Abstract
Thymidylate synthase (TS) catalyses the reductive methylation of dUMP to form dTMP, a reaction that is essential for maintenance of nucleotide pools during cell growth. Because the enzyme is indispensable for DNA replication in actively dividing cells, it is an important target for cytotoxic drugs used in cancer chemotherapy, including fluoropyrimidines (e.g. 5-fluorouracil and 5-fluoro-2′-deoxyuridine) and anti-folates (e.g. raltitrexed, LY231514, ZD9331 and BW1843U89). These drugs generate metabolites that bind to the enzyme's active site and inhibit catalytic activity, leading to thymidylate deprivation and cellular apoptosis. Ligand binding to TS results in stabilization of the enzyme and an increase in its intracellular concentration. Previously, we showed that degradation of the TS polypeptide is carried out by the 26 S proteasome in a ubiquitin-independent manner. Such degradation is directed by the disordered N-terminal region of the TS polypeptide, and is abrogated by ligand binding. In the present study, we have verified the ubiquitin-independent nature of TS proteolysis by showing that a ‘lysine-less’ polypeptide, in which all lysine residues were replaced by arginine, is still subject to proteasome-mediated degradation. In addition, we have mapped the structural determinants of intracellular TS degradation in more detail and show that residues at the N-terminal end of the molecule, particularly the penultimate amino acid Pro2, play an important role in governing the half-life of the enzyme. This region is capable on its own of destabilizing an evolutionarily distinct TS molecule that normally lacks this domain, indicating that it functions as a degradation signal. Interestingly, degradation of an intrinsically unstable mutant form of TS, containing a Pro→Leu substitution at residue 303, is directed by C-terminal, rather than N-terminal, sequences. The implications of these findings for the control of TS expression, and for the regulation of protein degradation in general, are discussed.
Keywords: enzyme degradation, fluoropyrimidine, proteasome, proteolysis, thymidylate synthase, ubiquitin-independent
Abbreviations: CHL, Chinese hamster lung; CH2H4PteGlu, N5,N10-methylene-5,6,7,8-tetrahydrofolic acid; FdUMP, 5′-fluoro-2′-deoxyuridylic acid; FdUrd, 5′-fluoro-2′-deoxyuridine; 5-FU, 5-fluorouracil; ODC, ornithine decarboxylase; TS, thymidylate synthase; P303L, TS P303L mutant; wtTS, wild-type TS
INTRODUCTION
Thymidylate synthase (TS; EC 2.1.1.45) is an S-phase enzyme that catalyses the reductive methylation of dUMP by CH2H4PteGlu (N5,N10-methylene-5,6,7,8-tetrahydrofolic acid) to generate dTMP and dihydrofolate [1]. This reaction is the sole de novo source of dTMP for DNA synthesis, making TS indispensable in actively dividing cells. Inhibition of the enzyme results in the depletion of thymidylate pools, elevation of dUTP levels and misincorporation of uracil into DNA. This leads to DNA fragmentation followed by apoptotic cell death [2,3]. As a consequence, TS has been a target for the development of anti-neoplastic agents. Fluoropyrimidine antimetabolites [e.g. 5-FU (5-fluorouracil) and FdUrd (5′-fluoro-2′-deoxyuridine)] are toxic to cells as a consequence of their being converted into the nucleotide analogue FdUMP (5-fluoro-2′-deoxyuridylic acid), which forms a stable, covalent inhibitory ternary complex composed of the enzyme, FdUMP, and the co-substrate CH2H4PteGlu at the active site [2,4–6]. The high electronegativity of the fluorine atom in FdUMP precludes the subsequent steps in the reaction, so that the ternary complex accumulates, and prolonged enzyme inhibition results. The fluoropyrimidines, particularly 5-FU, have long been the standard choice for treatment of colorectal cancer [7]. While TS is the primary target of these agents, incorporation of the analogue into RNA and DNA may also play significant roles in their antitumour activities; thus the mode of inhibition by these agents is not completely TS-specific [7]. In recent years, folic acid-based inhibitors of TS [e.g. ZD1694 (raltitrexed or Tomudex), AG337 and GW1843U89 (U89)] have been developed. These compounds inhibit TS with high degrees of specificity, and have shown activity in the management of head, neck, breast, stomach and colon cancers in several clinical trials [8].
Numerous studies with a variety of tumour cell lines and biopsied tissues from cancer patients have shown that TS levels are induced after treatment with TS inhibitors [9,10]. Such an elevation of enzyme levels is believed to have a direct effect on the efficacy of TS inhibitors, and may promote cellular resistance to them. While the phenomenon of ligand-mediated TS induction has been widely documented, its mechanism is incompletely understood. Chu et al. [11,12] proposed that TS binding to its own mRNA and repression of translation occur, and are relieved by ligand binding to the enzyme, leading to augmentation of TS synthesis. Other studies, however, have shown that the degradation rate of ligand-bound TS is lower than for the ligand-free enzyme, indicating that TS induction is a consequence of enzyme stabilization [13].
Recently, we showed that degradation of human TS is carried out by the 26 S proteasome in a ubiquitin-independent fashion [14]. The N-terminal region of the polypeptide was identified as a primary determinant of this degradation [14]. The region is extended by approx. 30 amino acids relative to that of the enzyme from Escherichia coli and is disordered in X-ray crystallographic structures [1,15,16]. Deletion of as few as six amino acids from the N-terminal end of the molecule elicits marked stabilization of the enzyme, with further deletions resulting in variable degrees of stability [14]. On the basis of these findings, we suggested that the disordered N-terminal domain mediates recognition by the proteasome and targets the polypeptide for proteolytic destruction.
In the present study, we have examined the determinants of intracellular TS degradation in more detail. We have utilized a ‘lysineless’ TS molecule to provide support for the ubiquitin-independent nature of TS proteolysis. In addition, we have carried out a more extensive dissection of the N-terminal domain of the TS polypeptide, particularly with regard to its role in mediating intracellular degradation. We demonstrate that amino acid residues at the very end of the polypeptide, including the penultimate residue Pro2, are central regulators of proteasomal degradation. We show that the domain is capable of destabilizing an evolutionarily distinct TS molecule, indicating that it functions as an independent degradation signal. Finally, we demonstrate that the intrinsic instability of a mutant form of TS is directed by the C-terminal, rather than the N-terminal, region of the molecule.
EXPERIMENTAL
Cell and tissue culture
All cells were maintained in Dulbecco's modified Eagle's medium (Cellgro) containing 4.5 g/l glucose supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Atlanta Biologicals) at 37 °C in a humidified 5% CO2 atmosphere. Cell line RJK88.13, a TS-deficient derivative of V79 CHL (Chinese hamster lung) cells [17], was maintained in the above medium supplemented with 10 μM thymidine. Where indicated, cells were treated with 100 nM FdUrd (with 10 μM folinic acid), 100 nM raltitrexed (Astra Zeneca) or 100 nM GW1843U89 (U89; Gilead) for 24 h prior to harvesting. Pretreatment with the proteasome inhibitors MG132 (25 μM; Sigma–Aldrich) or lactacystin (7.5 μM; Calbiochem) was for 4 h prior to the addition of cycloheximide (50 μg/ml; Sigma–Aldrich).
Plasmid construction, mutagenesis and transfection
Plasmid constructions were done by standard molecular biology techniques, and were directly verified by DNA sequencing. The sequences of primers used for mutagenesis and DNA sequencing are available upon request. Constructs were generated in the pSV2-CAT expression vector (Invitrogen) where the CAT (chloramphenicol acetyltransferase) gene had been removed, and was replaced by a full-length human TS cDNA, so that the cDNA is under the control of the SV40 (simian virus 40) promoter [14]. The template for site-directed mutagenesis of wtTS (wild-type TS) was plasmid pJZ205 [14], while the template for mutagenesis of the P303L mutant (designated P303L) was plasmid pTF489 [14].
A ‘lysine-less’ TS molecule was created by sequential mutagenesis using the Multi-QuikChange Mutagenesis kit (Stratagene) according to the manufacturer's instructions; primers contained AAA or AAG→CGG substitutions at each of the 15 lysine residues within the polypeptide, converting them into arginine. Plasmid pJZ205 was the template for mutagenesis; the resulting plasmid was denoted pSK641.
All other mutant plasmids were generated using the Quik-Change Site-Directed Mutagenesis kit (Stratagene) following the manufacturer's instructions. Construction of N-terminal deletion mutant wtTS-del(2–7) was described previously [14]. The del(2–7) mutation in combination with the P303L mutation was generated by digesting the wtTS-del(2–7) plasmid (pTF577) with HindIII and BglII, and cloning the mutated fragment into the corresponding sites of plasmid pTF489; the resulting plasmid was designated pTF600. To generate the wtTS-del(2–4), wtTS-del(2–3) and wtTS-del(2) mutants, primers containing an ATG→GGC substitution (destroying the translational initiation codon) and a GCC→ATG mutation at codon 4, a GTG→ATG mutation at codon 3 or a CCT→ATG mutation at codon 2 (creating new initiator codons) were used with plasmid pJZ205. The resulting mutant plasmids were designated pMP664, pSK690 and pSK689 respectively.
The V3R/A4D/G5D mutant of wtTS was produced with a primer that contained GTG→AGG, GCC→GAC and GGC→GAC substitutions at codons 3, 4 and 5 respectively; plasmid pJZ205 was the template, and the resulting plasmid was denoted pKW636. The V3R/A4R/G5R mutant was generated using a primer that contained GTG→GAC, GCC→GAC and GGC→GAC substitutions at codons 3, 4 and 5 respectively; the mutant plasmid was designated pSK669.
Substitutions at Pro2 in wtTS were generated using pJZ205 as the template. The P2G mutant was made by introducing a CCT→GGT change at codon 2, P2V by a CCT→GTT change, P2A by a CCT→GCT change, P2D by a CCT→GAT change, P2R by a CCT→CGT change, P2W by a CCT→TGG change and P2Y by a CCT→TAT change. The mutagenized plasmids were designated pSK676, pSK677, pSK691, pSK692, pSK693, pSK694 and pSK695 respectively.
Construction of N-terminally His6-tagged derivatives of wtTS and P303L, containing the tag between Met1 and Pro2, was described previously [14]. C-terminally His6-tagged TS molecules were produced by inserting a NotI restriction site between codons 311 and 312 at the C-terminus of both wtTS and the P303L mutant. Two complementary oligonucleotides containing six His codons with flanking EagI restriction sites were annealed and ligated into the NotI site. The ligated products were further digested with NotI and transformed into E. coli DH5α cells. TS molecules containing the His6 tag were identified by loss of the NotI site. The presence and correct orientation of the His6 tag were further verified by DNA sequencing. Plasmids containing the His6 tag at the C-terminal end of wtTS and the P303L mutant were designated pYX644 and pYX648 respectively.
A construct expressing the E. coli enzyme was generated by subcloning the 880 bp BfrBI–HpaI fragment of pMP608 (containing the full-length E. coli TS gene optimized for expression in mammalian cells [18]) into EcoRV–HpaI-digested pJZ205; the resulting plasmid was designated pSK670. To fuse the N-terminal region of human TS to E. coli TS, a PstI site at codons 31–32 of pJZ205 was converted into an ScaI site; the resulting plasmid was digested with ScaI, and the 2.2 kb fragment containing the human N-terminal region was ligated to the 2.3 kb ScaI fragment of pSK670. The final construct was designated pSK672.
Mutants containing deletions at the C-terminal end of both wtTS and the P303L mutant were generated by converting appropriate codons into termination codons. Plasmids pJZ205 and pTF489 were used as templates for mutagenesis. The del(313) mutant was generated by a GTT→TAG change at codon 313, the del(311–313) mutation by an ATG→TAG change at codon 311, the del(308–313) mutation by an AAA→TAG change at codon 308, the del(305–313) mutation by a CCA→TAG change at codon 305 and the del(300–313) mutation by a GGG→TAG change at codon 300. The del(305–307) deletion mutant was generated using complementary oligonucleotides lacking codons 305–307 for mutagenesis. Plasmid pZR560 contains the del(313) mutation in the P303L polypeptide [14], while plasmid pMP653 contains the del(313) mutation in wtTS. Plasmids pYX660, pMP654, pYX658, pYX659 and pMP661 contain the del(311–313), del(308–313), del(305–313) and del(300–313) mutations in wtTS respectively. Plasmids pYX657, pMP655 and PY656 contain the del(311–313), del(308–313) and del(305–313) mutations in the P303L mutant respectively. Plasmid pYX659 contains the del(300–313) mutant, while pMP661 and pMP662 contain the del(305–307) mutation in the wtTS and P303L mutant molecules respectively.
Transfections were performed using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. All plasmids were transfected into CHL cell line RJK88.13 in media containing 10 μM thymidine. Stable transfectants were selected in thymidine-free medium with or without 5 μM dipyridamole (Sigma), a nucleoside transport inhibitor. Stable transfectants were pooled and maintained in mass culture.
Determination of the half-life of the TS polypeptide
Cells were split into 60 mm plates, and cycloheximide was added to a concentration of 50 μg/ml 24 h later. At various times following addition of cycloheximide, cells were harvested and lysed by sonication (3×10 s) in NET2 buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl and 0.05% Nonidet P40) containing 10 mM dithiothreitol, 2 mM 2-mercaptoethanol, 5 mM PMSF, 200 μg/ml aprotinin, 100 μg/ml pepstatin and 50 μg/ml leupeptin. The crude lysates were centrifuged at 15000 g for 1 h at 4 °C, and the protein was quantified using the Bio-Rad assay reagent using BSA as a standard. Immunoblotting was performed by standard techniques, using a monoclonal anti-human TS antibody (1:1000 dilution) as probe [14]; in some experiments, a monoclonal antihuman TS antibody (1:100 dilution) was purchased from Abcam (Cambridge, MA, U.S.A.) and used as probe. As an internal control for equal loading, blots were stripped and reprobed with antibodies against α-actin (1:1000 dilution; Clone AC-40; Sigma). The antigen–antibody complexes were visualized by chemiluminescence using an ECL® kit (Amersham Biosciences). Films that were exposed within the linear range of the chemiluminescence were scanned, and bands were quantified using the Quantity-One software (Bio-Rad). All values were normalized to levels of actin, obtained by quantification of blots that were stripped and reprobed with an anti-α-actin antibody. In all experiments, parental enzymes to which the mutants were compared (i.e. either wtTS or the P303L enzyme) were run in parallel.
RESULTS
A ‘lysine-less’ derivative of human TS retains sensitivity to proteasome-mediated degradation
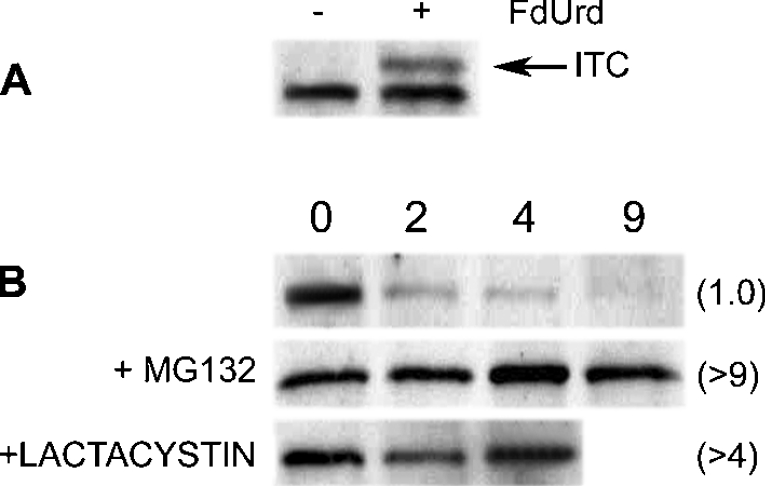
Most proteins targeted for degradation by the 26 S proteasome are ‘tagged’ with one or more polyubiquitin chains. However, our previous studies indicated that human TS is degraded in a ubiquitin-independent manner [14], representing one of only a few proteins to behave in such a fashion [19]. Because ubiquitin moieties are typically attached to proteins via internal lysine residues, the removal or modification of these residues generally leads to loss of ubiquitin ligation and resistance to proteasome-mediated degradation. This provides a further test for the role of ubiquitinylation in protein degradation. Therefore we measured the rate of degradation of a TS polypeptide in which all 15 lysine residues were converted into arginine. A plasmid that encodes such a ‘lysine-less’ TS polypeptide was transfected into TS-deficient cell line RJK88.13, and thymidine-independent derivatives were obtained. Immunoblotting and TS activity measurements indicated that the exogenous enzyme was fully capable of forming an inhibitory ternary complex following addition of FdUrd to the growth media (Figure 1A). Thus the structure and catalytic activity of the TS polypeptide were retained in a ‘lysine-less’ mutant. Importantly, cycloheximide chase experiments showed that the mutant had a half-life of approx. 1.0 h, and was stabilized by the proteasome inhibitors MG132 and lactacystin (Figure 1B). An in vitro reticulocyte lysate-based assay [20] verified that the ‘lysine-less’ TS polypeptide is stabilized by MG132 (results not shown).
Figure 1. A ‘lysine-less’ TS molecule is degraded by the proteasome.
A plasmid encoding a ‘lysine-less’ TS molecule was stably transfected into TS-deficient RJK88.13 cells using Lipofectamine™ 2000. (A) Transfected cells were either untreated (−) or treated (+) with 100 nM FdUrd and 10 μM folinic acid for 24 h. Total cell extracts were analysed by Western blotting using a monoclonal antibody against human TS. ITC indicates the band corresponding to the inhibitory ternary complex. (B) Transfected cells were treated for the indicated times in 50 μg/ml cycloheximide; where indicated, cells were pretreated with 25 μM MG132 or 7.5 μM lactacystin for 4 h prior to the addition of cycloheximide. Cells were harvested, and total cell extracts were analysed by Western blotting using monoclonal antibody against wtTS. Numbers in parentheses indicate the half-lives (h), as determined by scanning densitometry.
These results show that degradation of the TS molecule is not dependent on the presence of internal lysine residues. This finding is consistent with previous observations indicating that intracellular degradation of TS occurs in a ubiquitin-independent manner.
The N-terminal end of the TS polypeptide functions as a degradation signal
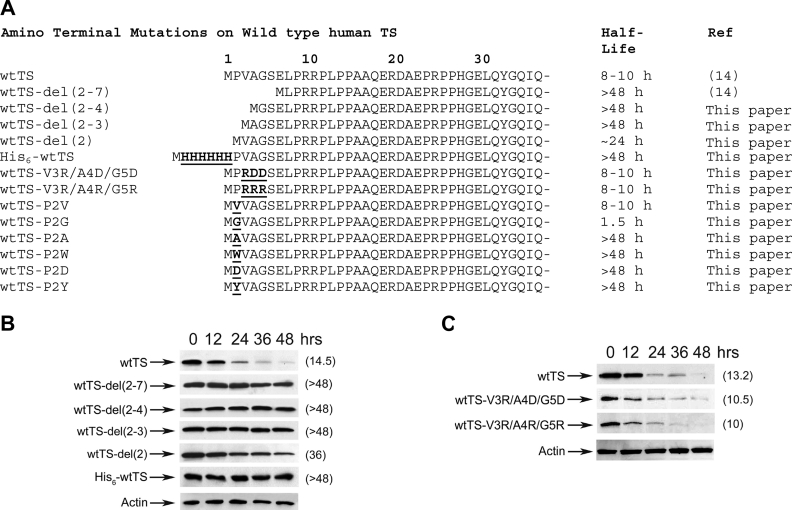
In earlier studies, we demonstrated that deletion of residues 2–7 at the N-terminus of TS increased the enzyme's half-life from approx. 10–12 h to >48 h [14]; deletion of residues 2–12, 2–21 or 2–29 resulted in relatively unstable proteins that were stabilized by ligands [14]. To further assess the role of the N-terminus in TS proteolysis, we measured the degradation rates of a number of additional mutant polypeptides containing alterations in this region. The various mutants that were examined are depicted in Figure 2(A).
Figure 2. Determinants for the degradation of wtTS are located at the N-terminal end of the molecule.
(A) Amino acid sequences of the various N-terminal mutants of wtTS are shown. Amino acid substitutions and insertions are indicated in boldface and are underlined. (B) RJK88.13 cells were stably transfected with plasmids encoding wtTS or mutants wtTS-del(2–7), wtTS-del(2–4), wtTS-del(2–2), wtTS-del(2) or His6–wtTS. Transfected cells were split into 60 mm plates, and cycloheximide treatment (50 μg/ml) was initiated 24 h later. Cells were harvested after the indicated times of cycloheximide exposure, and steady-state levels of wtTS were determined by Western-blot analysis of total cell extracts using a monoclonal antibody against human TS. As an internal control for equal loading, blots were stripped and steady-state levels of actin were analysed using an α-actin monoclonal antibody; a representative blot for actin is shown (Actin). Enzyme half-lives were estimated by scanning densitometry of the films, and are indicated in parentheses. (C) Plasmids encoding wtTS, wtTS-V3R/A4D/G5D and wtTS-V3R/A4R/G5R were stably transfected into RJK88.13 cells. Cells were treated, extracts obtained, and analysed by Western blotting as described in (B). Blots were stripped and reprobed with an α-actin monoclonal antibody to control for equal loading. A representative blot (Actin) is shown. Half-lives (h), estimated by scanning and quantification of the films, are indicated in parentheses.
wtTS exhibited a half-life of approx. 14 h following initiation of cycloheximide treatment (Figure 2B). In numerous experiments, we have consistently observed a 9–15 h half-life for this enzyme ([14], and results not shown). As observed previously [14], mutant wtTS-del(2–7), which is missing residues 2–7, was completely stable over the 48 h time period of the experiment (Figure 2B). Mutants wtTS-del(2–4) and wtTS-del(2–3), which lack residues 2–4 and 2–3 respectively, were also resistant to proteolysis (Figure 2B). Finally, wtTS-del(2), which is missing only the penultimate residue, had a half-life of approx. 24–36 h, indicating that it was more stable than the wild-type molecule, though not quite as stable as the other mutants (Figure 2B). Thus residues very near the N-terminus are major determinants of the polypeptide's sensitivity to intracellular proteolysis.
We assessed the impact of a His6 tag at the polypeptide's N-terminal end. Mutant His6–wtTS, which contains six His residues inserted between positions Met1 and Pro2 [14], was completely stable, having a half-life of >48 h (Figure 2B). Thus blocking the N-terminus with a His6 tag allowed the TS polypeptide to fully escape intracellular degradation, further substantiating the importance of this region in regulating the enzyme's metabolic stability.
Recognition of ubiquitinylated proteins by the proteasome requires hydrophobic ‘patches’ on the surface of the polyubiquitin moieties [21]. It is possible that the hydrophobic nature of residues 2–7 of TS might be critical to its interaction with the proteasome. To test this notion, we introduced charged amino acids into the region. Mutants wtTS-V3R/A4D/G5D and wtTS-V3R/A4R/G5R, both of which contain charged in place of hydrophobic residues at positions 3–5, exhibited half-lives of 11 and 10 h respectively, which are very similar to that for wtTS (Figure 2C). Thus hydrophobicity at the N-terminal end of the TS polypeptide does not appear to be required for degradation.
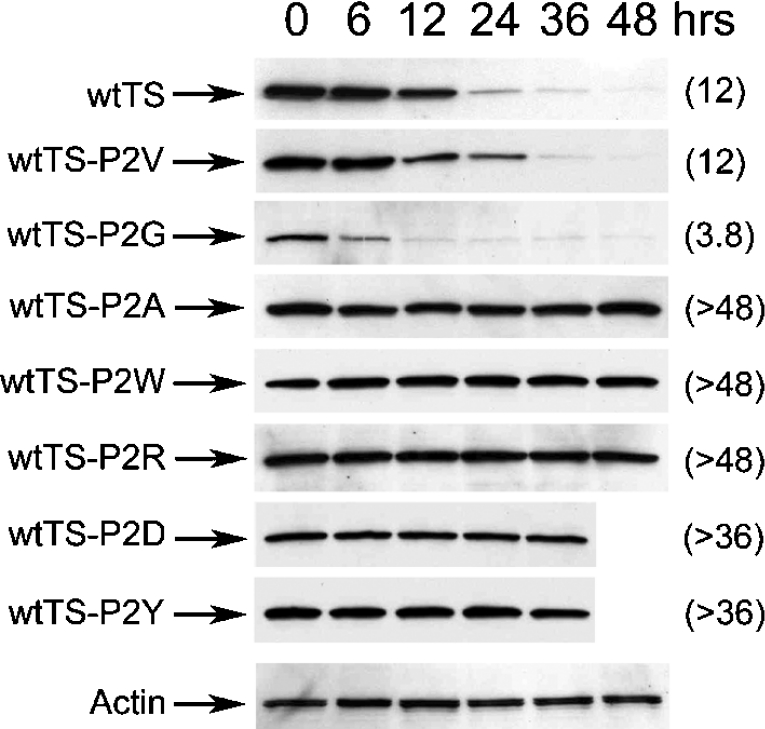
Pro2 plays an important role in TS degradation
The finding that wtTS-del(2) is stable relative to the wild-type enzyme indicated that Pro2 may be an important determinant of TS degradation. Therefore further experiments focused on this residue. A number of amino acid substitutions were introduced at position 2, and the half-lives of the resulting polypeptides were measured. Mutant wtTS-P2V, containing Val in place of Pro, exhibited a half-life of 12 h, which is very similar to wtTS (Figure 3). In contrast, mutant wtTS-P2G, containing Gly at this site, exhibited a half-life of 3.8 h, indicating that it was markedly less stable than wtTS (Figure 3). The reduced half-life of this mutant was confirmed in a separate experiment in which the analysis was conducted over a shorter time period following cycloheximide addition (results not shown). Five other mutants with substitutions at Pro2 (wtTS-P2A, -P2R, -P2D, -P2Y and -P2W) were found to be highly resistant to degradation, with half-lives greater than 36–48 h (Figure 3). Thus Pro2 substitutions can alter the activity of the N-terminal region in promoting TS proteolysis, indicating that the penultimate residue plays a significant role in regulating the enzyme's susceptibility to intracellular proteolysis.
Figure 3. Amino acid substitutions at the penultimate proline residue affect the half-life of wtTS.
RJK88.13 cells were stably transfected with plasmids encoding wtTS-P2V, wtTS-P2G, wtTS-P2A, wtTS-P2W, wtTS-P2R, wtTS-P2D or wtTS-P2Y. Cells were split into 60 mm plates, and cycloheximide treatment (50 μg/ml) was initiated 24 h later. Cells were harvested after the indicated times of cycloheximide exposure, and steady-state levels of wtTS were determined by Westernblot analysis of total cell extracts using a monoclonal antibody against human TS. As an internal control for equal loading, blots were stripped and steady-state levels of actin were analysed using an α-actin monoclonal antibody; a representative blot for actin is shown (Actin). Half-lives (h), estimated by scanning and quantification of the films, are indicated in parentheses.
The N-terminal domain of human TS functions as a degradation signal
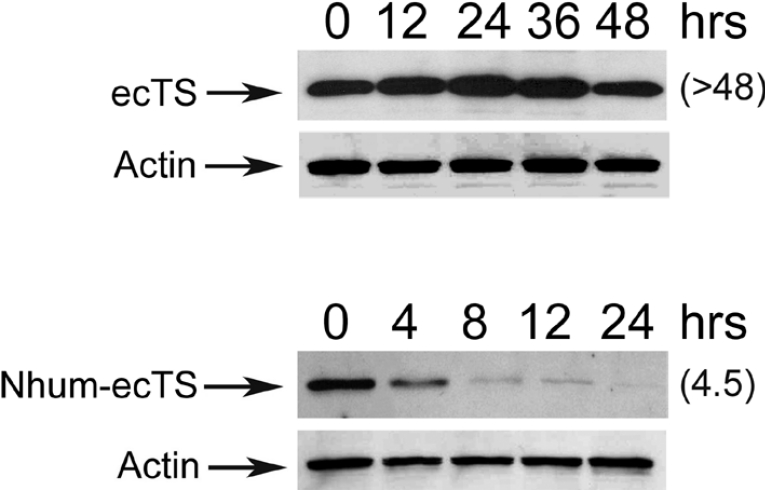
The N-terminal region of human TS is extended by approx. 30 amino acids relative to E. coli TS, so that Met1 of the latter corresponds to Glu30 of the former [1,15,16,22]. Making use of this species difference, we asked if the N-terminal domain of human TS is capable of modifying degradation of the E. coli polypeptide, which lacks the domain. A chimaeric molecule, denoted Nhum-wtTS and containing the first 30 residues of human TS ligated to the N-terminal end of E. coli TS, was generated. Its rate of degradation was measured and compared with that for the normal E. coli enzyme (denoted ecTS). As seen in Figure 4, ecTS was quite stable, exhibiting a half-life of >48 h, while the fusion protein had a half-life of only 4.5 h. Thus the presence of the N-terminal domain of human TS causes marked destabilization of the E. coli enzyme. This suggests that the first 30 residues of the human enzyme function as a degradation signal.
Figure 4. The N-terminal domain of human TS directs degradation of E. coli TS.
RJK88.13 cells were stably transfected with a plasmid encoding wild-type E. coli TS (ecTS) or a fusion protein containing the first 30 residues of human TS ligated to the N-terminal end of ecTS (Nhum-ecTS). Cells were split into 60 mm plates, and cycloheximide treatment (50 μg/ml) was initiated 24 h later. Cells were treated for the indicated times, and steady-state levels of wtTS were determined by Western-blot analysis of total cell extracts using a polyclonal antibody against ecTS. As an internal control for equal loading, blots were stripped and steady-state levels of actin were analysed using an α-actin monoclonal antibody (Actin). Half-lives (h), estimated by scanning and quantification of the films, are indicated in parentheses.
Degradation of the P303L mutant is governed by its C-terminus
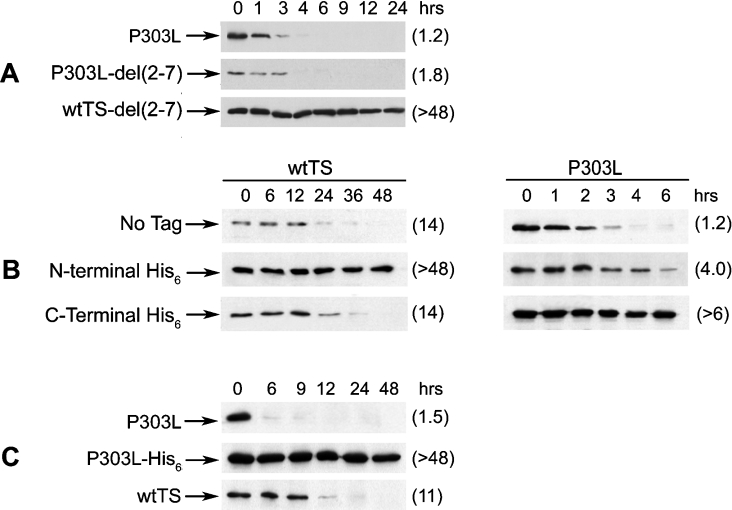
In previous studies, the P303L form of TS, which was originally identified in an FdUrd-resistant colon tumour cell line, was found to be metabolically unstable, having a half-life of approx. 1–3 h [23]. Like wtTS, it forms a stable ternary complex with FdUrd, and its degradation is carried out by the 26 S proteasome independent of ubiquitinylation [14]. To gain insight into the basis of the metabolic instability of the P303L enzyme, we tested the effects of various N-terminal mutations on its degradation. Surprisingly, deletion of residues 2–7 from the N-terminal end of the polypeptide, which had a dramatic effect on the stability of wtTS, had no measurable impact on the half-life of P303L (Figure 5A). Similarly, insertion of a His6 tag between residues Pro2 and Val3, which greatly stabilized wtTS, caused only a slight stabilization of the P303L form (Figure 5B). Thus the intracellular stability of the P303L enzyme is less sensitive than that of the wild-type enzyme to mutational alterations within the N-terminal region of the polypeptide, suggesting that other regions of the polypeptide may be important.
Figure 5. Determinants for the degradation of the P303L form of TS are located at the C-terminus.
(A) RJK88.13 cells were stably transfected with plasmids expressing P303L, P303L-del(2–7) or wtTS-del(2–7). Transfectants were treated with cycloheximide (50 μg/ml) for the indicated times, and total cell extracts were analysed by Western blotting. (B) RJK88.13 cells were stably transfected with plasmids expressing the following polypeptides: wtTS; P303L; wtTS containing either an N-terminal His6 tag (His6–wtTS) or a C-terminal His6 tag (wtTS–His6); P303L containing either an N-terminal His6 tag (His6–P303L) or a C-terminal His6 tag (P303L–His6). Cells were treated with cycloheximide for the indicated times, and total cell extracts were analysed for steady-state TS levels by Western blotting. (C) RJK88.13 cells stably transfected with plasmids expressing P303L, P303L–His6 or wtTS were treated with cycloheximide (50 μg/ml) for the indicated times, and total cell extracts were analysed for steady-state TS expression by Western blotting. Blots were probed with the monoclonal antibody against human TS. Following visualization, blots were stripped and then reprobed with α-actin monoclonal antibody as the control for equal loading (results not shown). Half-lives (h), estimated by scanning and quantification of the films, are indicated in parentheses.
Since Pro303 is only ten amino acids away from the C-terminus, and probably contributes to the structure of the C-terminal region [24], we postulated that this end of the molecule may be an important determinant of the unstable character of the P303L enzyme. We therefore analysed a series of molecules with alterations at the C-terminus. A His6 tag was inserted between residues 311 and 312 of the P303L enzyme (human TS is 313 residues in length); a similar His6 tag was inserted at a similar location within wtTS. The effects of the tags were different between the two polypeptides. The C-terminal His6 tag exerted no measurable impact on the half-life of wtTS, as both the tagged and untagged polypeptides had half-lives of 14 h (Figure 5B). In contrast, the half-life of P303L–His6 was increased relative to that of P303L (Figure 5B); indeed, the tagged protein was completely stable, having a half-life of >48 h (Figure 5C). Thus degradation of the P303L polypeptide is profoundly impaired by alterations at the C-terminal end of the molecule and is only slightly affected by alterations at the N-terminal end. This contrasts with the situation for wtTS and indicates that the intrinsic instability of the P303L mutant is primarily determined by the C-terminal end of the molecule.
To further define the residues that govern the stability of the P303L polypeptide, a number of C-terminal deletions were constructed and analysed. The deletions are depicted in Figure 6(A). Because an intact C-terminus is a prerequisite for covalent bond formation between TS and its substrates [25], none of these mutants is capable of forming an inhibitory ternary complex (results not shown). Mutants P303L-del(313), P303L-del(311–313) and P303L-del(308–313), which are missing one, three and six amino acids respectively, exhibited half-lives of 2.1–2.7 h, which are similar to the 2.3 h half-life of P303L itself (Figure 6B). On the other hand, P303L-del(305–313), which lacks the C-terminal nine residues, was markedly stabilized (Figure 6B). The half-life of wtTS was not affected by any of these deletions (results not shown). The results indicate that residues 305–307 are important in regulating degradation of the P303L form of TS.
Figure 6. Degradation of the P303L enzyme is determined by its C-terminal region.
(A) Amino acid sequences of the various C-terminal mutants of the P303L form of TS are shown. The P303L substitution and His6 insertion are indicated in boldface and underlined. Additional residues flanking the His6 sequence (Ser-Arg and Gly-Arg) occur as an inevitable consequence of insertion of a NotI restriction site for subcloning (see the Experimental section), and are also indicated in boldface and underlined. (B) RJK88.13 cells were transiently transfected with plasmids encoding the P303L mutant and its variants P303L-del(313), P303L-del(311–313), P303L-del(308–313), P303L-del(305–313), P303L-del(305–307) and P303L–His6. After 24 h, transfected cells were split into 60 mm plates, and cycloheximide treatment (50 μg/ml) was initiated 24 h later. Cells were harvested after the indicated times of cycloheximide exposure, and steady-state levels of the P303L polypeptide were determined by Western-blot analysis of total cell extracts, using the human TS monoclonal antibody. As a control, immunoblots were stripped and steady-state levels of actin were analysed using an α-actin monoclonal antibody. A representative blot is shown (Actin) to indicate equal loading. Half-lives (h), estimated by scanning and quantification of the films, are indicated in parentheses.
Interestingly, residues 305–307 comprise a Pro-Thr-Ile tripeptide motif that is similar to the Pro-Thr-Leu sequence that dictates degradation of the c-Fos protein in asynchronously growing cells [26]. Deletion of the motif from c-Fos results in stabilization of the protein [26]. We tested the possibility that the Pro-Thr-Ile sequence of TS on its own directs the stability of P303L by deleting it in the context of a full-length polypeptide. As shown in Figure 6(B), the half-life of P303L-del(305–307), which is missing only residues 305–307, was 1.8 h, which is nearly identical with that of P303L. Thus the Pro-Thr-Ile motif is not required for the high instability of the P303L enzyme.
Taken in total, our experiments indicate that altering the C-terminal region of TS disrupts degradation of the P303L enzyme, but has little or no effect on the wild-type polypeptide. This observation indicates that the structural determinants for P303L degradation are distinct from those for wtTS.
DISCUSSION
Recognition of substrates by the 26 S proteasome is typically accomplished through ligation of a polyubiquitin chain to one or more internal Lys residues or to the N-terminal amino group of a target substrate [21,27]. Once bound to the proteasome, the poly-ubiquitinylated substrates must be unfolded and inserted into the proteasome's catalytic chamber, where proteolysis occurs [28]. In previous work, we used a variety of strategies to demonstrate that TS is neither ubiquitinylated nor dependent on the ubiquitinylation pathway for its degradation [14]. In the present study, we demonstrate that a ‘lysine-less’ TS molecule, in which all Lys residues were replaced by Arg, is still efficiently degraded by the proteasome. Since Lys is the site of ubiquitin ligation in most molecules targeted for proteasomal degradation, this finding is consistent with the notion that TS proteolysis is ubiquitin-independent. It is important to recognize that the maintenance of normal protein degradation in ‘lysine-less’ mutants does not rule out the possibility that ubiquitin ligation occurs at the N-terminal amino group [27]. However, it is doubtful that TS is ubiquitinylated at its N-terminal end. First, MS measurements have identified N-acetylmethionine at the N-terminus of the rat enzyme [29,30], making ubiquitinylation at this site highly unlikely. Secondly, biochemical analysis has failed to show evidence of ubiquitin moieties attached to the TS polypeptide [14]. Finally, genetic ablation of the ubiquitin ligation pathway does not lead to stabilization of TS [14]. Along with the present results showing proteasome-mediated degradation of a ‘lysine-less’ TS molecule, the results are collectively consistent with the notion that TS degradation by the 26 S proteasome is ubiquitin-independent.
An increasing number of proteins have been shown to be degraded by the proteasome in an ubiquitin-independent manner, including ODC (ornithine decarboxylase) [31,32], p21Cip1 [27], IκBα (inhibitory κBα) [33], p53 [34], Rb [35], TCRα (T-cell receptor α) [36], c-Jun [37] and calmodulin [38]. For some of these, degradation is completely independent of ubiquitinylation, while for others, it occurs through both ubiquitin-dependent and -independent pathways. In the absence of ubiquitinylation, recognition of the proteasome may require either a ‘chaperone’-like accessory protein or a specific degradation signal within the target protein. The degradation of ODC occurs through a C-terminal domain that mimics a polyubiquitin chain in binding to the proteasome [31,32,39]. Proteasomal degradation of TS has several similarities to that of ODC. For both molecules, ubiquitin-independent degradation is mediated by amino acid residues at the termini of their respective polypeptide chains: the N-terminal end for TS and the C-terminal end for ODC. Deletion of as few as six amino acids from the C-terminus of ODC and of only one from the N-terminus of TS rescues the proteins from degradation, and stabilizes them. In addition, both the N-terminus of TS [15,16] and the C-terminus of ODC [40] are disordered in X-ray crystal structures, a shared feature that may be relevant to the roles of the terminal regions in governing susceptibility to degradation.
The N-terminal end of the TS polypeptide appears to act as a signal for ubiquitin-independent degradation, with the penultimate residue (Pro2) being particularly important. Deletion of this residue results in significant stabilization of the enzyme (Figure 2B). Furthermore, substituting Ala, Arg, Tyr, Trp or Asp in place of Pro2 leads to loss of degradation capacity, and resistance to intracellular proteolysis (Figure 3). Thus the nature of the penultimate amino acid is clearly an important determinant of TS degradation. However, it is not the sole determinant, and probably functions in the context of additional residues. For example, even though mutants wtTS-del(2–4) and wtTS-P2G contain Gly as their penultimate residues, the former is resistant to proteolysis, while the latter is not (see Figures 2B and 3). This suggests that the function of the penultimate residue in TS degradation depends on nearby residues.
The N-terminal domain of the human enzyme is capable on its own of conferring reduced stability to an evolutionarily distinct TS molecule (i.e. that derived from E. coli), indicating that the region functions as a degron [41]. Experiments are under way to determine if the region can modulate degradation of a completely unrelated polypeptide. While further work is necessary to precisely define the mechanisms regulating TS degradation, the importance of residues very near the end of the molecule leads us to speculate that post-translational processing may be involved. Indeed, as mentioned above, results of MS analyses have indicated that the rat TS polypeptide contains N-acetylmethionine at its N-terminus [29,30]. Acetylation of the N-terminus has been estimated to occur in 80–90% of cytoplasmic proteins in eukaryotic cells, making it a common mode of protein processing [42,43]. Interestingly, a protein's susceptibility to N-terminal acetylation is dependent on the primary sequence proximate to the N-terminal end [42,43]. While the physiological function of N-terminal acetylation is incompletely defined, effects on protein stability have been suggested [44–46]. It is possible that degradation of TS may depend on the acetylation status of its N-terminal residue. Chemical analyses of the N-terminus of wild-type and mutant forms of TS are under way, in order to test this possibility.
Previously, we postulated that the N-terminal domain of TS interacts with the proteasome by mimicking polyubiquitin chains [14]. This is similar to what has been demonstrated for the C-terminal region of ODC, which competes with ubiquitin for binding sites on the proteasome [32,39]. Since the proteasome recognizes target proteins through hydrophobic ‘patches’ on the surface of the polyubiquitin moieties [21], it was deemed possible that the hydrophobicity of residues 3–5 of TS is a critical determinant of degradation. However, results shown in Figure 2(C) indicate that this is not the case. Thus some other feature of the region may be important. One such feature may be the high degree of disorder exhibited by the domain. This disorder may facilitate the protein's interaction with the proteasome and entry into the proteolytic chamber. Indeed, recent studies by Prakash et al. [47] showed that efficient proteasomal degradation of tightly folded substrates is mediated by the presence of an unstructured region that serves as the site for initiation of degradation. The N-terminal domain of wtTS may play a similar role.
It was rather surprising to find that C-terminal, rather than N-terminal, residues determine proteasome-mediated degradation of the intrinsically unstable P303L mutant. Deletion of as few as two amino acids from the N-terminal end of wtTS, as well as ligation of a His6 tag between residues Met1 and Pro2, completely stabilized wtTS, yet had little or no effect on P303L. On the other hand, deletion of the last nine residues from the C-terminal end, or ligation of a His6 tag to the C-terminus, stabilized the P303L enzyme, while having no effect on wtTS. Thus the primary structural determinants for proteasomal degradation of wtTS and P303L are at opposite ends of the molecules. It is likely that directional translocation of the wtTS and P303L molecules into the proteasome's catalytic chamber is initiated at different ends of the polypeptides. Thus the accessibility of the termini, which probably differs between the wtTS and mutant TS molecules, is apt to be important for proteasome recognition.
The C-terminal domain of the P303L enzyme overrides the N-terminal region in regulating TS degradation, even though the latter domain is present and unaltered in the mutant polypeptide. The C-terminal region of TS exhibits two conformations, depending on the presence or absence of ligands. In the ligand-free enzyme, the region assumes an ‘open’ conformation that allows substrate access to the active site cavity [15]. Thus one might postulate that in the P303L enzyme, this domain is disordered, and provides access to factors essential for proteasomal recognition. Detailed structural analyses of the P303L mutant have not been carried out, owing to technical difficulties associated with expressing and purifying high amounts of this enzyme. However, the C-terminal domain of a mutant form of E. coli TS containing a Pro→Asp substitution at residue 254, which is analogous to residue 303 of the human enzyme, has been shown to have an altered conformation [24]. A similar alteration may occur in the P303L form of human TS as well, leading to an increase in the region's accessibility to the proteasomal machinery. Enhanced recognition of either the proteasome itself or an accessory molecule mediating proteasomal recognition would result in the molecule exhibiting a short half-life compared with wtTS.
Although the N- and C-termini of the TS polypeptide appear to function independently, it is possible that there may in fact be an interaction between them. Blocking the C-terminus of P303L with a His6 tag resulted in complete stabilization of the molecule. It might have been expected that the modified protein would behave like wtTS in using its N-terminal determinants to acquire a half-life of approx. 9–15 h. The fact that the C-terminally His6-tagged P303L enzyme exhibited such high stability raises the possibility that the Pro→Leu substitution not only caused the adoption of the C-terminus for mediating degradation, but also blocked the ability of the N-terminus to direct degradation. Further experiments will be needed to substantiate and extend this point.
In summary, our current view is that the N-terminal end of the wtTS polypeptide (or the C-terminal end of the P303L mutant) governs ubiquitin-independent targeting of the molecule to the proteasome, and initiates its entry into the proteolytic chamber. Complete proteolysis of the enzyme requires it to unfold and translocate through the pore that protects the chamber's entrance, a process that is likely to be a common mode of ubiquitin-independent targeting of substrates to the 26 S proteasome. For example, Zhang et al. [39] have shown that proteolysis of ODC begins at the C-terminal end, and progresses in the N-terminal direction. Ligand-mediated stabilization of TS may not directly involve the protein termini, but may be a consequence of increased thermodynamic constraints on unfolding of the polypeptide chain caused by bonding contacts between ligands and specific enzyme residues. Clearly, in vitro studies are needed to define the molecular components of the proteasomal system that are necessary for degradation of wtTS and the P303L mutant.
It is interesting to point out that utilization of degradation signals at protein termini is reminiscent of the ClpX and ClpA chaperones of E. coli, which recognize an amino acid ‘tag’ sequence at one end of their various substrates. This mediates the unfolding of these substrates and directs them to the site of proteolysis catalysed by ClpP [48]. A proteomic screen of targets for the ClpXP protease revealed various sequence motifs (two in the C-terminus and three in the N-terminus) that target protein substrates for degradation [49]. Regulation of TS degradation via signals located at different ends of the molecule suggests that mechanisms similar to those in E. coli have been retained in mammalian cells, at least for a certain subset of proteins.
Acknowledgments
We thank members of F. G. B.'s laboratory for stimulating discussions and assistance with the experiments. This work was supported by grants from the National Institutes of Health (CA 44013 and CA 80361).
References
- 1.Carreras C. W., Santi D. V. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 2.Santi D. V., McHenry C. S., Raines R. T., Ivanetich K. M. Kinetics and thermodynamics of the interaction of 5-fluoro-2′-deoxyuridylate with thymidylate synthase. Biochemistry. 1987;26:8606–8613. doi: 10.1021/bi00400a017. [DOI] [PubMed] [Google Scholar]
- 3.Harwood F. G., Kasibhatla S., Petak I., Vernes R., Green D. R., Houghton J. A. Regulation of FasL by NF-κB and AP-1 in Fas-dependent thymineless death of human colon carcinoma cells. J. Biol. Chem. 2000;275:10023–10029. doi: 10.1074/jbc.275.14.10023. [DOI] [PubMed] [Google Scholar]
- 4.Berger S. H., Berger F. G. Thymidylate synthase as a determinant of 5-fluoro-2′-deoxyuridine response in human colonic tumor cell lines. Mol. Pharmacol. 1988;34:474–479. [PubMed] [Google Scholar]
- 5.Bertino J. R. Chemotherapy of colorectal cancer: history and new themes. Semin. Oncol. 1997;24:S18-13–S18-17. [PubMed] [Google Scholar]
- 6.Berger S. H., Davis S. T., Barbour K. W., Berger F. G. The role of thymidylate synthase in the response to fluoropyrimidine-folinic acid combinations. Adv. Exp. Med. Biol. 1988;244:59–69. doi: 10.1007/978-1-4684-5607-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Longley D. B., Harkin D. P., Johnston P. G. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 8.Takemura Y., Jackman A. L. Folate-based thymidylate synthase inhibitors in cancer chemotherapy. Anticancer Drugs. 1997;8:3–16. doi: 10.1097/00001813-199701000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Van der Wilt C. L., Pinedo H. M., Smid K., Peters G. J. Elevation of thymidylate synthase following 5-fluorouracil treatment is prevented by the addition of leucovorin in murine colon tumors. Cancer Res. 1992;52:4922–4928. [PubMed] [Google Scholar]
- 10.Gorlick R., Metzger R., Danenberg K. D., Salonga D., Miles J. S., Longo G. S., Fu J., Banerjee D., Klimstra D., Jhanwar S., et al. Higher levels of thymidylate synthase gene expression are observed in pulmonary as compared with hepatic metastases of colorectal adenocarcinoma. J. Clin. Oncol. 1998;16:1465–1469. doi: 10.1200/JCO.1998.16.4.1465. [DOI] [PubMed] [Google Scholar]
- 11.Chu E., Koeller D. M., Casey J. L., Drake J. C., Chabner B. A., Elwood P. C., Zinn S., Allegra C. J. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu E., Allegra C. J. The role of thymidylate synthase in cellular regulation. Adv. Enzyme Regul. 1996;36:143–163. doi: 10.1016/0065-2571(95)00004-6. [DOI] [PubMed] [Google Scholar]
- 13.Kitchens M. E., Forsthoefel A. M., Rafique Z., Spencer H. T., Berger F. G. Ligand-mediated induction of thymidylate synthase occurs by enzyme stabilization. Implications for autoregulation of translation. J. Biol. Chem. 1999;274:12544–12547. doi: 10.1074/jbc.274.18.12544. [DOI] [PubMed] [Google Scholar]
- 14.Forsthoefel A. M., Pena M. M., Xing Y. Y., Rafique Z., Berger F. G. Structural determinants for the intracellular degradation of human thymidylate synthase. Biochemistry. 2004;43:1972–1979. doi: 10.1021/bi035894p. [DOI] [PubMed] [Google Scholar]
- 15.Phan J., Steadman D. J., Koli S., Ding W. C., Minor W., Dunlap R. B., Berger S. H., Lebioda L. Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J. Biol. Chem. 2001;276:14170–14177. doi: 10.1074/jbc.M009493200. [DOI] [PubMed] [Google Scholar]
- 16.Schiffer C. A., Clifton I. J., Davisson V. J., Santi D. V., Stroud R. M. Crystal structure of human thymidylate synthase: a structural mechanism for guiding substrates into the active site. Biochemistry. 1995;34:16279–16287. doi: 10.1021/bi00050a007. [DOI] [PubMed] [Google Scholar]
- 17.Nussbaum R. L., Walmsley R. M., Lesko J. G., Airhart S. D., Ledbetter D. H. Thymidylate synthase-deficient Chinese hamster cells: a selection system for human chromosome 18 and experimental system for the study of thymidylate synthase regulation and fragile X expression. Am. J. Hum. Genet. 1985;37:1192–1205. [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw D., Berger F. G., Spencer H. T. Retroviral expression of Escherichia coli thymidylate synthase cDNA confers high-level antifolate resistance to hematopoietic cells. Hum. Gene Ther. 2001;12:51–59. doi: 10.1089/104303401450960. [DOI] [PubMed] [Google Scholar]
- 19.Hoyt M. A., Coffino P. Ubiquitin-free routes into the proteasome. Cell. Mol. Life Sci. 2004;61:1596–1600. doi: 10.1007/s00018-004-4133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman C. S., Pegg A. E. Polyamine analogues inhibit the ubiquitination of spermidine/spermine N1-acetyltransferase and prevent its targeting to the proteasome for degradation. Biochem. J. 2001;358:137–145. doi: 10.1042/0264-6021:3580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy L. W., Finer-Moore J. S., Montfort W. R., Jones M. O., Santi D. V., Stroud R. M. Atomic structure of thymidylate synthase: target for rational drug design. Science. 1987;235:448–455. doi: 10.1126/science.3099389. [DOI] [PubMed] [Google Scholar]
- 23.Kitchens M. E., Forsthoefel A. M., Barbour K. W., Spencer H. T., Berger F. G. Mechanisms of acquired resistance to thymidylate synthase inhibitors: the role of enzyme stability. Mol. Pharmacol. 1999;56:1063–1070. doi: 10.1124/mol.56.5.1063. [DOI] [PubMed] [Google Scholar]
- 24.Fantz C., Shaw D., Jennings W., Forsthoefel A., Kitchens M., Phan J., Minor W., Lebioda L., Berger F. G., Spencer H. T. Drug-resistant variants of Escherichia coli thymidylate synthase: effects of substitutions at Pro-254. Mol. Pharmacol. 2000;57:359–366. [PubMed] [Google Scholar]
- 25.Perry K. M., Carreras C. W., Chang L. C., Santi D. V., Stroud R. M. Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2′-deoxyuridine 5′-monophosphate and 5,10-methylenetetrahydrofolate. Biochemistry. 1993;32:7116–7125. doi: 10.1021/bi00079a007. [DOI] [PubMed] [Google Scholar]
- 26.Acquaviva C., Brockly F., Ferrara P., Bossis G., Salvat C., Jariel-Encontre I., Piechaczyk M. Identification of a C-terminal tripeptide motif involved in the control of rapid proteasomal degradation of c-Fos proto-oncoprotein during the G(0)-to-S phase transition. Oncogene. 2001;20:7563–7572. doi: 10.1038/sj.onc.1204880. [DOI] [PubMed] [Google Scholar]
- 27.Bloom J., Amador V., Bartolini F., DeMartino G., Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell (Cambridge, Mass.) 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 28.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciesla J., Weiner K. X., Weiner R. S., Reston J. T., Maley G. F., Maley F. Isolation and expression of rat thymidylate synthase cDNA: phylogenetic comparison with human and mouse thymidylate synthases. Biochim. Biophys. Acta. 1995;1261:233–242. doi: 10.1016/0167-4781(95)00008-5. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen-Lane J., Maley G. F., Chu E., Maley F. High-level expression of human thymidylate synthase. Protein Expression Purif. 1997;10:256–262. doi: 10.1006/prep.1997.0750. [DOI] [PubMed] [Google Scholar]
- 31.Coffino P. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2001;2:188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- 32.Zhang M., Pickart C. M., Coffino P. Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate. EMBO J. 2003;22:1488–1496. doi: 10.1093/emboj/cdg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krappmann D., Wulczyn F. G., Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκBα in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 34.Asher G., Lotem J., Sachs L., Kahana C., Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalejta R. F., Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H., Kaung G., Kobayashi S., Kopito R. R. Cytosolic degradation of T-cell receptor α chains by the proteasome. J. Biol. Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 37.Jariel-Encontre I., Pariat M., Martin F., Carillo S., Salvat C., Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J. Biol. Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 38.Tarcsa E., Szymanska G., Lecker S., O'Connor C. M., Goldberg A. L. Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26 S proteasomes without ubiquitination. J. Biol. Chem. 2000;275:20295–20301. doi: 10.1074/jbc.M001555200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M., MacDonald A. I., Hoyt M. A., Coffino P. Proteasomes begin ornithine decarboxylase digestion at the C terminus. J. Biol. Chem. 2004;279:20959–20965. doi: 10.1074/jbc.M314043200. [DOI] [PubMed] [Google Scholar]
- 40.Almrud J. J., Oliveira M. A., Kern A. D., Grishin N. V., Phillips M. A., Hackert M. L. Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding. J. Mol. Biol. 2000;295:7–16. doi: 10.1006/jmbi.1999.3331. [DOI] [PubMed] [Google Scholar]
- 41.Varshavsky A. Naming a targeting signal. Cell (Cambridge, Mass.) 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 42.Polevoda B., Sherman F. Nα-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 43.Polevoda B., Sherman F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 2003;308:1–11. doi: 10.1016/s0006-291x(03)01316-0. [DOI] [PubMed] [Google Scholar]
- 44.Matsuura S., Arpin M., Hannum C., Margoliash E., Sabatini D. D., Morimoto T. In vitro synthesis and posttranslational uptake of cytochrome c into isolated mitochondria: role of a specific addressing signal in the apocytochrome. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4368–4372. doi: 10.1073/pnas.78.7.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hershko A., Heller H., Eytan E., Kaklij G., Rose I. A. Role of the α-amino group of protein in ubiquitin-mediated protein breakdown. Proc. Natl. Acad. Sci. U.S.A. 1984;81:7021–7025. doi: 10.1073/pnas.81.22.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson G. G., Kronert W. A., Bernstein S. I., Chapman V. M., Smith K. D. Altered turnover of allelic variants of hypoxanthine phosphoribosyltransferase is associated with N-terminal amino acid sequence variation. J. Biol. Chem. 1988;263:9079–9082. [PubMed] [Google Scholar]
- 47.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 48.Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 49.Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]