaldehyde | 7.62 | 4.90±1.04 (570) | 29.96±6.85 (560) | 11.64±3.45 (570) | 28.01±7.66 (570) | 4.20±0.34 (555) | 2.30±0.49 (550) |
| p-Cyanophenol | 7.90 | 4.43±0.62 (570) | 26.21±5.68 (578) | 3.61±1.13 (560) | 44.96±18.46 (580) | 6.72±0.96 (550) | 10.11±2.62 (560) |
| p-Hydroxyacetophenone | 8.05 | 4.00±0.10 (571) | 35.84±8.73 (574) | 7.10±2.59 (575) | 28.20±6.51 (580) | 3.51±0.81 (570) | 8.10±1.92 (580) |
| p-Chlorophenol | 9.34 | * | * | * | * | 17.25±3.53 (604) | 44.98±14.00 (622) |
| p-Fluorophenol | 9.43 | * | * | * | * | 51.15±24.57 (629) | 14.35±1.71 (620) |
| p-Methoxyphenol | 10.20 | * | * | * | * | 94.26±56.83 (650) | 92.19±28.94 (686) |
| p-Cresol | 10.30 | * | * | * | * | 29.47±12.98 (642) | 68.46±14.10 (645) |
In contrast with GST–SYE4, the GST–SYE1 and GST–SYE3 proteins failed to give long-wavelength charge transitions at pH 8.0 when titrated with ligands that have a pKa of the free phenol equal to, or larger than, 9.34. It is known from the crystal structure of p-hydroxybenzaldehyde-bound OYE1 [28] that side chains of the active-site residues His191 and Asn194 (OYE1 numbering) form stabilizing hydrogen bonds with the p-hydroxy group, while the hydroxy group of Tyr375 forms a hydrogen bond with the aldehyde moiety. These stabilizing hydrogen bonds result in a decrease of the pKa of the bound phenol with several pH units [25,28], which is evident in the fact that the H191N and H191N/N194H mutant enzymes do not form CT transitions at pH 7.0 when the pKa of the free phenol is 8.0 or greater [38]. Also, all phenols tested were found to bind much more weakly in both mutants than in wild-type OYE1. This means that small changes in the ligand-binding environment can have important repercussions on ligand-binding properties. It is therefore reasonable to assume that small differences are present in the active-site structure between SYE1, SYE3 and SYE4. Although Tyr375 is conserved in all plant and yeast OYEs, except in Schizosaccharomyces pombe, it is replaced by a phenylalanine in SYE1 and SYE3, and by the non-aromatic residue isoleucine in SYE4. Also, SYE4 resembles OYE1 in having an additional aromatic residue (Phe279) in its active site.
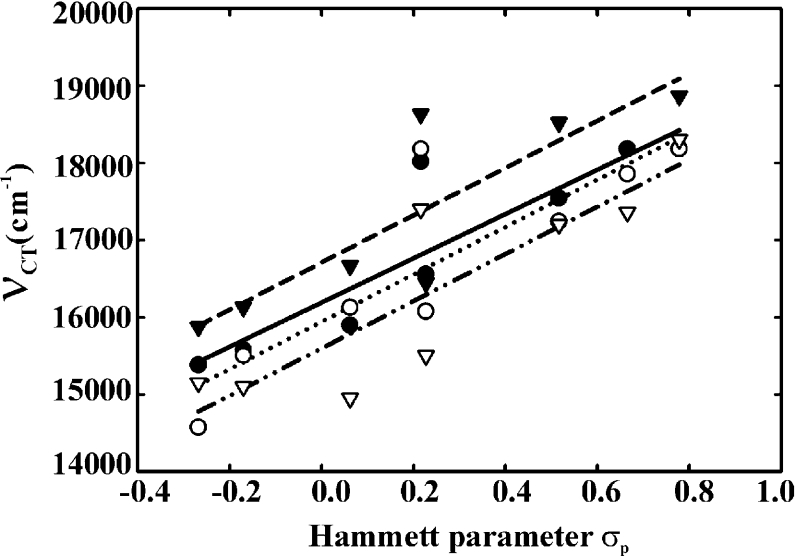
For all six SYE proteins, a linear correlation could be demonstrated between the long-wavelength transition energies for the ligand–SYE complexes and the Hammett para-constants (σp, linear free-energy parameter) of the different ligands. The reciprocal of each available wavelength maximum (νCT, wave number) was plotted against the σp of the different ligands [40,41]. The fits of the linear regressions performed with SigmaPlot all had positive slopes (correlation coefficients of 0.903 and 0.863 for SYE4 and GST–SYE4 respectively), as observed for a correlation between an acceptor compound and a series of donor compounds. As shown in Figure 3, the correlation coefficients of the GST–SYE4 CT complexes were very similar to those of the OYE1 [25] and LeOPR CT [12] complexes.
Figure 3. Linear correlation between the Hammett p-constant (σp) of p-phenols and the long-wavelength transition energies (νCT) of the GST–SYE4, LeOPR and OYE1 CT complexes.
The Hammett p-constant values of the following compounds were reported in [40]: p-methoxyphenol (−0.268), p-cresol (−0.170), p-fluorophenol (0.062), p-hydroxybenzaldehyde (0.216), p-chlorophenol (0.227), p-hydroxyacetophenone (0.516), p-cyanophenol (0.665) and p-nitrophenol (0.778). The wave numbers of the absorbance maxima of the CT complexes are represented as ● (SYE4), ○ (GST–SYE4), ▼ (LeOPR) [12] and ▽ (OYE1) [25]. The fits of the linear regressions performed with SigmaPlot version 8.0 (Jandel Scientific) are as follows: broken line, SYE4; dotted line, GST–SYE4; solid line, LeOPR; and broken/dotted line, OYE1.
Photoreduction
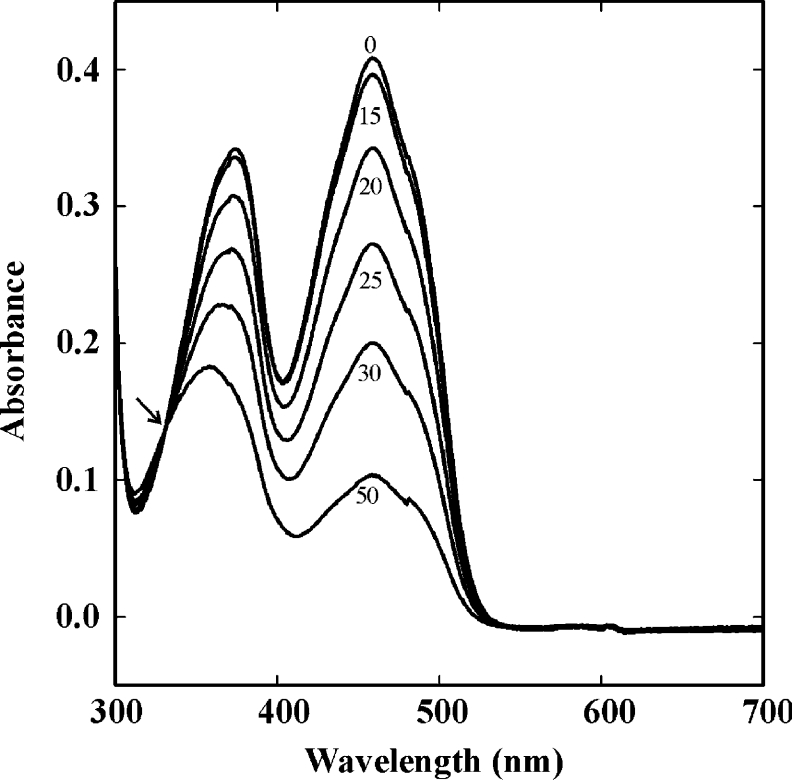
Photoreduction of the untagged SYE1, SYE3 and SYE4 proteins was performed by illumination with VIS light and by including EDTA as electron donor in the buffer used. The observed absorption spectra were similar for all proteins; the experiment performed with SYE1 is represented in Figure 4. The reduction of the flavin cofactor (absorbance maximum at 458, 459 and 462 nm for SYE1, SYE3 and SYE4 respectively) was not accompanied by a concomitant increase at 380 nm, meaning that there was no production of the red anionic flavin semiquinone. Also, long-wavelength spectral changes characteristic of the blue neutral semiquinone were not observed. Without any measurable semiquinone formation, the flavin of the SYE proteins thus appears to be reduced directly to the dihydroflavin form. This is also the case for other bacterial OYEs [23,42,43], which is in contrast with yeast OYE and LeOPR, in which the photoreduction is a two-step process [44,12]. An isosbestic point (at 332 nm for SYE1; see Figure 4) was observed upon reduction, which again indicates that only two absorbing species, i.e. oxidized and two-electron-reduced protein, were present. Re-oxidation of the proteins following exposure to air was slow; the intermediate spectra during re-oxidation passed through the same isosbestic points as during photoreduction.
Figure 4. Photoreduction of SYE1.
The effect of white-light irradiation on SYE1 in the presence of 5 mM EDTA was determined under anaerobic conditions. Spectra were recorded between 300 and 700 nm at the indicated times (0, 15, 20, 25, 30 and 50 min). The arrow indicates the isosbestic point.
Cofactor preference
All six tagged and untagged SYE proteins were found to catalyse the oxidation of both NADH and NADPH in the presence of molecular oxygen (Table 2). As with the other OYE family members, the NAD(P)H oxidation rates of the SYE proteins accelerated significantly in the presence of an α,β-unsaturated compound (NEM) as alternative electron acceptor [12,23]. Each protein showed a distinct preference for either of the two reductants. The oxidation rates for SYE4 and GST–SYE4 were highest with NADPH, while tagged and untagged SYE1 preferred NADH. The oxidation rates of the SYE3 proteins were slightly higher with NADPH in the presence of molecular oxygen as the sole electron acceptor, yet the NADPH oxidation rates in the presence of NEM were very low compared with the NADH oxidation rates, indicating a preference for NADH over NADPH. Only two family members with a preference for NADH are known so far, i.e. GTN reductase [18] and MR [19].
Table 2. Comparison of the rate of NAD(P)H oxidation by the different GST–SYE proteins.
Assays were performed in 200 mM sodium phosphate buffer, pH 7.3, in the presence of molecular oxygen either with (+) or without (−) 5 mM NEM. The rate of oxidation is expressed as the number of mol of NAD(P)H oxidized/min per mol of enzyme flavin.
| Assay condition | ||||
|---|---|---|---|---|
| −NEM | +NEM | |||
| Protein | NADH | NADPH | NADH | NADPH |
| SYE1 | 11.0 | 1.5 | 2159 | <1 |
| GST–SYE1 | 13.2 | 5.2 | 984 | <1 |
| SYE3 | <1 | 2.6 | 2190 | 28 |
| GST–SYE3 | <1 | 5.4 | 5089 | 61 |
| SYE4 | 3.8 | 14.1 | 61 | 572 |
| GST–SYE4 | 8.6 | 23.1 | 106 | 1565 |
Catalytic activity
The specific activities of the different tagged and untagged SYE proteins were compared using a range of potential substrates, i.e. NEM, 2-cyclohexen-1-one, acrolein, fumaric acid, maleic acid, NG, picric acid, progesterone (inhibitor) and 1,4-androstadiene-3,17-dione. Experiments were carried out using the reductant of preference for each protein (NADPH in the case of SYE4 and GST–SYE4, and NADH in the case of SYE1, GST–SYE1, SYE3 and GST–SYE3). The values of the apparent kinetic constants kcat and Km are summarized in Table 3. The tagged and untagged forms of the three SYE proteins were able to accept the same selection of oxidants. None of the SYE proteins, however, accepted fumaric acid, maleic acid, picric acid, progesterone or 1,4-androstadiene-3,17-dione as substrate. The catalytic efficiencies of the tagged forms were generally lower compared with the corresponding native SYE proteins. A comparable difference in catalytic activity was also seen between the GST-tagged and untagged form of YqjM from B. subtilis [45]. The lower catalytic efficiency of GST–YqjM was mainly reflected in the kcat values, which were, depending on the substrate tested, 4–10-fold lower than for YqjM. This is in contrast with the SYE proteins, where the difference, for the greatest part, was displayed by Km values generally 1.6–10-fold higher in the tagged forms than in the untagged forms. The catalytic efficiencies of all SYE proteins were highest for NEM and were significantly lower for 2-cyclohexen-1-one and acrolein, yet the apparent kinetic constants for NEM and 2-cyclohexen-1-one were of the same order of magnitude as those determined for LeOPR [12] and YqjM [23,45]. We have shown for the first time that bacterial OYE family members can reduce acrolein, a lipid peroxidation product that is formed endogenously in eukaryotic cells. Among all α,β-unsaturated aldehydes, acrolein is by far the strongest electrophile and is therefore highly toxic to the cell.
Table 3. Steady-state kinetic properties of the different GST–SYE proteins.
Apparent kinetic constants were derived from steady-state kinetic analyses using the software SigmaPlot. Enzymatic activity was assayed spectrophotometrically at 340 nm by determining the rate of NAD(P)H oxidation, i.e. the number of mol of NAD(P)H oxidized/s per mol of enzyme flavin, in the presence of various substrate concentrations. Assays were performed under strict anaerobic conditions in 1 ml of 200 mM sodium phosphate buffer, pH 7.3, and 60–100 μM NAD(P)H as reductant. NADPH was used in the experiments with GST–SYE4, while NADH was used in the assays with the tagged and untagged SYE1 and SYE3 proteins. The enzyme concentration was varied according to the observed reaction rate. No activity was detected with fumaric acid, maleic acid, picric acid, progesterone and 1,4-androstadiene-3,17-dione. NR, no reaction. Data represent means±S.D. for three separate experiments.
| Substrate | Protein | kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) |
|---|---|---|---|---|
| NEM | SYE1 | 36.00±0.82 | 51.1±5.2 | 0.70 |
| GST–SYE1 | 16.39±1.51 | 10.9±2.7 | 1.50 | |
| SYE3 | 36.50±0.96 | 27.5±3.8 | 1.33 | |
| GST–SYE3 | 84.81±4.11 | 204±34 | 0.42 | |
| SYE4 | 9.54±0.63 | 1.6±0.5 | 5.96 | |
| GST–SYE4 | 26.08±1.25 | 16.7±3.9 | 1.56 | |
| 2-Cyclohexen-1-one | SYE1 | 13.19±0.94 | 544±138 | 0.0242 |
| GST–SYE1 | 12.82±1.64 | 1837±517 | 0.0070 | |
| SYE3 | 9.97±1.13 | 578±113 | 0.0172 | |
| GST–SYE3 | 7.72±0.89 | 2954±746 | 0.0026 | |
| SYE4 | 4.75±0.42 | 54.7±16.1 | 0.0868 | |
| GST–SYE4 | 7.13±0.38 | 42.7±7.4 | 0.1670 | |
| Acrolein | SYE1 | 22.31±3.24 | 588±139 | 0.0380 |
| GST–SYE1 | 15.18±1.33 | 1231±215 | 0.0123 | |
| SYE3 | 15.16±0.99 | 593±157 | 0.0256 | |
| GST–SYE3 | 2.35±0.13 | 317±87 | 0.0074 | |
| SYE4 | 11.66±0.44 | 109±20 | 0.1070 | |
| GST–SYE4 | 31.56±3.64 | 578±96 | 0.0546 | |
| NG | SYE1 | NR | NR | NR |
| GST–SYE1 | NR | NR | NR | |
| SYE3 | NR | NR | NR | |
| GST–SYE3 | NR | NR | NR | |
| SYE4 | 6.84±0.13 | 26.4±5.6 | 0.26 | |
| GST–SYE4 | 9.85±0.88 | 42.2±10.9 | 0.23 |
The catalytic properties of SYE4 were found to be different from those of the other SYE proteins. SYE4 and GST–SYE4 have a slightly different substrate profile as they also showed activity towards NG. Also, SYE4 and GST–SYE4 were, in general, found to be characterized by higher catalytic efficiencies than the other SYE proteins, which exhibit more comparable kinetic values. We note that an orange coloration was observed with SYE4 after prolonged incubation (16 h) with NADPH and picric acid. This indicates the formation of the hydride-Meisenheimer complex and suggests that SYE4 is capable of hydride addition to the aromatic ring of picric acid. This characteristic has so far only been seen with PETN reductase [46], xenobiotic reductase B [47] and E. coli NEM reductase [48], which all form TNT (2,4,6,-trinitrotoluene) hydride addition products.
Immunodetection of SYE expression under oxidative stress conditions
As it has been shown that YqjM from B. subtilis is rapidly induced under conditions of oxidative stress [23], we analysed whether the SYE proteins are expressed under similar conditions. The polyclonal antiserum raised against pure recombinant SYE1 was first assayed for cross-reactivity with the other SYE proteins. Western blots of the pure proteins, crude extracts of the different overexpression cultures (including samples with insoluble GST–SYE2) and the appropriate controls were probed with the SYE1 antibodies. The results indicated that the antibodies could be used for the detection of all four SYE proteins. This was confirmed by ProteinChip analysis in which the pure SYE1, SYE3 and SYE4 proteins were retained by the antibodies immobilized on a pre-activated PS20 ProteinChip array. Also, analysis using the SELDI ProteinChip System showed that the molecular masses of the captured SYE proteins corresponded to the calculated masses.
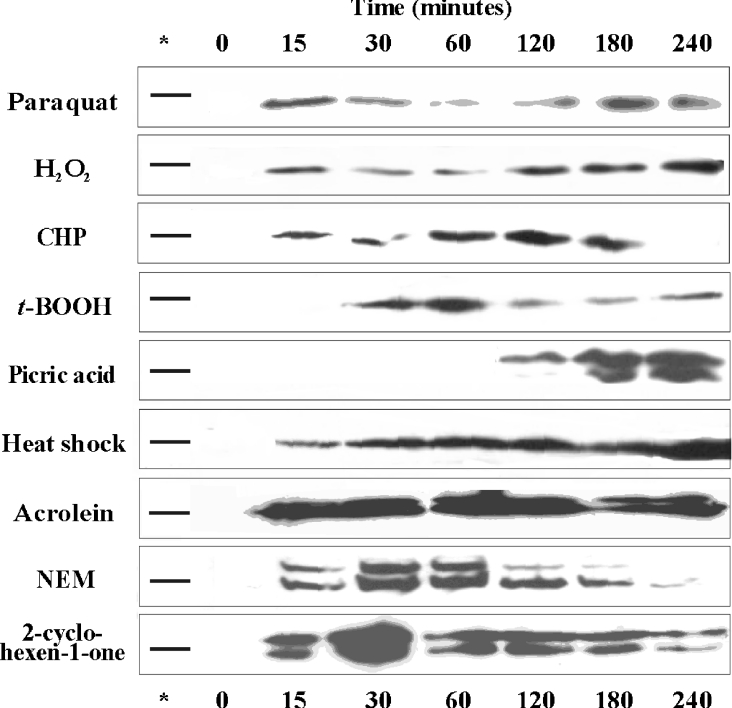
In contrast with B. subtilis YqjM [23], no basal expression of any of the SYE proteins was observed under normal growth conditions, nor in cultures entering the stationary phase. However, a labelled band near the 37 kDa marker protein appeared rapidly in the presence of compounds generating oxidative stress conditions (Figure 5). The up-regulated protein was reasoned to be SYE4 (calculated mass 39 kDa) as the other SYE proteins have higher molecular masses of 39.6 (SYE1), 40 (SYE2) and 41 (SYE3) kDa. The identity of SYE4 was confirmed by peptide mass fingerprinting following partial purification of the labelled protein. The induction pattern was similar using 2.5 mM paraquat (induction of O2− production), 0.2 mM H2O2, 0.05 mM CHP and 0.55 mM t-BOOH; although the expression level was quite low, induction occurred already within the 15-min (paraquat, H2O2 and CHP) or 30-min (t-BOOH) time interval. SYE4 was also rapidly induced after heat-shock treatment (temperature shift from 28 to 42 °C), yet the expression level was approx. 2-fold higher than under the conditions mentioned above. This result is in agreement with a recent DNA microarray study by Gao et al. [49], in which the genomic response of S. oneidensis to a temperature upshift from 30° to 42 °C was evaluated over a period of 25 min: only the sye4 gene was up-regulated, whereas the other sye genes were not. Induction with 0.4 mM picric acid, a TNT analogue that enhances O2− and H2O2 production [50], occurred only after 2 h. At this point, the colour of the culture immediately shifted from yellow to orange. As seen in Figure 5, the induction signal had a double-band pattern. The band of higher molecular mass was first thought to be one of the other SYE proteins. However, the two labelled proteins were partially purified and were both identified as non-truncated SYE4 by N-terminal sequence analysis and peptide mapping. As a similar double-band pattern was sometimes observed on Coomassie-Blue-stained SDS/PAGE gels during purification of recombinant SYE4, we assume that the upper band represents FMN-bound protein and the lower band represents FMN-free apoprotein. Although the FMN group is bound non-covalently to proteins of the OYE family, it appears to be bound very tightly to SYE4, as it was not fully removed after boiling of the samples and during gel electrophoresis. This is consistent with the observations made during sample preparation for TLC analysis. In contrast to B. subtilis YqjM, SYE4 expression was not induced with 1.5 mM NG, a compound also shown to increase the production of reactive oxygen species [51]. As higher concentrations of peroxides were needed for the up-regulation of SYE4 compared with YqjM, it is possible that higher concentrations of NG are also required. In conclusion, none of the SYE proteins is expressed under normal growth conditions. Only SYE4 was detected in vivo, i.e. under conditions of oxidative stress. This indicates an involvement in the oxidative stress response for SYE4, but not for the other three SYE proteins.
Figure 5. Induction of S. oneidensis SYE4 under different growth conditions.
The expression level of SYE4 was assayed with Western blot analysis after heat-shock treatment (temperature upshift from 28° to 42 °C) or after exposure to 2.5 mM paraquat, 0.2 mM H2O2, 0.05 mM CHP, 0.55 mM t-BOOH, 0.4 mM picric acid, 85 μM acrolein, 25 μM NEM or 10 mM 2-cyclohexen-1-one. Cells were harvested at the indicated time intervals and crude extracts were probed with antisera against SYE1. The position of the 37 kDa marker protein is indicated in the lane marked with an asterisk.
Induction of SYE expression in the presence of substrates
As the SYE proteins are able to use acrolein, NEM and 2-cyclohexen-1-one as substrate, expression was also assayed in the presence of these compounds (85 μM, 25 μM and 10 mM respectively). All three substrates resulted in a very rapid and strong induction of the SYE4 protein (Figure 5). A similar double-band pattern was observed, as seen with picric acid. This direct substrate induction yielded expression levels that were at least 2–3-fold higher than under the oxidative-stress-generating conditions mentioned above, even with NEM concentrations as low as 25 μM. This can be explained by the fact that all three highly electrophilic compounds can cause oxidative stress in two different ways: either directly by the induction of disulphide bonds within and between proteins, or indirectly by depletion of reduced glutathione. Given the observation that yeast OYE can repair disulphide bonds in oxidized actin [33], it is possible that bacterial OYEs are also recruited as a cellular defence mechanism against oxidative injury. A study is currently under way in our laboratory to define the exact role of SYE4.
Conclusions
Several members of the OYE family have been characterized structurally and biochemically, but the family consists of a still-increasing number of, as yet, uncharacterized enzymes. Considerable differences between the enzymatic activities towards a diverse range of substrates have been found, which indicates either that the true physiological substrate has not yet been found or that the metabolic function varies between organisms. The bacterium S. oneidensis is exceptional in encoding four proteins (SYE1–SYE4) with high sequence similarity to OYE, yet the mutual identities between these enzymes (38–52%) are significantly lower compared with the identities beween the different OYE isoforms in yeasts and plants (69–91%). This provides a unique opportunity for detailed and comparative analysis, as in the present study, in order to gain a better insight into the possible functional role of bacterial OYEs.
An important finding from our work was the biochemical divergence between the SYE proteins. Although SYE1 and SYE3 were found to have comparable characteristics, SYE4 has different properties regarding cofactor preference, catalytic efficiency and substrate specificity. This suggests a differentiation in physiological function as well, which was shown by the results of an expression analysis. Only SYE4 was induced in oxidative stress conditions and in the presence of the substrates acrolein, NEM and 2-cyclohexen-1-one. Despite the fact that SYE1 and SYE3 also showed catalytic activity with these compounds, they were not induced under the conditions tested. Although the sye1, sye2 and sye3 genes may therefore seem redundant, these genes are conserved in, for example, Photobacterium and Vibrio species (NCBI databases), implying that they are bona fide genes. The SYE proteins thus appear to have a different physiological function and/or they may be induced by another factor. The latter is plausible because sye4 is preceded by a tetR repressor gene, while sye1, sye2 and sye3 are preceded by a lysR regulator sequence. Our results have important implications for the search for the true physiological role of OYE family members, as there may not be a specific conserved functional role for these enzymes. At least one possible function can be assigned to bacterial OYE family members, i.e. an involvement in oxidative stress response, but our results suggest strongly that multiple physiological roles may exist for the different OYE homologues within a single organism. Hence gaining a profound insight into the true functions of this class of proteins has become even more challenging.
Acknowledgments
This work was funded by the STWW (Strategic Technologies for Welfare and Well-being) programme of the IWT (Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie)-Flanders (project 174IWT20) and by the Concerted Research Actions programmes GOA (Geconcerteerde Onderzoeksactie) 12050198 and GOA B/05036. The authors thank Isabel Vandenberghe for performing the protein sequence analyses and Elke Lecocq for peptide mass fingerprint mapping.
References
- 1.Raine A. R., Scrutton N. S., Mathews F. S. On the evolution of alternate core packing in eightfold β/α-barrels. Protein Sci. 1994;3:1889–1892. [Google Scholar]
- 2.Warburg O., Christian W. Über das gelbe oxydationsferment. Biochem. Z. 1933;263:228–229. [Google Scholar]
- 3.Madani N. D., Malloy P. J., Rodriguez-Pombo P., Krishnan A. V., Feldman D. Candida albicans estrogen-binding protein gene encodes an oxidoreductase that is inhibited by estradiol. Proc. Natl. Acad. Sci. U.S.A. 1994;91:922–926. doi: 10.1073/pnas.91.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franklund C. V., Baron S. F., Hylemon P. B. Characterization of the baiH gene encoding a bile acid-inducible NADH:flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J. Bacteriol. 1993;175:3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd G., Mathews F. S., Packman L. C., Scrutton N. S. Trimethylamine dehydrogenase of bacterium W3A1: molecular cloning, sequence determination and overexpression of the gene. FEBS Lett. 1992;308:271–276. doi: 10.1016/0014-5793(92)81291-s. [DOI] [PubMed] [Google Scholar]
- 6.Stott K., Saito K., Thiele D. J., Massey V. Old Yellow Enzyme: the discovery of multiple isozymes and a family of related proteins. J. Biol. Chem. 1993;268:6097–6106. [PubMed] [Google Scholar]
- 7.Miranda M., Ramirez J., Guevara S., Ongay-Larios L., Pena A., Coria R. Nucleotide sequence and chromosomal localization of the gene encoding the Old Yellow Enzyme from Kluyveromyces lactis. Yeast. 1995;11:459–465. doi: 10.1002/yea.320110509. [DOI] [PubMed] [Google Scholar]
- 8.Niino Y. S., Chakraborty S., Brown B. J., Massey V. A new old yellow enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:1983–1991. doi: 10.1074/jbc.270.5.1983. [DOI] [PubMed] [Google Scholar]
- 9.Schaller F., Weiler E. W. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana: structural and functional relationship to yeast Old Yellow Enzyme. J. Biol. Chem. 1997;272:28066–28072. doi: 10.1074/jbc.272.44.28066. [DOI] [PubMed] [Google Scholar]
- 10.Schaller F., Hennig P., Weiler E. W. 12-Oxophytodienoate-10,11-reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12-oxophytodienoic acid. Plant Physiol. 1998;118:1345–1351. doi: 10.1104/pp.118.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biesgen C., Weiler E. W. Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta. 1999;208:155–165. doi: 10.1007/s004250050545. [DOI] [PubMed] [Google Scholar]
- 12.Strassner J., Fürholz A., Macheroux P., Amrhein N., Schaller A. A homolog of Old Yellow Enzyme in tomato: spectral properties and substrate specificity of the recombinant protein. J. Biol. Chem. 1999;274:35067–35073. doi: 10.1074/jbc.274.49.35067. [DOI] [PubMed] [Google Scholar]
- 13.Schaller F., Biesgen C., Mussig C., Altmann T., Weiler E. W. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 2000;210:979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- 14.Komduur J. A., Leão A. N., Monastyrska I., Veenhuis M., Kiel J. A. K. W. Old yellow enzyme confers resistance of Hansenula polymorpha towards allyl alcohol. Curr. Genet. 2002;41:401–406. doi: 10.1007/s00294-002-0321-z. [DOI] [PubMed] [Google Scholar]
- 15.Kubata B. K., Kabututu Z., Nozaki T., Munday C. J., Fukuzumi S., Ohkubo K., Lazarus M., Maruyama T., Martin S. K., Duszenko M., Urade Y. A key role for old yellow enzyme in the metabolism of drugs by Trypanosoma cruzi. J. Exp. Med. 2002;196:1241–1251. doi: 10.1084/jem.20020885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobajima H., Takeda M., Sugimori M., Kobashi N., Kiribuchi K., Cho E. M., Akimoto C., Yamaguchi T., Minami E., Shibuya N., et al. Cloning and characterization of a jasmonic acid-responsive gene encoding 12-oxophytodienoic acid reductase in suspension-cultured rice cells. Planta. 2003;216:692–698. doi: 10.1007/s00425-002-0909-z. [DOI] [PubMed] [Google Scholar]
- 17.French C. E., Nicklin S., Bruce N. C. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2. J. Bacteriol. 1996;178:6623–6627. doi: 10.1128/jb.178.22.6623-6627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snape J. R., Walkley N. A., Morby A. P., Nicklin S., White G. F. Purification, properties, and sequence of glycerol trinitrate reductase from Agrobacterium radiobacter. J. Bacteriol. 1997;179:7796–7802. doi: 10.1128/jb.179.24.7796-7802.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French C. E., Bruce N. C. Bacterial morphinone reductase is related to Old Yellow Enzyme. Biochem. J. 1995;312:671–678. doi: 10.1042/bj3120671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohde B. H., Schmid R., Ullrich M. S. Thermoregulated expression and characterization of an NAD(P)H-dependent 2-cyclohexen-1-one reductase in the plant pathogenic bacterium Pseudomonas syringae pv. glycinea. J. Bacteriol. 1999;181:814–822. doi: 10.1128/jb.181.3.814-822.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blehert D. S., Fox B. G., Chambliss G. H. Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J. Bacteriol. 1999;181:6254–6263. doi: 10.1128/jb.181.20.6254-6263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura K., Tomioka Y., Suzuki H., Yonezawa M., Hishinuma T., Mizugaki M. Molecular cloning of the nemA gene encoding N-ethylmaleimide reductase from Escherichia coli. Biol. Pharm. Bull. 1997;20:110–112. doi: 10.1248/bpb.20.110. [DOI] [PubMed] [Google Scholar]
- 23.Fitzpatrick T. B., Amrhein N., Macheroux P. Characterization of YqjM, an Old Yellow Enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J. Biol. Chem. 2003;278:19891–19897. doi: 10.1074/jbc.M211778200. [DOI] [PubMed] [Google Scholar]
- 24.Vaz A. D., Chakraborty S., Massey V. Old Yellow Enzyme: aromatization of cyclic enones and the mechanism of a novel dismutation reaction. Biochemistry. 1995;34:4246–4256. doi: 10.1021/bi00013a014. [DOI] [PubMed] [Google Scholar]
- 25.Abramovitz A. S., Massey V. Interaction of phenols with Old Yellow Enzyme: physical evidence for charge-transfer complexes. J. Biol. Chem. 1976;251:5327–5336. [PubMed] [Google Scholar]
- 26.Stewart R. C., Massey V. Potentiometric studies of native and flavinsubstituted Old Yellow Enzyme. J. Biol. Chem. 1985;260:13639–13647. [PubMed] [Google Scholar]
- 27.Lim L. W., Shamala N., Mathews F. S., Steenkamp D. J., Hamlin R., Xuong N. H. Three-dimensional structure of the iron-sulfur flavoprotein trimethylamine dehydrogenase at 2.4 Å resolution. J. Biol. Chem. 1986;261:15140–15146. [PubMed] [Google Scholar]
- 28.Fox K. M., Karplus P. A. Old yellow enzyme at 2 Å resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure. 1994;2:1089–1105. [PubMed] [Google Scholar]
- 29.Barna T. M., Khan H., Bruce N. C., Barsukov I., Scrutton N. S., Moody P. C. E. Crystal structure of pentaerythritol tetranitrate reductase: ‘flipped’ binding geometries for steroid substrates in different redox states of the enzyme. J. Mol. Biol. 2001;310:433–447. doi: 10.1006/jmbi.2001.4779. [DOI] [PubMed] [Google Scholar]
- 30.Breithaupt C., Strassner J., Breitinger U., Huber R., Macheroux P., Schaller A., Clausen T. X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE. Structure. 2001;9:419–429. doi: 10.1016/s0969-2126(01)00602-5. [DOI] [PubMed] [Google Scholar]
- 31.Barna T., Messiha H. L., Petosa C., Bruce N. C., Scrutton N. S., Moody P. C. E. Crystal structure of bacterial morphinone reductase and properties of the C191A mutant enzyme. J. Biol. Chem. 2002;277:30976–30983. doi: 10.1074/jbc.M202846200. [DOI] [PubMed] [Google Scholar]
- 32.Kitzing K., Fitzpatrick T. B., Wilken C., Sawa J., Bourenkov G. P., Macheroux P., Clausen T. The 1.3 Å crystal structure of the flavoprotein YqjM reveals a novel class of old yellow enzymes. J. Biol. Chem. 2005;280:27904–27913. doi: 10.1074/jbc.M502587200. [DOI] [PubMed] [Google Scholar]
- 33.Haarer B. K., Amberg D. C. Old yellow enzyme protects the actin cytoskeleton from oxidative stress. Mol. Biol. Cell. 2004;15:4522–4531. doi: 10.1091/mbc.E04-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Heidelberg J. F., Paulsen I. T., Nelson K. E., Gaidos E. J., Nelson W. C., Read T. D., Eisen J. A., Seshadri R., Ward N., Methe B., et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nature Biotech. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 35.Tatusova T. A., Madden T. L. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 36a.Higgins D., Thompson J., Gibson T., Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36b.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Marshall S. J., Krause D., Blencowe D. K., White G. F. Characterization of glycerol trinitrate reductase (NerA) and the catalytic role of active-site residues. J. Bacteriol. 2004;186:1802–1810. doi: 10.1128/JB.186.6.1802-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown B. J., Deng Z., Karplus P. A., Massey V. On the active site of Old Yellow Enzyme: role of histidine 191 and asparagine 194. J. Biol. Chem. 1998;273:32753–32762. doi: 10.1074/jbc.273.49.32753. [DOI] [PubMed] [Google Scholar]
- 39.Abramovitz A. S., Massey V. Purification of intact old yellow enzyme using an affinity matrix for the sole chromatographic step. J. Biol. Chem. 1976;251:5321–5326. [PubMed] [Google Scholar]
- 40.Jaffe H. H. A reexamination of the Hammett equation. Chem. Rev. 1953;53:191–261. [Google Scholar]
- 41.Hansch C., Leo A., Taft R. W. A survey of Hammett substituent constants and resonance and field parameters. Chem. Rev. 1991;91:165–195. [Google Scholar]
- 42.Blehert D. S., Knoke K. L., Fox B. G., Chambliss G. H. Regioselectivity of nitroglycerin denitration by flavoprotein nitroester reductases purified from two Pseudomonas species. J. Bacteriol. 1997;179:6912–6920. doi: 10.1128/jb.179.22.6912-6920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig D. H., Moody P. C., Bruce N. C., Scrutton N. S. Reductive and oxidative half-reactions of morphinone reductase from Pseudomonas putida M10: a kinetic and thermodynamic analysis. Biochemistry. 1998;37:7598–7607. doi: 10.1021/bi980345i. [DOI] [PubMed] [Google Scholar]
- 44.Massey V., Stankovich M., Hemmerich P. Light-mediated reduction of flavoproteins with flavins as catalysts. Biochemistry. 1978;17:1–8. doi: 10.1021/bi00594a001. [DOI] [PubMed] [Google Scholar]
- 45.Fitzpatrick T. B., Auweter S., Kitzing K., Clausen T., Amrhein N., Macheroux P. Structural and functional impairment of an Old Yellow Enzyme homologue upon affinity tag incorporation. Protein Expression Purif. 2004;36:280–291. doi: 10.1016/j.pep.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 46.French C. E., Nicklin S., Bruce N. C. Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl. Environ. Microbiol. 1998;64:2864–2868. doi: 10.1128/aem.64.8.2864-2868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pak J. W., Knoke K. L., Noguera D. R., Fox B. G., Chambliss G. H. Transformation of 2,4,6-trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C. Appl. Environ. Microbiol. 2000;66:4742–4750. doi: 10.1128/aem.66.11.4742-4750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams R. E., Rathbone D. A., Scrutton N. S., Bruce N. C. Biotransformation of explosives by the old yellow enzyme family of flavoproteins. Appl. Environ. Microbiol. 2004;70:3566–3574. doi: 10.1128/AEM.70.6.3566-3574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao H., Wang Y., Liu X., Yan T., Wu L., Alm E., Arkin A., Thompson D. K., Zhou J. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 2004;186:7796–7803. doi: 10.1128/JB.186.22.7796-7803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong L. Y., Jiang Q. C., Qu Q. S. Formation of superoxide radical and hydrogen peroxide enhanced by trinitrotoluene in rat liver, brain, kidney, and testicle in vitro and monkey liver in vivo. Biomed. Environ. Sci. 1989;2:72–77. [PubMed] [Google Scholar]
- 51.Sydow K., Daiber A., Oelze M., Chen Z., August M., Wendt M., Ullrich V., Mülsch A., Schulz E., Keaney J. F., Jr, et al. Central role of mitochondrial aldehyde dehydrogenase and reactive oxygen species in nitroglycerin tolerance and cross-tolerance. J. Clin. Invest. 2004;113:482–489. doi: 10.1172/JCI19267. [DOI] [PMC free article] [PubMed] [Google Scholar]