Abstract
Dissipation of excess light energy in plant photosynthetic membranes plays an important role in the response of plants to the environment, providing short-term balancing between the intensity of sunlight and photosynthetic capacity. The carotenoid zeaxanthin and the photosystem II subunit PsbS play vital roles in this process, but the mechanism of their action is largely unexplained. Here we report that the isolated photosystem II subunit PsbS was able to bind exogenous zeaxanthin, the binding resulting in a strong red shift in the absorption spectrum, and the appearance of characteristic features in the resonance Raman spectrum and a distinct circular dichroism spectrum, indicating pigment–protein, as well as specific pigment–pigment, interaction. A strong shift in the absorption spectrum of PsbS phenylalanine residues after zeaxanthin binding was observed. It is concluded that zeaxanthin binding to PsbS is the origin of the well known energy dissipation-related 535-nm absorption change that we showed in vivo to arise from activation of 1–2 molecules of this pigment. The altered properties of zeaxanthin and PsbS that result from this interaction provide the first direct indication about how they regulate energy dissipation.
Plants possess a variety of features that enable them to adjust to changes in the intensity of light encountered under natural conditions (1, 2). These features serve not only to optimize photosynthesis but to provide protection from the damaging effects of absorbed radiation. Dissipation of excess excitation absorbed by the light-harvesting antenna of plant photosystem II (PSII) provides a dynamic response to short-term changes in light intensity (3, 4). It has been shown to be an important regulatory mechanism that impacts plant fitness (5). Energy dissipation is detected as the nonphotochemical quenching of chlorophyll fluorescence (qE) and is regulated by the transthylakoid pH gradient (ΔpH) and the xanthophyll cycle, the reversible deepoxidation of violaxanthin into zeaxanthin (3, 4, 6). Zeaxanthin accumulates under conditions of excess light because of the activation of violaxanthin deepoxidase by ΔpH generated under these conditions (6). In order for zeaxanthin to carry out its role in qE, it is bound to one or more of the proteins that constitute the LHCII (light-harvesting complexes of PSII)–PSII macromolecular complex (7, 8). This binding, which has the characteristics of an allosteric regulator of qE (9, 10), depends on or is stimulated by the increase in thylakoid ΔpH (11). Formation of qE is correlated with an absorption change, wavelength maximum ≈535 nm (12, 13), which we showed recently to be caused by an unusually large red shift in the absorption spectrum of one to two molecules of zeaxanthin to ≈525 nm (14). This absorption change was considered to arise from binding to the specific site in PSII involved in regulating qE, which was suggested to be on the PsbS subunit. PsbS has an obligatory role in qE (15), and it therefore has been suggested to be the site at which protons and/or zeaxanthin bind. To date, however, the evidence that PsbS can bind any pigments is still controversial (16, 17); there is certainly no evidence that PsbS can bind zeaxanthin, and therefore the identity of this key binding site of zeaxanthin is unknown.
One obstacle to the elucidation of the role of PsbS is its extreme hydrophobicity, which leads to problems of aggregation when isolated by using established methods of fractionation of thylakoid membranes (17). Here we describe a rapid procedure for preparing PsbS from spinach PSII membranes that yields a pure, soluble protein, amenable to the study of pigment binding in vitro. It was found that the PsbS preparation, when mixed with zeaxanthin, was able to bind this pigment. Moreover, after interaction with PsbS, zeaxanthin showed a strong red shift in its absorption spectrum and the appearance of a characteristic resonance Raman (RR) spectrum almost identical to those observed after qE formation in vivo. Hence, the activated state that gives rise to the 535-nm change, and which is a key event in the dissipation of excitation energy in plants, has been reconstituted in vitro.
Methods
PSII membrane fragments were prepared from spinach thylakoid membranes essentially as described (18). For PsbS preparation, the PSII membrane fragments (2 mg of total chlorophyll) were extracted with 1% n-dodecyl β-d-maltoside (DM), incubated on ice for 50 min, stirred occasionally, and centrifuged at 10,000 × g for 20 min. The pellet was extracted with 0.5% sodium cholate (Sigma), pH 7.0/250 mM NaCl (19), incubated in the dark on ice with vigorous stirring for 5 min, and centrifuged at 10,000 × g for 10 min. The supernatant was passed through a Sephadex G-25 column, and eluted fractions containing PsbS were stored in a solution of elution buffer (25 mM Hepes, pH 8.0/0.01% DM). The protein concentration was determined from the absorption spectra of the sample by using the OD of phenylalanine at 275 nm and taking into account the phenylalanine-to-protein ratio in PsbS. Protein samples were solubilized and separated by 15% denaturing SDS/PAGE using standard procedures (20). Proteins were transferred to a poly(vinylidene difluoride) membrane (Hybond-P, Amersham Pharmacia) in a Mini Trans-Blot transfer cell (Bio-Rad) at 30 mA for 12 h. Membranes were probed with an antibody raised in rabbit against a 12-aa synthetic peptide (GDRGRFVDEPTT) that was completely specific to spinach PsbS. The primary antibody was detected by using a horseradish peroxidase-labeled secondary antibody using an ECL Plus kit (Amersham Pharmacia). Chemiluminescence was detected on Hyperfilm ECL (Amersham Pharmacia) photographic film and developed for 20 min. Zeaxanthin was purified from orange peppers and dried onto the surface of a glass tube under a stream of N2. Binding of zeaxanthin to PsbS was achieved by adding 1 ml of 9 μM PsbS solution to a glass tube containing 14 nmol of dried zeaxanthin and mixing for 30 s with sonication in an ultrasonic bath. Sucrose-gradient centrifugation was carried out as described with a DM concentration of 0.3% (8). Absorption spectra were recorded by using a Cary 500 spectrophotometer at room temperature. Circular dichroism (CD) spectra were obtained by using a Jasco J810 spectropolarimeter. RR spectra were recorded as described (14).
Results
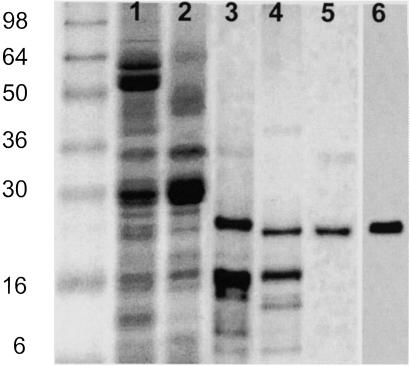
Extraction of PSII membrane fragments (Fig. 1, lane 2) with DM leaves a pellet enriched in a protein that has an apparent molecular mass of 22 kDa (lane 3). Treatment of this sample with cholate, shown to be useful for extracting PsbS (19), brings about solubilization of many of the proteins in the DM pellet, with some apparent enrichment of this 22-kDa protein (lane 4). This cholate extract was passed through a Sephadex G-25 column, yielding a sample that was assessed to be 90% pure in this protein from observation of the silver-stained gel (lane 5). The protein reacts with an antibody raised against a PsbS peptide (lane 6) that does not crossreact with any other PSII protein. We conclude that this represents a highly purified preparation of PsbS. The absorption spectrum of this PsbS preparation shows that it contained no bound pigment; if the absorption spectrum shown in Fig. 2A is extended into the visible region, there is no recorded absorption from either carotenoid or chlorophyll. In contrast, a sample of LHCII at the same protein concentration would give an OD of ≈20 in the red region of the spectrum.
Fig 1.
Purification of PsbS. Silver-stained SDS polyacrylamide gel showing spinach thylakoids (lane 1), PSII membrane fragments (lane 2), pellet after extraction with DM (lane 3), supernatant after extraction with cholate (lane 4), and after passing down a Sephadex G-25 column (lane 5). A Western blot of lane 5 with anti-PsbS antibody is shown in lane 6. Molecular weight markers (See Blue, Invitrogen) are shown in the far-left lane.
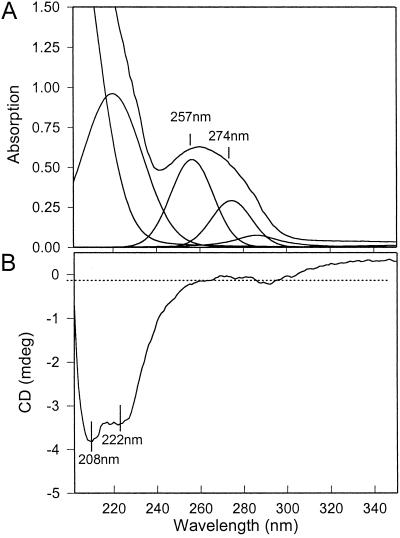
Fig 2.
Absorption (A) and CD (B) spectra of the preparation of purified PsbS. The absorption spectrum was deconvoluted, revealing the bands at 257 and 274 nm arising from phenylalanine and tyrosine, respectively.
Spectroscopic analysis was used to further establish the purity of the PsbS preparation. The published amino acid sequence of spinach PsbS indicates the presence of 17 phenylalanines and 1 tyrosine (21, 22). By using the extinction coefficients at 257 and 274 nm of 0.19 × 105 and 1.25 × 105 M−1⋅m−1, respectively, a theoretical absorption ratio of phenylalanine to tyrosine of 2.5 is predicted. The absorption spectrum of the PsbS preparation was deconvoluted to show the contributions from phenylalanine and tyrosine (Fig. 2A), and these data gave a phenylalanine/tyrosine ratio of 2.0. A very minor contribution from tryptophan (not present in the PsbS sequence) at 288 nm is evident, but given the large extinction coefficient of this amino acid, the level of contamination by other proteins would appear to be minimal. The CD spectrum of the PsbS preparation shows the presence of a secondary α-helical structure with characteristic minima at 208 and 222 nm (Fig. 2B), providing evidence that the protein was not denatured and suggesting that it had been isolated in a native form.
We tested whether purified PsbS would bind zeaxanthin. For this, zeaxanthin deposited as a dry film on the surface of a glass tube was washed with a solution of PsbS. (This procedure was found to be more satisfactory than mixing zeaxanthin dissolved in ethanol with the PsbS preparation, although qualitatively similar results were obtained with both methods.) The mixture then was analyzed by sucrose-gradient centrifugation, and it was found that 60% of the zeaxanthin was located in the PsbS-containing band at ≈0.3 M sucrose. Very little free carotenoid was detected. It was concluded that PsbS was able to bind zeaxanthin to form a PsbS–zeaxanthin complex. We estimated the molar ratio of zeaxanthin/PsbS to be ≈2:1 in this complex.
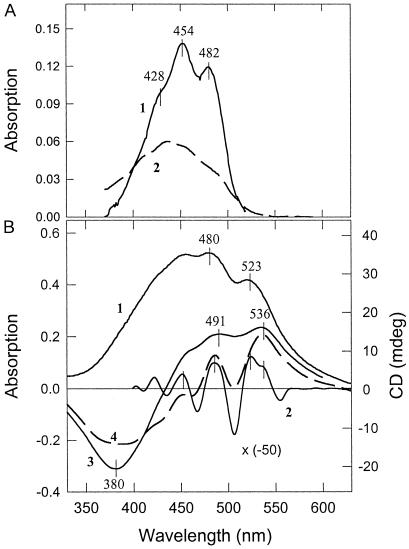
Zeaxanthin dissolved in ethanol has a characteristic absorption spectrum with maxima at 482, 454, and 428 nm (Fig. 3A, trace 1). The positions of these maxima can be shifted depending on the polarity of the solvent, the 0–0 transition at 482 nm in ethanol shifting to 511 nm in CS2 (14). When zeaxanthin and PsbS are mixed, the absorption spectrum of zeaxanthin was altered dramatically; there was a strong red shift, with the 0–0 absorption maximum position appearing at ≈523 nm (Fig. 3B, trace 1). The derivative spectrum shows maxima at 525, 487, and 450 nm, with evidence of a secondary band at ≈536 nm (Fig. 3B, trace 2). It should be noted that mixing of zeaxanthin with the aqueous buffer minus PsbS causes a quite different effect (Fig. 3A, trace 2): the appearance of the slightly blue-shifted bands, arising probably from some aggregation of zeaxanthin, similar to that described (23). A spectrum the same as trace 2 was obtained when zeaxanthin was mixed with BSA dissolved in buffer (not shown). Under the same reconstitution conditions, there was no evidence of formation of a complex between violaxanthin and PsbS; there was no difference between the absorption spectra of violaxanthin mixed with PsbS and with the detergent buffer alone.
Fig 3.
(A) Absorption spectra of zeaxanthin in ethanol (1) and in detergent buffer used in reconstitution experiments (2). (B) 1, Absorption spectrum of PsbS reconstituted with zeaxanthin; 2, second derivative of spectrum 1; 3, CD spectrum of PsbS reconstituted with zeaxanthin; 4, simulated CD spectrum of two excitonically coupled zeaxanthin molecules with the higher and lower exciton components at 507 and 536 nm, respectively.
The PsbS–zeaxanthin complex displayed a CD spectrum not found for either constituent alone. There were positive bands at 491 and 536 nm and a strong negative band at 380 nm (Fig. 3B, trace 3). These negative and positive symmetrical features can be attributed to the negative and positive Cotton effects of excitonically coupled pigments (24), suggesting that two zeaxanthin molecules are interacting. Indeed, because the CD spectrum has a differential nature, it can be modeled by using two shifted-absorption spectra. Fig. 3B (trace 4) represents the difference between spectrum 1 and the same spectrum with a 15-nm blue shift. It is clear that both the negative and positive parts resemble those of the CD spectrum (trace 3). The only difference is that the positive group of bands is better resolved, and there is a clear minimum at 507 nm, the higher excitonic component. This difference could be due to a presence of the band at 525 nm, which is seen clearly in the second derivative absorption spectrum (trace 2). This transition could belong to a monomeric zeaxanthin bound to the protein. These features of the CD spectrum demonstrate that PsbS can exert strong effects on the properties of zeaxanthin, indicative of binding.
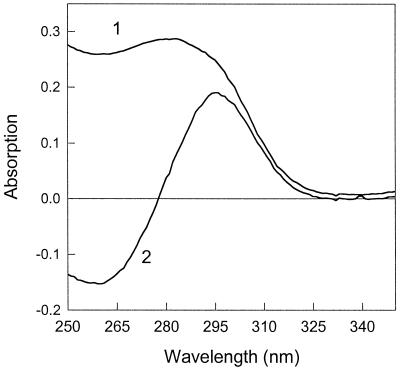
It was noted also that the absorption spectrum of PsbS in the UV region was altered by the presence of zeaxanthin; the absorption band at 258 nm, arising mainly from phenylalanine (see Fig. 2A), was shifted to 280 nm (Fig. 4, spectrum 1). The conserved nature of the corresponding difference spectrum shows that the change can be explained by a red shift in a population of phenylalanine residues in the PsbS sample (Fig. 4, spectrum 2). This is further evidence of a rather specific interaction between zeaxanthin and PsbS.
Fig 4.
UV absorption spectrum of PsbS reconstituted with zeaxanthin (1) and a (PsbS+zeaxanthin) − (PsbS only) absorption difference spectrum (2).
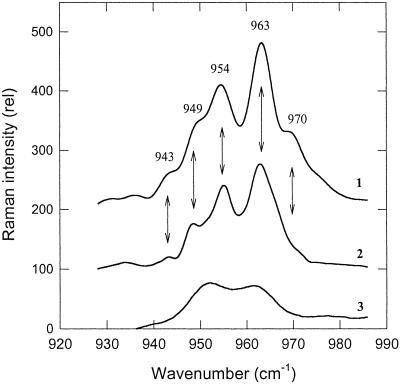
In vivo, the appearance of the red-shifted zeaxanthin was associated with characteristic features in the RR spectrum (14). The ν4 region of the RR spectrum arises from wagging vibrations of various C–H groups (25), and bound red-shifted zeaxanthin in vivo displays five new transitions not found in zeaxanthin dissolved in detergent or solvent. This zeaxanthin “fingerprint” is shown in Fig. 5 (spectrum 1), in comparison with the rather featureless spectrum of zeaxanthin in detergent micelles (spectrum 3). The in vitro PsbS–zeaxanthin complex displayed an almost identical RR spectrum to that associated with qE (spectrum 2). The two major bands at 954 and 963 cm−1 were the same in vivo and for the PsbS–zeaxanthin complex, although the minor bands varied slightly in intensity and position in the two spectra.
Fig 5.
RR spectra of zeaxanthin in ν4. 1, qE-activated zeaxanthin absorbing at 535 nm (modified from ref. 14). 2, zeaxanthin bound to PsbS obtained as described for Fig. 3; and 3, zeaxanthin in detergent-lipid micelles purified on a sucrose gradient (modified from ref. 14). The arrows indicate the five main transitions associated with zeaxanthin activation.
Discussion
The dissipation of excitation energy in PSII, qE, correlates strongly to an absorption change at 535 nm (12, 13). Recently we showed that the 535-nm change could be explained by a strong red shift in the absorption spectrum of one to two molecules of zeaxanthin (14). The appearance of red-shifted zeaxanthin absorbing at 523–525 nm, compared with ≈505 nm in the absence of qE, gives rise to a band at 535 nm in the qE difference spectrum. Here we provide evidence to support the hypothesis that this “activated” zeaxanthin is bound to PsbS, because we successfully reconstituted a zeaxanthin–PsbS complex that shows a similarly strong red shift to ≈523 nm. This shift was found to be sufficient to give rise to a 535-nm band in a difference spectrum calculated by subtracting an absorption spectrum of “nonactivated” zeaxanthin, simulated by using a spectrum recorded in CS2 as described (14). Therefore, these data represent the first direct link between zeaxanthin and PsbS, which both play vital roles in qE (6, 15). The data also provide an explanation of why the 535-nm change is absent in the npq4 mutant of Arabidopsis, which lacks this protein (15).
The zeaxanthin–PsbS complex has several important features. In addition to the large red shift in the absorption spectrum of zeaxanthin, there is a strong CD spectrum of zeaxanthin with a conserved nature and an alteration in the UV-absorption spectrum of the protein. These features indicate that PsbS provides a highly polarizing environment that strongly affects the photophysical properties of zeaxanthin and, conversely, that zeaxanthin binding alters the structure of PsbS, precisely the helical regions, which contain all the phenylalanine residues. Specifically, it is in the predicted fourth helix, present in PsbS but not in the Lhcb proteins (21, 22), that numerous phenylalanine residues are found, suggesting that this region of the protein provides the binding site for zeaxanthin. Violaxanthin does not appear to bind to this site. In vivo, zeaxanthin is bound to all the Lhcb proteins (7, 8), and in vitro zeaxanthin can induce fluorescence quenching in these proteins (26–28). However, thus far we have not observed any red shifts in the absorption spectra of zeaxanthin bound to these proteins (29). It is interesting that in Chlamydomonas, mutation of the Lhcbm1 protein results in a loss of qE (30). It is possible that in this organism, the much weaker qE (compared with higher plants) is controlled by zeaxanthin binding to LHCII.
The features of the CD spectrum of the zeaxanthin–PsbS complex can be interpreted as resulting from two processes. First, there is an excitonic interaction between two bound zeaxanthin molecules (507- and 535-nm components of a splitting), consistent also with the upper estimate of two molecules bound (both in vitro and in vivo). Second, there is binding of a single pigment with absorption shifted to ≈520 nm. The presence of these two effects indicates some heterogeneity in the binding stoichiometry in the complex. The enhancement of the RR spectrum of zeaxanthin provides further compelling evidence of a strong and specific interaction between zeaxanthin and PsbS, which establishes a particular configuration of the bound zeaxanthin. Moreover, the changes in the absorption and RR spectrum after reconstitution of the zeaxanthin–PsbS complex have the same features found in vivo, suggesting that the binding of zeaxanthin to PsbS in vitro is mimicking a similar process occurring in vivo.
These conclusions add considerably to the understanding of the chain of events that lead to energy dissipation in PSII under excess light conditions. The two factors that control this process are the thylakoid ΔpH and the deepoxidation of violaxanthin to zeaxanthin. The interaction between these factors, in which zeaxanthin is an allosteric activator of qE (9, 10), suggests that they bind to the same protein or protein complex. Recently, evidence has been obtained that certain carboxyl amino acids on PsbS are essential for qE formation (31). Here we show that PsbS can bind zeaxanthin, and the features of this binding are consistent with a similar binding associated with in vivo qE (14), suggesting that the qE-associated activation of zeaxanthin arises because of its binding to PsbS. We should point out that zeaxanthin binding to PsbS in vitro was not pH-dependent (data not shown). We suggest that in vivo, binding is prevented by structural constraints imposed by the local environment in the thylakoid membrane. A conformational change in PsbS induced by protonation of these carboxyl amino acids would be necessary then for zeaxanthin binding.
The subsequent event, how the protonated PsbS–zeaxanthin complex is able to interact with the antenna chlorophyll to bring about dissipation of excited states, of course, is not resolved. However, given the abundant evidence of the intrinsic quenching capabilities of isolated light-harvesting proteins (26–28) and the similarities between this process and in vivo qE (3), the most favored hypothesis is that an interaction between PsbS and one or more of these proteins controls the dissipation process (32). We suggest that this interaction is controlled in turn by the protonation and binding of zeaxanthin to PsbS. The successful reconstitution of PsbS and zeaxanthin reported here should enable further testing of this hypothesis.
Acknowledgments
This work was supported by a grant from the United Kingdom Biotechnology and Biological Sciences Research Council (to P.H.) and grants from the Joint Infrastructure Fund and Joint Research Equipment Initiative (to the Department of Molecular Biology and Biotechnology, University of Sheffield).
Abbreviations
PSII, photosystem II
ΔpH, transthylakoid proton gradient
qE, nonphotochemical quenching dependent on ΔpH
LHCII, light-harvesting complexes of PSII
RR, resonance Raman
DM, n-dodecyl β-d-maltoside
CD, circular dichroism
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Björkman O. & Demmig-Adams, B. (1995) in Ecophysiology of Photosynthesis: Ecological Studies, eds. Schulze, E. D. & Caldwell, M. M. (Springer, Berlin), pp. 14–47.
- 2.Horton P., Murchie, E. H., Ruban, A. V. & Walters, R. G. (2001) Novartis Found. Symp. 236, 117-134. [DOI] [PubMed] [Google Scholar]
- 3.Horton P., Ruban, A. V. & Walters, R. G. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655-684. [DOI] [PubMed] [Google Scholar]
- 4.Niyogi K. K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333-360. [DOI] [PubMed] [Google Scholar]
- 5.Külheim C., Ägren, J. & Jansson, S. (2002) Science 297, 91-93. [DOI] [PubMed] [Google Scholar]
- 6.Demmig-Adams B. (1990) Biochim. Biophys. Acta 1020, 1-24. [Google Scholar]
- 7.Ruban A. V., Young, A. J., Pascal, A. A. & Horton, P. (1994) Plant Physiol. 104, 227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruban A. V., Lee, P. J., Wentworth, M. W., Young, A. J. & Horton, P. (1999) J. Biol. Chem. 274, 10458-10465. [DOI] [PubMed] [Google Scholar]
- 9.Ruban A. V., Wentworth, M. & Horton, P. (2001) Biochemistry 40, 9896-9901. [DOI] [PubMed] [Google Scholar]
- 10.Wentworth M., Ruban, A. V. & Horton, P. (2001) Biochemistry 40, 9902-9908. [DOI] [PubMed] [Google Scholar]
- 11.Gilmore A. M., Shinkarev, V. P., Hazlett, T. L. & Govindjee, (1998) Biochemistry 37, 13582-13593. [DOI] [PubMed] [Google Scholar]
- 12.Ruban A. V., Young, A. J. & Horton, P. (1993) Plant Physiol. 102, 741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilger W. & Björkman, O. (1994) Planta 193, 238-246. [Google Scholar]
- 14.Ruban A. V., Pascal, A. A., Robert, B. & Horton, P. (2002) J. Biol. Chem. 277, 7785-7789. [DOI] [PubMed] [Google Scholar]
- 15.Li X.-P., Björkman, O., Shih, C., Grossman, A. R., Rosenquist, M., Jansson, S. & Niyogi, K. K. (2000) Nature 403, 391-395. [DOI] [PubMed] [Google Scholar]
- 16.Funk C., Schroder, W. P., Napowotzki, A., Tjus, S. E., Renger, G. & Andersson, B. (1995) Biochemistry 34, 11133-11141. [DOI] [PubMed] [Google Scholar]
- 17.Dominici P., Caffarri, S., Armenante, F., Ceoldo, S., Crimi, M. & Bassi, R. (2002) J. Biol. Chem. 277, 22750-22758. [DOI] [PubMed] [Google Scholar]
- 18.Berthold D. A., Babcock, G. T. & Yocum, C. F. (1981) FEBS Lett. 134, 231-234. [Google Scholar]
- 19.Bowlby N. R. & Yocum, C. F. (1993) Biochim. Biophys. Acta 1144, 271-277. [Google Scholar]
- 20.Laemmli U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 21.Kim S., Sandusky, P., Bowlby, N. R., Aebersold, R., Green, B. R., Vlahakis, S., Yocum, C. F. & Pichersky, E. (1992) FEBS Lett. 314, 67-71. [DOI] [PubMed] [Google Scholar]
- 22.Wedel N., Klein, R., Ljungberg, U., Andersson, B. & Herrmann, R. G. (1992) FEBS Lett. 314, 61-66. [DOI] [PubMed] [Google Scholar]
- 23.Ruban A. V., Horton, P. & Young, A. J. (1993) J. Photochem. Photobiol. B 21, 229-234. [Google Scholar]
- 24.Zsila F., Bikadi, Z. & Simonyi, M. (2002) Tetrahedron: Assymetry 13, 273-283. [Google Scholar]
- 25.Robert B. (1999) in The Photochemistry of Carotenoids, eds. Frank, H., Young, A., Britton, G. & Cogdell, R. (Kluwer, Dordrecht, The Netherlands), pp. 189–201.
- 26.Ruban A. V., Young, A. J. & Horton, P. (1994) Biochim. Biophys. Acta 1186, 123-127. [Google Scholar]
- 27.Ruban A. V., Young, A. J. & Horton, P. (1996) Biochemistry 35, 674-678. [DOI] [PubMed] [Google Scholar]
- 28.Wentworth M., Ruban, A. V. & Horton, P. (2000) FEBS Lett. 471, 71-74. [DOI] [PubMed] [Google Scholar]
- 29.Ruban A. V., Pascal, A., Lee, P. J., Robert, B. & Horton, P. (2002) J. Biol. Chem. 277, 42937-42942. [DOI] [PubMed] [Google Scholar]
- 30.Elrad D., Niyogi, K. K. & Grossman, A. R. (2002) Plant Cell 14, 1801-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X.-P., Phippard, A., Pasari, J. & Niyogi, K. K. (2002) Funct. Plant Biol. 29, 1131-1139. [DOI] [PubMed] [Google Scholar]
- 32.Horton P., Ruban, A. V. & Wentworth, M. (2000) Philos. Trans. R. Soc. London B 355, 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]