Abstract
The most dramatic phase change in plants is the transition from vegetative to reproductive growth. This flowering process is regulated by several interacting pathways that monitor both the developmental state of the plants and environmental cues such as light and temperature. The flowering-time genes FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), together with the floral meristem identity gene LEAFY (LFY), are three essential regulators integrating floral signals from multiple pathways in Arabidopsis thaliana. Part of the crosstalk among these genes is mediated by a putative transcription factor, AGAMOUS-LIKE 24 (AGL24). This gene is gradually activated in shoot apical meristems during the floral transition and later located in the whole zone of both inflorescence and floral meristems. Loss and reduction of AGL24 activity by double-stranded RNA-mediated interference result in late flowering, whereas constitutive overexpression of AGL24 causes precocious flowering. The correlation between the level of AGL24 accumulation and the alteration of flowering time suggests that AGL24 is a dosage-dependent flowering promoter. Analysis of AGL24 expression in various flowering-time mutants shows that it is regulated in several floral inductive pathways. Further genetic analyses of epistasis indicate that AGL24 may act downstream of SOC1 and upstream of LFY.
Multiple genetic pathways in response to developmental and environmental signals coordinate the transition from vegetative to reproductive development in Arabidopsis thaliana (1, 2). The autonomous pathway responds to endogenous signals from specific developmental states, whereas the photoperiod and vernalization pathways monitor environmental conditions, such as light and temperature. The pathway mediated by gibberellic acid (GA) plays a particularly promotive role in flowering under noninductive photoperiods, especially in Arabidopsis. Analysis of flowering mutants and natural variation in different ecotypes in Arabidopsis has revealed >80 loci that are related to the control of flowering time. To date, at least 20 genes that affect flowering time have been identified and assigned to distinct genetic pathways by the investigation of mutant phenotypes and epistatic relationships (3–5).
Recent striking advances have shown that the flowering-time genes FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), in parallel with the meristem identity gene LEAFY (LFY), are important regulators integrating floral inductive signals from multiple promotion pathways (6, 7). FT and LFY mostly regulate independently of each other downstream from CONSTANS (CO), a critical gene that promotes flowering in response to long days (7–10). Although several pathways may simultaneously play roles in the regulation of FT and LFY, it is clear that FT, but not LFY, is one of the early targets of CO in the photoperiod pathway (7). SOC1 is another essential integrator positively regulated by not only the redundant vernalization and autonomous pathways but also the photoperiod pathway (7, 11). It has been demonstrated that SOC1 is also one early target of CO with possible interactions with FT and LFY (7, 11). In the suggested scenario (11), LFY acts at least in part downstream of SOC1, whereas FT plays a role in the regulation of SOC1. So far, substantial gaps still remain in our understanding of the contexts in which these basic regulators integrate floral signals and crosstalk to act synergistically in the control of flowering time.
Double-stranded RNA (dsRNA)-mediated interference, which is the approach that simultaneously expresses both antisense and sense fragments of a specific gene in transgenic organisms, has provided consistent and efficient suppression of target genes in plants (12–14). Compared with other classical methods for reverse genetic screening of mutants, it is possible to generate a series of specific mutants with gradually reduced gene expression by the application of RNA interference (RNAi). Furthermore, genetic interference by RNAi in plants was shown to be stably heritable (14), thus facilitating the further investigation of gene functions and interactions by applying the RNAi materials with genetic methods.
By the generation of dsRNA-mediated interference with expression of the AGL24 gene, we created both agl24 loss- and reduction-of-function mutants. Molecular genetic studies of AGL24 in this study suggest that AGL24 is a basic dosage-dependent promoter acting downstream of SOC1 in the regulation of flowering time in Arabidopsis.
Materials and Methods
Plant Materials.
A. thaliana ecotype Columbia was grown in long days (LD, 16 h light/8 h dark) or short days (SD, 16 h light/8 h dark), at 23 ± 2°C. The late-flowering mutants co-1, gi-1, ft-1, fve-3, and soc1 are in the Columbia background, and co-2, gi-3, ft-1, fve-1, fpa-1, fca-1, and soc1 are in the Landsberg erecta background. soc1 introgressed into Ler and ft-1 into Columbia was provided by I. Lee (Seoul National University, Seoul, South Korea) and D. Weigel (Salk Institute, San Diego), respectively. FRI FLC line is a Columbia near-isogenic line described previously (15). agl24 is an En transposon line in the Columbia background, which was provided by M. Yanofsky (University of California, San Diego). agl24 LFY:GUS was generated by crossing the Landsberg erecta LFY:GUS line (DW 150-304) with the Ler near-isogenic agl24 line (HY17), which was obtained by three backcrosses of the agl24 Columbia line into Ler.
Analysis of AGL24 Expression.
For GA treatment, exogenous GA (100 μM) was sprayed onto wild-type Columbia plants grown under SDs with visible floral buds for two consecutive days. The inflorescence apices were harvested with and without GA treatment for the RT-PCR analysis. For vernalization treatment, seeds were sown on Murashige and Skoog (Life Technologies, Grand Island, NY) agar plates and incubated at 4°C under low-light levels for 6 weeks. Although no obvious circadian modulation of AGL24 RNA accumulation was found, all of the samples were harvested at the same time of day. The amplified RT-PCR products were fractionated on agarose gels, transferred onto nylon membranes (Roche Molecular Biochemicals), and hybridized with the appropriate digoxigenin-labeled DNA probes (Roche Molecular Biochemicals). Signal intensity was detected by autoradiography and determined with Gel-Pro image analyzer (Media Cybernetics, Silver Spring, MD).
Primers used for RT-PCR were as follows: AGL24-SP1 (5′-GGATGAGAATAAGAGACTGAGGGATAAAC-3′) and AGL24-SP2 (5′-GACCCAATAACACGTACAATATCTGAAA-C-3′) for AGL24; AP1-P1 (5′-GCACCTGAGTCCGACGTC-3′) and AP1-P2 (5′-GCGGCGAAGCAGCCAAGG-3′) for APETALA1 (AP1); FLC-P1 (5′-GAGAAGCCATGGGAAGAAAAAAACTAG-3′) and FLC-P2 (5′-TTAAGGTGGCTAATTAAGTAGTGGGAG-3′) for FLC; LFY-P1 (5′-TGAAGGACGAGGAGCTT-3′) and LFY-P2 (5′-TTGCCACGTGCCACTTC-3′) for LEAFY (LFY); and TUB-P1 (5′-ATCCGTGAAGAGTACCCAGAT-3′) and TUB-P2 (5′-TCACCTTCTTCATCCGCAGTT-3′) for β-tubulin (TUB2). RT-PCR analysis was repeated three times using samples that were collected separately.
For Northern blot analysis, total RNA was fractionated on 1% glyoxal-agarose gels and transferred to positively charged nylon membranes (Roche Molecular Biochemicals) by capillary blotting. RNA gel blots were hybridized and detected as described (16).
Construction of 35S:AGL24.
The entire AGL24 cDNA was amplified by RT-PCR on the total RNA extracted from Columbia inflorescence stems and cloned into pGEM-T Easy Vector (Promega, Madison, WI) to yield pHY1. The primers used were AGL24-G1 (5′-AGAACAGTAGTGAAGGAGAGATCTGGTAA-3′) and AGL24-G2 (5′-ATTTGTGGGCTTCCATCGAAGTCAACTCT-3′). The AGL24 cDNA with blunt ends was subsequently obtained from the pHY1 by PCR amplification with Vent DNA polymerase (New England Biolabs) and AGL24-G1 and SP6 primers. The resulting PCR products were cut with SacI to produce 3′ cohesive end and cloned into the SmaI and SacI sites of pBI121 binary vector (CLONTECH) downstream of the cauliflower mosaic virus 35S promoter in the place of β-glucuronidase gene (GUS). The construct was sequenced to eliminate selection of PCR-introduced mutations.
Construction of AGL24 dsRNA Interference (AGL24-RNAi) Plasmid.
For the construction of AGL24-RNAi plasmid, the GUS fragment containing nucleotides 787–1,809 was used as a loop linker between the AGL24 3′ end-specific fragments in the antisense and sense orientations (14). The 540-bp AGL24 gene-specific region was produced by PCR amplification with the primers AGL24-SP2 and AGL24-SP3 (5′-GTCGAAGACAAAACCAAGCAGCTACG-3′). The GUS loop was amplified by the primers GUS1 (5′-GATATCTACCCGCTTCGC-3′) and the GUS2-Sense linker (5′-CTTGGTTTTGTCTTCGACTCATTGTTTGCCTCCCT-3′). The linked GUS:Sense fragment was subsequently generated by PCR amplification on both the templates of the AGL24 gene-specific region and GUS loop using the primers GUS1 and AGL24-SP2. This fragment was then cloned into the SmaI and SacI sites of pBI121 vector (CLONTECH) downstream of the cauliflower mosaic virus 35S promoter and instead of GUS gene (pHY5). AGL24 gene-specific sequence in the antisense orientation with XbaI and BamHI sites on respective ends was created by PCR mutations, digested with the corresponding enzymes, and cloned into the XbaI and BamHI sites of the pHY5 vector upstream of the GUS:Sense insert. The sequence of this AGL24-RNAi plasmid was also confirmed to eliminate selection of possible mutations introduced by PCR.
Plant Transformation.
The binary vectors, harboring the cassettes of 35S:AGL24 and AGL24-RNAi, were introduced into Agrobacterium tumefaciens LBA4404 by triparental mating. Arabidopsis ecotype Columbia was transformed by using the floral dip method (17). Transformants were subsequently selected on half-strength Murashige and Skoog medium containing 50 mg/liter kanamycin.
In Situ Hybridization.
For synthesis of the antisense and sense AGL24 RNA probes, the 3′ end gene-specific region was amplified with AGL24-SP1 and AGL24-SP2 primers, introduced into the pGEM-T Easy vector (Promega), and transcribed in vitro using the digoxygenin (DIG) RNA Labeling kit (Roche Molecular Biochemicals). Tissues were fixed in a solution of formaldehyde/acetic acid/ethanol (3:5:60 vol/vol) at 4°C overnight. The fixed materials were dehydrated, cleared, and embedded in paraffin. Microtome sections (8 μm thick) were mounted on Superfrost Plus slides (Fisher Scientific). In situ hybridization and immunological detection were performed as described by Yu et al. (18).
GUS Staining.
In situ localization of GUS activity was performed according to the method of Sieburth and Meyerowitz (19). Tissues were prefixed in 90% acetone on ice for 20 min and stained at 37°C overnight in a solution containing 50 mM sodium phosphate (pH 7.0), 10 mM EDTA, 2 mM 5-bromo-4-chloro-3-indoyl glucuronide, 1 mM potassium ferricyanide, and 1 mM potassium ferrocyanide. Stained tissues were cleared of chlorophyll in an ethanol series. Except for tissues used for direct observation, other tissues were fixed, and embedded in paraffin. Ten-micrometer sections were made, briefly incubated in xylene and photographed on a microscope (TMS-F, Nikon).
Results and Discussion
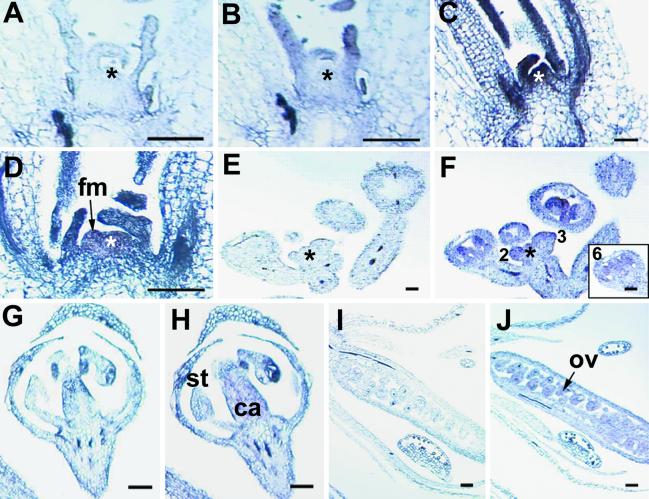
AGL24 encodes a typical MADS-domain protein containing the conserved MADS-box at its N-terminal end and the relatively conserved K-box located between residues 88 and 156 (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Sequence alignment showed that in Arabidopsis, AGL24 has the highest similarity to the SHORT VEGETATIVE PHASE (SVP) gene, which is a negative regulator of the floral transition in Arabidopsis (20). In situ localization of AGL24 transcripts showed that AGL24 was expressed in the whole zone of the vegetative shoot apical meristem and emerging leaf primordia, as well as the provascular strands of relatively old leaves (Fig. 1 A–C). During floral transition, high levels of AGL24 mRNA were detected in both the shoot apex and the emerging floral meristem (Fig. 1D). At a later stage, AGL24 mRNA was distributed throughout the inflorescence meristem and the stage 2–3 flowers (Fig. 1 E and F). In the stage 6 flower, the AGL24 gene expression was mostly confined to the carpel and stamen primordia (Fig. 1F). The accumulation of AGL24 mRNA was obvious in the carpel (Fig. 1 G and H) and ovules (Fig. 1 I and J) in the developing flower. These expression patterns suggest that AGL24 might play roles in the regulation of flowering time and the development of floral organs.
Fig 1.
In situ localization of AGL24 expression in wild-type plants. Sections (B–D, F, H, and J) are hybridized with the antisense probe, and control sections (A, E, G, and I) are hybridized with the sense probe. Asterisks indicate shoot apical meristem. (A and B) An 8-day-old seedling. (C) A 12-day-old seedling. (D) A 16-day-old seedling. (E and F) An inflorescence apex and a stage 6 flower (Inset). (G and H) A stage 8 flower. (I and J) A stage 15 flower. fm, floral meristem; st, stamen; ca, carpel; ov, ovule. (Bars = 100 μm.)
AGL24 Is a Dosage-Dependent Promoter of Flowering.
To investigate the function of the AGL24 gene, we introduced AGL24 dsRNA-expressing constructs into Arabidopsis with Agrobacterium-mediated transformation. We isolated a total of 36 transformants, among which 30 plants showed different degrees of late-flowering phenotype. These plants were grouped into two categories: reduction- and loss-of function mutants. The AGL24 dsRNA interference (AGL24-RNAi) strong mutants show typical late flowering under both LD and SD conditions, which is comparable to agl24 loss-of-function mutants (Table 1; Fig. 2C, D, G, and H; and R. M. Amasino, personal communication). It is noteworthy that the AGL24-RNAi weak mutants show intermediate reduction-of-function phenotypes between wild-type and loss-of-function plants corresponding to the endogenous levels of AGL24 mRNA (Figs. 2 A–H and 3D; Table 2, which is published as supporting information on the PNAS web site), which is similar to the semidominant nature of agl24 mutants (Table 1). These data demonstrate that AGL24 is a dosage-dependent promoter of flowering. This suggestion is further supported by the results of constitutive expression of AGL24 in transgenic Arabidopsis plants. Among 47 35S:AGL24 transgenic plants, 35 plants demonstrated early flowering under both LDs and SDs (Table 1; Fig. 2 A–H). Also, we observed that the up-regulated level of AGL24 transcripts was closely related to the degree of precocious flowering (Fig. 3D; Table 3, which is published as supporting information on the PNAS web site). Simultaneously, although we have not observed obvious defects in flower development in RNAi mutant lines, overexpression of AGL24 caused some phenotypic aberrations in flower shape and size, which indicates its potential function in flower development as revealed by its expression patterns.
Table 1.
Comparison of flowering times of transgenic and mutant plants
| Genotype | Rosette leaves | n |
|---|---|---|
| LD | ||
| Columbia wild type | 16.3 ± 0.9 | 35 |
| 35S:AGL24 (no. 3-6) | 14.7 ± 1.0 | 18 |
| 35S:AGL24 (no. 8-4) | 11.9 ± 0.8 | 32 |
| AGL24-RNAi (no. 18-3) | 19.2 ± 0.7 | 27 |
| AGL24-RNAi (no. 35-17) | 23.2 ± 1.4 | 29 |
| agl24 | 23.4 ± 1.5 | 37 |
| agl24/+ | 18.7 ± 1.7 | 9 |
| 35S:SOC1 | 4.4 ± 0.3 | 32 |
| 35S:SOC1 agl24 | 9.8 ± 0.6 | 35 |
| soc1 | 29.8 ± 1.7 | 19 |
| 35S:AGL24 (no. 8-4) soc1 | 18.8 ± 0.9 | 18 |
| 35S:FT | 3.7 ± 0.6 | 34 |
| 35S:FT agl24 | 5.6 ± 0.7 | 29 |
| 35S:LFY | 12.3 ± 1.2 | 19 |
| 35S:LFY agl24 | 13.2 ± 1.5 | 27 |
| SD | ||
| Columbia wild type | 45.6 ± 2.3 | 45 |
| 35S:AGL24 (no. 3-6) | 36.6 ± 1.7 | 23 |
| 35S:AGL24 (no. 8-4) | 25.7 ± 1.5 | 33 |
| AGL24-RNAi (no. 18-3) | 52.3 ± 1.1 | 26 |
| AGL24-RNAi (no. 35-17) | 65.0 ± 0.8 | 43 |
| agl24 | 64.7 ± 0.9 | 46 |
| soc1 | 75.6 ± 0.7 | 33 |
All of the plants are of the same Columbia background.
Flowering time is presented as the number of rosette leaves on the main shoot when the inflorescence was ≈3 cm in length.
LD, 16 h light/8 h dark; SD, 8 h light/16 h dark.
Fig 2.
Phenotypes of transgenic plants in Columbia background. (A–D) Transgenic plants under long days. At 30 days after germination, an early flowering 35S:AGL24 line (A) is compared with a wild-type plant (B), which has not bolted. At 40 days, a bolting (black arrow) AGL24-RNAi weak line (C) is compared with an AGL24-RNAi strong line (D), whose inflorescence apex is only visible (arrowhead). (E–H) Transgenic plants under short days. At 75 days, a 35S:AGL24 transgenic line (E) and a wild-type plant (F) are flowering with 32 and 45 rosette leaves, respectively. Simultaneously, a weak line (G) is just bolting (arrow) and the inflorescence apex in a strong line (H) is only visible (arrowhead). (I–M) Localization of LFY:GUS activity in Landsberg erecta plants. In 9-day-old agl24 (I) and wild-type (J) plants at the similar developmental stage under long days, there is a stronger GUS expression in the leaf primordia surrounding the shoot apical meristem in a wild-type plant than in agl24. At 12 days, strong LFY:GUS expression is observed in both the shoot apex undergoing floral transition and the emerging floral meristem (fm) in a wild-type plant (L), whereas relatively weak GUS expression is present in the leaf primordia surrounding the shoot apex in agl24 (K). LFY:GUS expression is detectable in the inflorescence meristem and young flowers (stages 2 and 3) in agl24 (M), which is comparable to its expression in a wild-type plant. (Bars = 100 μm.)
Fig 3.
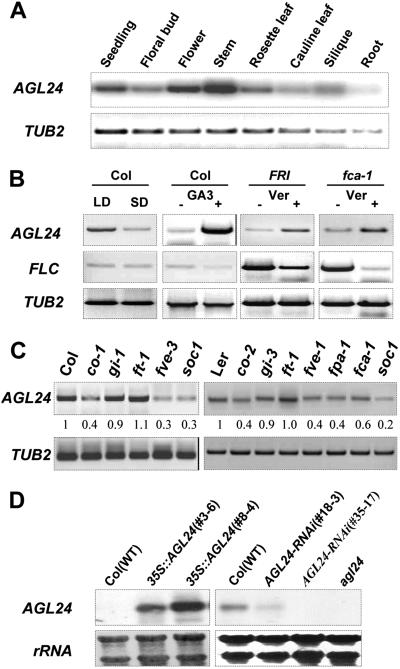
Expression of AGL24. (A–C) RT-PCR analysis of AGL24 expression. The β-tubulin gene (TUB2) was amplified as a quantitative control. (A) AGL24 expression in different organs. (B) Effect of photoperiod, GA, and vernalization on the expression of AGL24 and FLC. (C) AGL24 expression in late-flowering mutants in Columbia (Left) and Landsberg erecta (Right) backgrounds. The numbers indicate the relative expression levels of AGL24. (D) Northern blot analysis of AGL24 expression in transgenic plants. For 35S:AGL24 lines, total RNA was isolated from roots. 35S:AGL24 transgenic lines 3-6 and 8-4 represent weak and strong overexpression of AGL24, respectively. For AGL24 double-stranded interference (AGL24-RNAi) mutants, total RNA was isolated from stems. AGL24-RNAi 18-3 and 35-17 represent weak and strong mutant lines, respectively. The rRNAs stained by methylene blue indicate the amount of total RNA loaded in each lane.
AGL24 Acts in Part Downstream of CONSTANS (CO) and SOC1.
AGL24 mRNA is present in all of the tissues with the strongest expression in stems (Fig. 3A). The expression of AGL24 gradually increases under both LDs and SDs, with the overall levels delayed and reduced levels in SDs (Fig. 3B; Fig. 6, which is published as supporting information on the PNAS web site). Simultaneously, the elevation of AGL24 expression was obvious upon the treatment of GA and vernalization (Fig. 3B). Therefore, it is most likely that AGL24 functions downstream of several floral promotion pathways, including the autonomous, vernalization, photoperiod, and GA pathways (4, 21). This scenario was confirmed by the observation of the relatively reduced expressions of AGL24 in various loss-of-function mutant backgrounds in the key genes of the different promotion pathways (Fig. 3C). However, the only exception is the almost unchanged AGL24 expression in ft-1 mutant. Because CO, FT, and SOC1 are three essential components in the photoperiod promotion pathway, we examined the details of the AGL24 accumulation in their loss-of-function mutants. AGL24 expression is significantly reduced and delayed in both co-1 and soc1 seedlings, whereas its expression profiles are similar in wild-type and ft-1 plants under LDs (Figs. 6 and 7, which are published as supporting information on the PNAS web site). Because FT and SOC1 act in independent pathways downstream of CO (6–9, 11), our results indicated that AGL24 might function in part downstream of CO and SOC1. This suggestion of the epistatic relation between SOC1 and AGL24 is consistent with the reduced AGL24 expression in most of the mutants examined (Fig. 3C), because SOC1 is an essential regulator downstream of CO integrating several promotion pathways (7, 11).
Our conclusions regarding the contribution of AGL24 in the flowering promotion pathways were supported by several lines of genetic evidence (Table 1). First, 35S:AGL24 could partially, though not totally, rescue the late-flowering phenotype of soc1, which is consistent with the suggested epistatic relation between SOC1 and AGL24. Second, whereas loss of AGL24 could significantly suppress the precocious flowering in 35S:SOC1 plants, constitutive expression of SOC1 in agl24 still caused earlier flowering than in the wild type. Thus, it is likely that AGL24 is not the only downstream effector of SOC1. It is also clear that both AGL24 gain- and loss-of function plants demonstrate less severe alterations of flowering time than those of SOC1 (Table 1). In addition, in situ hybridization revealed that AGL24 and SOC1 had especially different expression patterns in the emerging floral primordia (Fig. 1 E and F) (7, 11). Together, these results support a hierarchy from SOC1 to AGL24 with the involvement of other unknown flowering-time regulators in parallel with and upstream of AGL24. Third, although agl24 could slightly attenuate the precocious flowering phenotype in 35S:FT plants, constitutive expression of FT almost completely suppressed the agl24 phenotype. This means that the photoperiod pathway via FT could compensate for the effect of loss of AGL24 function. Strikingly, the mutants with the loss and gain of SOC1 or AGL24 functions were still sensitive to photoperiod (Table 1) (7, 11, 22), which is in sharp contrast to the phenomena demonstrated by the plants with the altered activities of CO and FT (7–9, 21). Therefore, despite FT and SOC1 functions in concert downstream of CO, FT should have a much greater contribution to the photoperiod pathway than the gene cascade from SOC1 to AGL24. Nevertheless, the incomplete suppression of agl24 phenotype by 35S:FT is consistent with the previous suggestion that FT also plays a role in the regulation of SOC1 (11), thus influencing the AGL24 activity, though FT may not directly or mainly regulate AGL24.
AGL24 Acts Upstream of LFY.
LFY plays an important role in promoting the floral transition (23, 24). To determine the possible relation between AGL24 and LFY, we introduced the LFY:β-glucuronidase (GUS) transgene into agl24 mutants and compared the mimic LFY expression in agl24 and wild-type backgrounds (25).
Our results demonstrated that the delay of the floral transition in agl24 is coupled with the delayed and reduced profile of LFY expression. At the similar developmental stage, there is a much stronger LFY:GUS activity in wild-type plants than in agl24 (Fig. 2 I and J; Fig. 8A, which is published as supporting information on the PNAS web site). Particularly, the up-regulation of LFY:GUS was delayed during floral transition in agl24 (Fig. 2 K and L; Fig. 8A). These results are in agreement with the totally reduced LFY expression in agl24 examined by RT-PCR during floral commitment (Fig. 8B), suggesting that AGL24 affects the transcriptional induction of LFY. This was supported strongly by the genetic evidence showing the almost complete compensation of the late-flowering phenotype of agl24 by 35S:LFY (Table 1). Therefore, LFY functions at least in part downstream of AGL24. It has been shown that FT and LFY may act independently of each other to regulate in concert the flowering process (9, 24, 26). Meanwhile, LFY is a possible regulator in part downstream of SOC1 (11). Together, this evidence indicates that SOC1 partially regulates the LFY activity via AGL24, which is independent of FT. Also, it has been clear that GA promotes flowering by activating the LFY gene (25). Because SOC1 and AGL24 are possible regulators in the GA promotion pathway (Fig. 3B) (27), and the up-regulation of AGL24 expression is dramatically reduced upon GA treatment in soc1 background (data not shown), the transcriptional regulation from SOC1 to LFY may integrate floral signals from multiple pathways including the GA promotion pathway (Figs. 4 and 8).
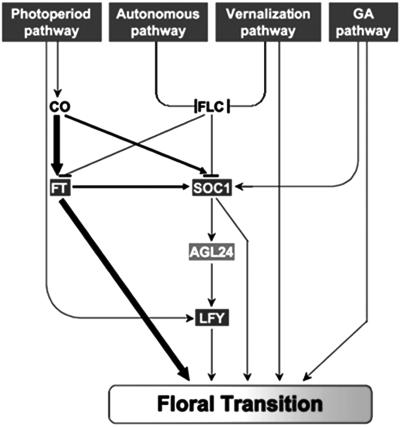
Fig 4.
Integration of floral signals is mediated by AGL24. Floral transition in Arabidopsis is regulated by multiple promotion pathways. Two flowering-time genes, FT and SOC1, along with the floral meristem identity gene LFY, are three essential regulators integrating floral signals from different pathways. AGL24 acts downstream of SOC1 and upstream of LFY. Arrows and T bars represent promotion and repression effects, respectively. The thickness of arrows indicates the relative contribution of CO and FT to the regulation of downstream genes.
To test if loss of AGL24 activity affects the function of LFY in the establishment of floral meristem fate and the activation of floral homeotic genes (28), we monitored LFY:GUS activity in the inflorescence meristem in agl24 (Fig. 2M). The results showed that the activity of LFY:GUS in the inflorescence meristem and young floral primordia is comparable to what is observed in a wild-type plant (24). Therefore, although AGL24 regulates the induction of LFY expression during floral transition, loss of AGL24 activity does not influence the expression of LFY in the subsequent stages of flower development.
In conclusion, AGL24 is a dosage-dependent promoter in regulating floral transition in Arabidopsis. In support of the suggestion of the presence of substantial interaction among three important integrators LFY, FT, and SOC1 (6, 11), our study reveals that AGL24 is a possible mediator acting in the regulatory cascade from SOC1 to LFY, which is only slightly affected by FT. The genetic branch via FT has a greater contribution to the LD photoperiod promotion pathway than the genetic pathway from SOC1 to AGL24. It is obvious that the level of AGL24 accumulation still gradually increases in soc1 mutants in response to the age of plants (Fig. 6), indicating that regulation of AGL24 is partially independent of SOC1. Meanwhile, we have suggested that genetically redundant gene(s) may act parallel to AGL24 downstream of SOC1. Thus, the linear hierarchy from SOC1 to AGL24 may only represent one typical regulatory pathway among their related networks leading to the control of flowering time. With the linear pathway clarified here, we will further elucidate if there are direct regulatory relationships among SOC1, AGL24, and LFY.
It is interesting to note that the MADS-box gene family, which is divided into repressors (FLC, FLM/MAF1, and SVP) and promoters (FUL, SOC1, and AGL24), has been one of the major classes of transcription factors mediating the antagonism between the promotive and repressive pathways during floral transition (7, 11, 15, 20, 29–32). The transcriptional cascades among MADS-box genes, just like the genetic pathway from FLC to SOC1 to AGL24, which is reminiscent of the proposed subtle regulatory hierarchy of MADS-box genes controlling flower development (33, 34), may represent one of the essential characteristics in the complex regulatory networks in the flowering-time control.
Supplementary Material
Acknowledgments
Special thanks to M. Yanofsky and R. M. Amasino for exchanging information on AGL24 and the gift of agl24 seed. We thank V. Sundaresan, T. Ito, E. Ziegelhoffer, J. Peng, and P. Lakshmanan for critical reading of the manuscript, and D. Weigel, I. Lee, M. A. Blázquez, and the National Science Foundation-supported Arabidopsis Biological Resource Center for materials. This work was supported by both Research Grant R-154-000-125-112 and a postdoctoral fellowship (to H.Y.) from the National University of Singapore.
Abbreviations
dsRNA, double-stranded RNA
RNAi, RNA interference
GA, gibberellic acid
GUS, β-glucuronidase
LD, long day
SD, short day
References
- 1.Koornneef M., Alonso-Blanco, C., Peeters, A. J. M. & Soppe, W. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345-370. [DOI] [PubMed] [Google Scholar]
- 2.Simpson G. G., Gendall, A. R. & Dean, C. (1999) Annu. Rev. Cell Dev. Biol. 15, 519-550. [DOI] [PubMed] [Google Scholar]
- 3.Mouradov A., Cremer, F. & Coupland, G. (2002) Plant Cell 14,(Suppl.), S111-S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koornneef M., Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C. J. & Peeters, A. J. M. (1998) Genetics 148, 885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy Y. Y. & Dean, C. (1998) Plant Cell 10, 1973-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araki T. (2001) Curr. Opin. Plant Biol. 4, 63-68. [DOI] [PubMed] [Google Scholar]
- 7.Samach A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F. & Coupland, G. (2000) Science 288, 1613-1616. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi Y., Kaya, H., Goto, K., Iwabuchi, M. & Araki, T. (1999) Science 286, 1960-1962. [DOI] [PubMed] [Google Scholar]
- 9.Kardailsky I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., Chory, J., Hurrison, M. J. & Weigel, D. (1999) Science 286, 1962-1965. [DOI] [PubMed] [Google Scholar]
- 10.Simon R., Igeño, M. I. & Coupland, G. (1996) Nature 384, 59-62. [DOI] [PubMed] [Google Scholar]
- 11.Lee H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., Lee, J. S., Kwon, Y. M. & Lee, I. (2000) Genes Dev. 14, 2366-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voinnet O., Vain, P., Angell, S. & Baulcombe, D. C. (1998) Cell 95, 177-187. [DOI] [PubMed] [Google Scholar]
- 13.Waterhouse P. M., Graham, M. W. & Wang, M. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang C. F. & Meyerowitz, E. M. (2000) Proc. Natl. Acad. Sci. USA 97, 4985-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaels S. D. & Amasino, R. M. (1999) Plant Cell 11, 949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H. & Goh, C. J. (2000) Plant Physiol. 123, 1325-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clough S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 18.Yu H., Yang, S. H. & Goh, C. J. (2000) Plant Cell 12, 2143-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieburth L. E. & Meyerowitz, E. M. (1997) Plant Cell 9, 355-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmann U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H. & Huijser, P. (2000) Plant J. 21, 351-360. [DOI] [PubMed] [Google Scholar]
- 21.Koornneef M., Hanhart, C. J. & van der Veen, J. H. (1991) Mol. Gen. Genet. 229, 57-66. [DOI] [PubMed] [Google Scholar]
- 22.Onouchi H., Igeño, I., Périlleux, C., Graves, K. & Coupland, G. (2000) Plant Cell 12, 885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weigel D. & Nilsson, O. (1995) Nature 377, 495-500. [DOI] [PubMed] [Google Scholar]
- 24.Blázquez M. A., Soowal, L. N., Lee, I. & Weigel, D. (1997) Development (Cambridge, U.K.) 124, 3835-3844. [DOI] [PubMed] [Google Scholar]
- 25.Blázquez M. A., Green, R., Nilsson, O., Sussman, M. R. & Weigel, D. (1998) Plant Cell 10, 791-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson O., Lee, I., Blázquez, M. A. & Weigel, D. (1998) Genetics 150, 403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borner R., Kampmann, G., Chandler, J., Gleissner, R., Wisman, E., Apel, K. & Melzer, S. (2000) Plant J. 24, 591-600. [DOI] [PubMed] [Google Scholar]
- 28.Parcy F., Nilsson, O., Busch, M. A., Lee, I. & Weigel, D. (1998) Nature 395, 561-566. [DOI] [PubMed] [Google Scholar]
- 29.Mandel M. A. & Yanofsky, M. F. (1995) Plant Cell 7, 1763-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheldon C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J. & Dennis, E. S. (1999) Plant Cell 11, 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scortecci K. C., Michaels, S. D. & Amasino, R. M. (2001) Plant J. 26, 229-236. [DOI] [PubMed] [Google Scholar]
- 32.Ratcliffe O. J., Nadzan, G. C., Lynne Reuber, T. & Riechmann, J. L. (2001) Plant Physiol. 126, 122-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rounsley S. D., Ditta, G. S. & Yanofsky, M. F. (1995) Plant Cell 7, 1259-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrándiz C., Gu, Q., Martienssen, R. & Yanofsky, M. F. (2000) Development (Cambridge, U.K.) 127, 725-734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.