Abstract
Signal-induced activation of the transcription factor serum response factor (SRF) requires alterations in actin dynamics. SRF activity can be inhibited by ectopic expression of β-actin, either because actin itself participates in SRF regulation or as a consequence of cytoskeletal perturbations. To distinguish between these possibilities, we studied actin mutants. Three mutant actins, G13R, R62D, and a C-terminal VP16 fusion protein, were shown not to polymerize in vivo, as judged by two-hybrid, immunofluorescence, and cell fractionation studies. These actins effectively inhibited SRF activation, as did wild-type actin, which increased the G-actin level without altering the F:G-actin ratio. Physical interaction between SRF and actin was not detectable by mammalian or yeast two-hybrid assays, suggesting that SRF regulation involves an unidentified cofactor. SRF activity was not blocked upon inhibition of CRM1-mediated nuclear export by leptomycin B. Two actin mutants were identified, V159N and S14C, whose expression favored F-actin formation and which strongly activated SRF in the absence of external signals. These mutants seemed unable to inhibit SRF activity, because their expression did not reduce the absolute level of G-actin as assessed by DNase I binding. Taken together, these results provide strong evidence that G-actin, or a subpopulation of it, plays a direct role in signal transduction to SRF.
INTRODUCTION
Serum response factor (SRF) is a transcription factor that regulates many immediate-early and muscle-specific genes. Deletion of SRF in ES cells leads to alterations in cellular morphology and adhesion, and is lethal in mice at gastrulation owing to defects in mesoderm formation (Arsenian et al., 1998; Weinhold et al., 2000; Schratt et al., 2002). SRF activity is controlled by the Rho family of small GTPases (Hill et al., 1995), and recent studies have revealed a close connection between SRF activation and actin polymerization. Downstream of RhoA, both the ROCK-LIMK-cofilin and the mDia effector pathways can promote both F-actin accumulation and SRF activity (Sotiropoulos et al., 1999; Tominaga et al., 2000; Copeland and Treisman, 2002; Geneste et al., 2002). The ability of LIMK and mDia mutants to activate SRF correlates with their ability to promote F-actin accumulation, and interfering derivatives of these proteins can inhibit the activation of SRF by extracellular signals (Sotiropoulos et al., 1999; Tominaga et al., 2000; Copeland and Treisman, 2002; Geneste et al., 2002). Alterations in actin dynamics are required for RhoA-mediated SRF activation, which is inhibited upon treatment of cells with the G-actin binding drug latrunculin or C2 toxin (Sotiropoulos et al., 1999). The RhoA-actin pathway controls a subset of SRF target genes, including the immediate-early genes β-actin, vinculin, and srf, and the muscle-specific SM22 and SM α-actin genes (Sotiropoulos et al., 1999; Gineitis and Treisman, 2001; Mack et al., 2001).
Several lines of evidence suggest that actin itself is intimately involved in the control of SRF. Stabilization of F-actin by the actin-binding drug jasplakinolide is sufficient to activate SRF in the absence of extracellular stimuli, whereas overexpression of actin inhibits SRF (Sotiropoulos et al., 1999). Moreover, SRF can be activated by overexpression of the actin-binding protein profilin, and this is blocked by profilin mutations that prevent actin binding (Sotiropoulos et al., 1999; Geneste, unpublished observation). It seems that SRF activity reflects decreased G-actin level rather than increased F-actin level, because SRF activity is potentiated by several actin-binding drugs that do not promote F-actin formation, such as cytochalasin D and swinholide. However, direct evidence for the participation of unpolymerized actin in the control of SRF activity has remained elusive, although previous studies detected actin in association with chromatin remodeling machines (Zhao et al., 1998; Shen et al., 2000; reviewed by Rando et al., 2000).
Actin is an ATPase that cycles between monomeric (G-actin) and polymerized (F-actin) states (Holmes et al., 1990; Kabsch et al., 1990). The four subdomains of actin form two lobes, separated by a deep cleft that binds nucleotide and a divalent cation, and the molecule adopts differing conformations according to whether ATP or ADP is bound (Kabsch et al., 1990; Chik et al., 1996; Otterbein et al., 2001). Nucleotide binding is not required for polymerization per se but stabilizes the molecule (Kabsch et al., 1990; De La Cruz et al., 2000). Instead, the binding and hydrolysis of ATP effectively directs monomer addition to the barbed end of the filament (Pollard et al., 2000). Although ATP hydrolysis on F-actin is rapid, the conformational changes that promote its interaction with the depolymerizing/severing factor cofilin occur only upon release of the phosphate, which occurs slowly, thus determining the lifetime of the filament (Belmont et al., 1999a; Pollard et al., 2000).
Mutant actins have provided extensive insights into F-actin structure and the role of nucleotide binding and hydrolysis (Chen et al., 1993, 1995; Chen and Rubenstein, 1995; Belmont and Drubin, 1998; Belmont et al., 1999a; Schuler et al., 1999). In this work, we used site-directed mutagenesis of β-actin to investigate the relationship between actin and SRF activation. We show that actins that cannot polymerize are effective inhibitors of signaling to SRF, but that it is unlikely that this involves physical interaction with SRF. We also show that two actin mutants that enhance F-actin formation can activate SRF-dependent transcription when overexpressed. These results present direct evidence for participation of monomeric actin in the signaling pathway to SRF.
MATERIALS AND METHODS
Plasmids
A synthetic promoter comprising three copies of the c-fos SRF binding site with Xenopus type 5 actin TATA box and transcription start site (Mohun et al., 1987; Hill et al., 1995; Geneste et al., 2002) was inserted into pGL3 (Promega, Madison, WI) to create the SRF reporter 3D.ALuc (Copeland and Treisman, 2002; Geneste et al., 2002). Other plasmids were as follows: pMLV.SRF.VP16 and SRF198/210 (Hill et al., 1994); pRL-TK (Renilla Luciferase controlled by thymidine kinase promoter; Promega); and pMLV-NLexA (Marais et al., 1993). pEF.FLAG-actin (Sotiropoulos et al., 1999) was used to generate β-actin derivatives. The actin double NES mutant changed 170-ALPHAILRLDL-180 to ALPHAILRADA and 211-DIKEKLCYVAL-221 to DIKEKLCYAAA. For two-hybrid assays of pGBT9, pGAD424 and pACT2 derivatives carrying actin, profilin, cofilin, and SRF sequences were used. pADH-SRF derivatives contain SRF sequences inserted into a modified pGBT9 lacking GAL4 sequences. pGADplink-SRF contain SRF sequences inserted into pGADplink, a pGAD424 derivative in which GAL4 sequences are replaced by the β-globin 5′ untranslated region and polylinker sequences (Treisman, unpublished data). Yeast reporter plasmids 4xSRF-LacZ and 5xGal4-LacZ are were constructed by insertion of the appropriate binding sites into pLG178 (Dalton and Treisman, 1992).
Yeast Two-Hybrid Tests
Direct yeast two-hybrid tests with Saccharomyces cerevisiae strain HF7c (CLONTECH, Palo Alto, CA) were essentially done as described previously (Sahai et al., 1998). One microgram of each bait and activation fusion plasmid together with 100 μg of carrier DNA (salmon sperm; Stratagene, La Jolla, CA) were added to 100 μl of a concentrated suspension of exponentially grown yeast cells. Transformation was achieved overnight by adding 500 μl of PLATE (40% PEG 3350, 100 mM LiAc, 10 mM Tris pH 7.5, 0.1 mM EDTA) and 20 μl of 1 M dithiothreitol. Transformants were selected on yeast nitrogen base-agar plates lacking uracil, tryptophan, and leucin. Protein interactions were scored by growth of three independent colonies on plates additionally lacking histidine. Strong interactions were semiquantified by adding 3-aminotriazole (Sigma-Aldrich, St. Louis, MO) at a concentration of 2, 5, or 30 mM. LacZ reporter assays were performed with colonies streaked on nylon filters according to standard protocols (CLONTECH). For one-hybrid tests, the diploid strain S62/His3 was used, generated from the haploid S62L strain (Dalton and Treisman, 1992) and the HIS3 strain that contains an integrated HIS3 gene driven by four SRF binding sites.
Transfections and Gene Expression Assays
Transient transfections were carried out using LipofectAMINE (Invitrogen, Carlsbad, CA). NIH3T3 cells (3–4 × 105 cells/6-cm dish) were transfected with 100 ng of p3D.A-Luc, 200 ng of pRL-TK, and 500 ng of SRF-VP16, made to a total of 2.3 μg with expression plasmid pEF-FLAG or derivatives. Cells were maintained in 0.5% fetal calf serum (FCS) for 40 h unless indicated otherwise before stimulation with 15% serum or cytochalasin D (Calbiochem, La Jolla, CA) for 7 h. Pretreatments with leptomycin B (gift from E. Nishida, Kyoto University, Japan) were for 30 min. Firefly luciferase activity was normalized to either protein content or to Renilla luciferase activity (as indicated) and expressed as a percentage of the activity of reporter activation by SRF-VP16 performed in parallel. Figures show mean ± SEM of at least three independent experiments. RNase protection assays were as described previously (Hill et al., 1995; Sotiropoulos et al., 1999), and immunoblotting of cell lysates was by standard techniques.
Actin Fractionation
Transfected cells were scraped, washed in phosphate-buffered saline (PBS), and then lysed in 0.75 ml of actin lysis buffer (50 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 20 mM HEPES, pH 7.9); 100,000-g supernatant and pellet fractions were then prepared. Supernatants were mixed directly with SDS-PAGE loading buffer, whereas pellets were resuspended in 0.75 ml of actin lysis buffer mixed with SDS-PAGE loading buffer and sonicated. Equal amounts were separated by 10% SDS-PAGE and detected by immunoblotting with anti-actin (AC-40; Sigma-Aldrich), anti-FLAG (M5; Sigma-Aldrich), or anti-hemagglutinin (HA) (12CA5; Roche Applied Science, Indianapolis, IN) monoclonal antibodies, with visualization by anti-mouse secondary antibodies coupled to horseradish peroxidase (DAKO, Glostrup, Denmark) and enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ). Jasplakinolide treatment was for 90 min by using 0.3 μM drug (Molecular Probes, Eugene, OR).
Immunofluorescence and FACS Analysis
Immunofluorescence staining was as described previously (Sotiropoulos et al., 1999; Tran Quang et al., 2000). Staining conditions were as follows: M2 anti-FLAG (Sigma-Aldrich), 1:100; rabbit polyclonal antiserum AGA-1 anti-LexA (Cancer Research UK antibody facility), 1:100; rhodamine-phalloidin (Molecular Probes), 1:200; 6-((7-amino-4-methylcoumarin-3-acetyl)amino)hexanoic acid-conjugated anti-rabbit (DAKO), 1:100; and fluorescein isothiocyanate (FITC)-conjugated anti-mouse (DAKO), 1:100. Triton X-100 extraction of soluble cytoplasmic proteins before fixation was as described previously (Algrain et al., 1993; Tran Quang et al., 2000). Micrographs were taken using a Zeiss Axioplan II microscope with Plan-Neofluar 63× objective, appropriate filters and a Quantix charge-couple device camera (Photometrics, Tucson, AZ), with SmartCapture 2 software (Applied Imaging, Newcastle, United Kingdom).
Fluorescence-activated cell sorting (FACS) quantitation of F-actin content (Bleul et al., 1996) or G-actin content was as described previously (Geneste et al., 2002). Cells (1 × 106/10-cm dish) were transfected with 2 μg of actin mutants and 2 μg of pEF.Fplink vector, maintained overnight in DMEM/10% FCS followed by 24 h in DMEM/0.5% FCS before trypsinization and fixation by using 4% paraformaldehyde. After permeabilization (10 min in PBS/0.2% Triton X-100) and staining with anti-FLAG (1:200 in PBS, 5% FCS, 0.05% Tween 20 for 1 h at room temperature), cells were washed in PBS containing 0.05% Tween 20 and incubated with Cy3-conjugated anti-mouse (1:500; Jackson Immunoresearch Laboratories), and FITC-phalloidin or FITC-DNase I (1:200; Molecular Probes). FACS analysis was performed using the FACScalibur (BD Biosciences, San Jose, CA) with CellQuest 3.1 software. The median FITC fluorescence intensity of viability-gated Cy3-positive cells (approx. 10,000) was measured relative to that of Cy3-negative cells from the same population. Gates for Cy3 were established using a mock-transfected control cell population.
RESULTS
Strategy for Identification of Nonpolymerizable Actin Mutants
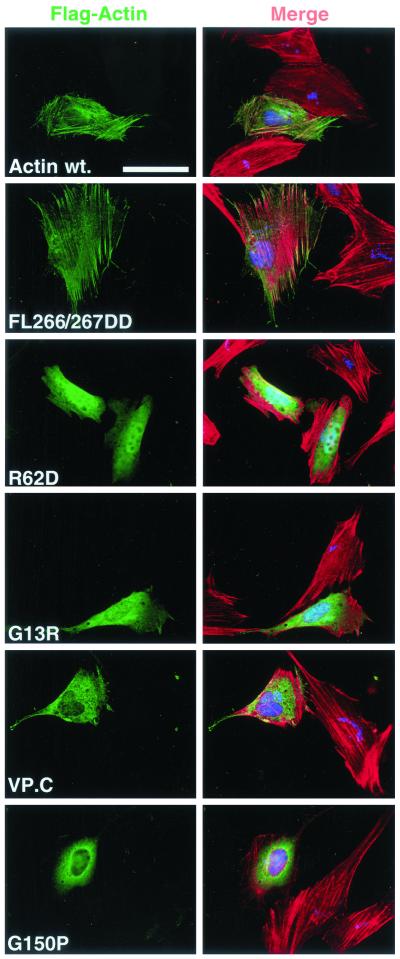
Many studies have identified mutations in yeast and vertebrate actins that exhibit defects in polymerization in vitro and that give rise to lethal or conditional phenotypes in vivo. We examined three types of mutation in human β-actin: those that alter surface-exposed residues implicated in intersubunit interactions in F-actin or interdomain interactions in actin monomer; mutants that affect the architecture or function of the nucleotide binding pocket; and fusion proteins containing substantial extraneous polypeptide sequences located at the N or C termini. The mutant actins were expressed transiently in NIH3T3 cells as N-terminally FLAG-tagged derivatives and their colocalization with cellular F-actin analyzed by indirect immunofluorescence. FLAG-actin entered phalloidin-stainable structures indistinguishable from those formed by endogenous actin (Figure 1, top), which are resistant to detergent extraction before staining (Figure 2). A number of the mutants were also tested in a yeast two-hybrid assay both for homotypic interaction and heterotypic interaction with wild-type actin (Table 1). Mutants exhibiting apparent defects in these assays were analyzed further using biochemical approaches. To exclude the possibility that mutant actins that fail to interact with cellular F-actin represent mutants that irreversibly associate with the actin chaperonin CCT, we also compared their properties with those of actin G150P, which irreversibly binds CCT in vitro (McCormack et al., 2001a).
Figure 1.
Actins R62D, G13R, VP.C, and G150P do not colocalize with cellular F-actin structures. NIH3T3 cells were transiently transfected with expression plasmids encoding FLAG-actin or mutant derivatives together with the nuclear-localized LexA derivative NLexA as transfection marker. Transfected cells were formaldehyde fixed and stained as described in MATERIALS AND METHODS. Left, transfected actins (anti-FLAG; green). Right, merge of transfected actins (anti-FLAG; green), total F-actin (rhodamine-phalloidin; red) and transfection marker (anti-LexA; blue). Bar, 50 μm.
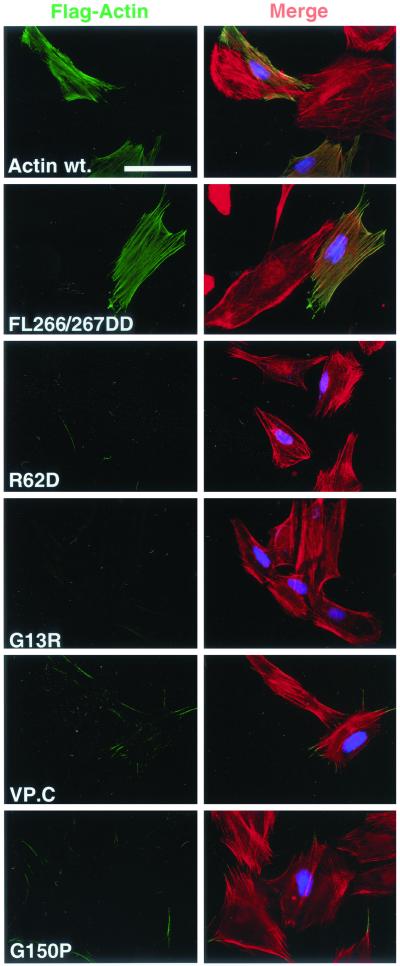
Figure 2.
Actins R62D, G13R, VP.C, and G150P can be quantitatively removed from cells by detergent extraction. NIH3T3 cells were transiently transfected with expression plasmids encoding FLAG-actin or mutant derivatives as in Figure 1, but the cells were subjected to brief extraction with 0.5% Triton X-100 before fixation and staining. Left, transfected actins (anti-FLAG; green). Right, merge of transfected actins (anti-FLAG; green), total F-actin (rhodamine-phalloidin; red) and transfection marker (anti-LexA; blue). Bar, 50 μm.
Table 1.
Yeast two-hybrid analysis of interactions between mutant actins, wild-type actin, profilin, and cofilin
| Gal4-DBD-actin fusion | Interaction partner
|
|||
|---|---|---|---|---|
| Wild-type actin | Mutant actin | Profilin | Cofilin | |
| pGBT9 | − | na | − | − |
| Actin wt. | + | na | − | + |
| FL266/267GG | + | + | + | + |
| L267D | + | − | + | + |
| FL266/267DD | − | − | + | + |
| G13R | − | − | + | − |
| S14A | + | nd | − | + |
| S14C | + | nd | − | + |
| G15R | − | nd | + | + |
| V159N | + | nd | − | + |
For actin and profilin interactions, + indicates growth on plates containing 2 mM aminotriazole; for cofilin interactions, + indicates growth on plates without aminotriazole. No pGBT9 derivatives were sufficient for growth alone. na, not applicable; nd, not done.
Actins R62D, G13R, and Actin Fusion VP.C Do Not Colocalize with F-Actin
We first tested mutants at residues F266 and L267, which are predicted to form a “hydrophobic plug” mediating contact between adjacent monomers in the actin filament (Holmes et al., 1990). Mutations that decrease the hydrophobicity of these side chains in yeast actin reduce polymerization in vitro, and lower affinity for nucleotide (Chen et al., 1993; Kuang and Rubenstein, 1997). β-Actin FL266/267GG showed unchanged interaction with actin in the two-hybrid assay (Table 1), in contrast to yeast actin (Kuang and Rubenstein, 1997). Introduction of charged residues at these positions had a greater effect (cf. Chen et al., 1993): mutant L267D showed normal interaction with wild-type actin but reduced homotypic interaction, whereas FL266/267DD reduced homotypic and heterotypic interactions to undetectable levels. However, all of these mutants showed identical properties to wild-type actin in the immunofluorescence assay (Figures 1 and 2; our unpublished data). Next, we examined the charge reversal mutant R62D, likely to disrupt a salt bridge between subdomains 2 and 4 (Kabsch et al., 1990; Otterbein et al., 2001). This mutation blocks interaction between human β-actin and the CAP protein (McCormack et al., 2001b) and in yeast the KR61/62AA mutation is lethal (Wertman et al., 1992). On transient expression in NIH3T3 cells, actin R62D accumulated to similar levels to wild-type actin (Figure 4A). When examined by immunofluorescence actin R62D showed even distribution throughout both cytoplasm and nucleus, and no colocalization with F-actin (Figure 1). Actin R62D could be completely removed from cells by detergent extraction before staining, leaving endogenous F-actin structures intact (compare Figures 1 and 2).
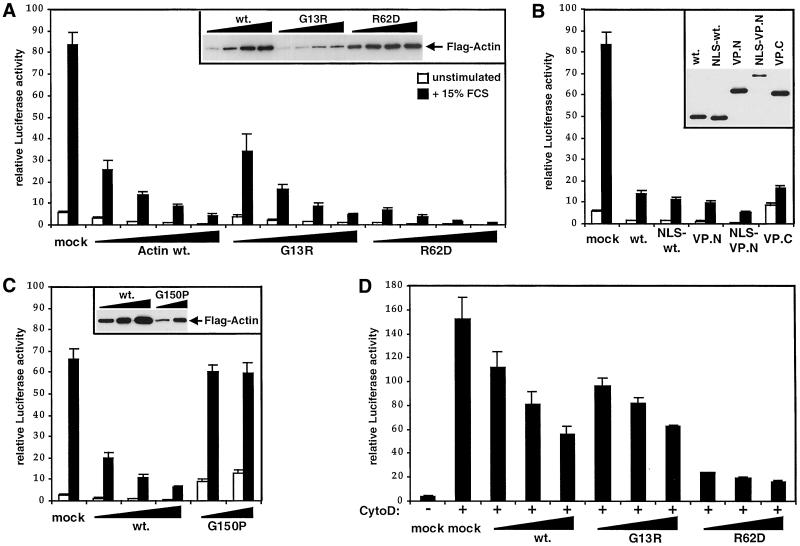
Figure 4.
Nonpolymerizable actin mutants inhibit SRF activation. (A) Actins G13R and R62D block serum-induced SRF activation. NIH3T3 cells were transfected with 0.2, 0.5, 1.0, or 2.0 μg of expression plasmid encoding FLAG-tagged wild-type actin, actin G13R, actin R62D, or empty vector, together with the SRF reporter 3D.ALuc. After serum stimulation, luciferase activities were measured and expressed relative to activation by SRF-VP16, taken as 100. Inset shows a FLAG immunoblot of cell extracts. Data show mean of at least three independent experiments with SEM indicated by the bars. (B) Actin/VP-16 fusion proteins inhibit SRF activation. NIH3T3 cells were transfected with expression plasmids encoding FLAG-tagged wild-type actin, NLS-actin, actin VP.N, actin NLS-VP.N (0.5 μg each), or actin VP.C (2 μg), or empty vector, together with the SRF reporter gene 3D.ALuc, and reporter activity was analyzed as in A. (C) CCT-binding mutant actin G150P does not inhibit SRF activation. Cells were transfected with the indicated mutants and 3D.ALuc as in A, except that activities were normalized to a cotransfected tk-luc reference plasmid rather than total recovered protein to minimize variations arising from the toxicity of actin G150P. (D) Nonpolymerizable actins inhibit SRF activation by cytochalasin D. Cells were transfected with actin expression plasmids (0.5, 1.0, or 2.0 μg) and 3D.ALuc and analyzed as in C, with stimulation by 10 μM cytochalasin D before analysis of reporter gene activity.
We next examined mutations in the nucleotide binding pocket. We reasoned that mutations in the evolutionarily conserved tripeptide G13-S/T14-G15 might lead to polymerization defects, because these residues are involved both in nucleotide binding and the conformational changes that occur upon ATP hydrolysis (Kabsch et al., 1990; Chen et al., 1995; Otterbein et al., 2001). First, we examined a novel mutant, G13R, arising from a polymerase chain reaction error. This mutant failed to interact detectably with either itself or wild-type actin in the two-hybrid assay (Table 1). On transient expression in NIH3T3 cells, actin G13R accumulated to a much lower level than the wild-type protein (Figure 4A). In the immunofluorescence assay, actin G13R did not colocalize with phalloidin-stainable F-actin; although present throughout the cell, actin G13R did not accumulate in the nucleus to the same extent as actin R62D (Figure 1). Like actin R62D, actin G13R was completely extracted from the cells by detergent, leaving endogenous F-actin structures intact (Figure 2). We examined two further mutants in this region, actin S14A and G15R. The yeast actin S14A mutation reduces affinity for ATP some 50-fold and confers temperature sensitivity in vivo (Chen et al., 1995; Chen and Rubenstein, 1995), whereas the G15R mutation has pathological effects both in yeast and in human skeletal muscle α-actin (Belmont et al., 1999b; Nowak et al., 1999). Neither of these mutations affected the ability of β-actin to colocalize with endogenous actin in the immunofluorescence assay (our unpublished data; Figure 6A), although G15R did show reduced interaction with wild-type actin in the two-hybrid assay.
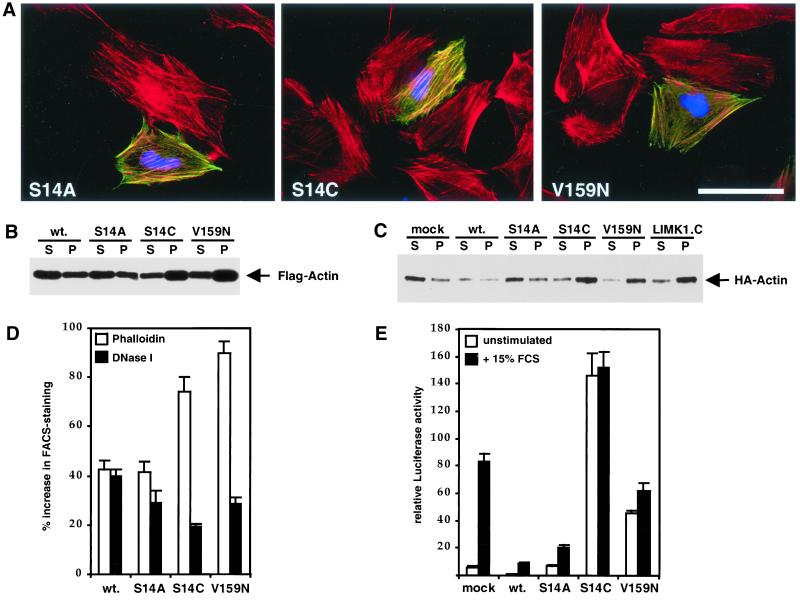
Figure 6.
Actins V159N and S14C stabilize F-actin and activate SRF. (A) Actins V159N and S14C colocalize with endogenous F-actin structures. NIH3T3 cells were transiently transfected with expression plasmids encoding FLAG-actin or mutant derivatives and subjected to brief extraction with 0.5% Triton X-100 before fixing and staining as in Figure 2. The Figure shows a merge of transfected actins (anti-FLAG; green), total F-actin (rhodamine-phalloidin; red), and transfection marker (anti-LexA; blue). Bar, 50 μm. (B) Actins V159N and S14C accumulate preferentially in the Triton X-100–insoluble fraction. Extracts of cells expressing the indicated FLAG-tagged actin mutants were prepared and separated into 100,000-g supernatant (S) and pellet (P) fractions as described in MATERIALS AND METHODS; equal amounts were separated by SDS-PAGE and FLAG-actin in each fraction was detected by immunoblotting with anti-FLAG antibodies. (C) Actins V159N and S14C copolymerize with wild-type actin and decrease its detergent solubility. Extracts were prepared from cells expressing the indicated actins, or the catalytic domain of LIMK1, together with HA-tagged wild-type actin and fractionated as in B before detection of HA-actin in each fraction by immunoblotting with anti-HA antibodies. (D) Expression of actins V159N and S14C increases F:G-actin ratio. Cells were transfected with expression plasmids encoding the indicated actins and fixed and stained for the FLAG epitope and either F-actin, with phalloidin, or G-actin, with DNase I. The levels of F-actin or G-actin in the transfected population were then quantitated relative to the untransfected population by using FACS, as described in MATERIALS AND METHODS. Bars represent the increase in mean relative F-actin (±SEM, n = 4) or G-actin (± half range, n = 2) contents. (E) Expression of actins V159N or S14C activates SRF in the absence of extracellular signals. NIH3T3 cells were transfected with expression plasmids encoding the indicated FLAG-tagged actins together with the SRF reporter gene 3D.ALuc as described in MATERIALS AND METHODS. After serum stimulation, luciferase activities were measured and expressed relative to activation by SRF-VP16, taken as 100. Data show mean of at least four independent experiments with SEM indicated by the bars.
Fusion of substantial polypeptide sequences at the actin N or C terminus can have deleterious effects on actin function in vivo (Doyle and Botstein, 1996; Westphal et al., 1997). We therefore constructed two fusion proteins, VP.N and VP.C, which contain the transcriptional activation domain of the herpes simplex virus protein VP16 at their N and C terminus, respectively. Actin VP.N behaved identically to wild-type actin in the immunofluorescence assay (our unpublished data). In contrast, actin VP.C, which was poorly expressed, did not colocalize with endogenous F-actin, was detergent extractable, and did not affect endogenous F-actin structures (compare Figures 1 and 2). Taken together, the data presented in this section show that actins R62D, G13R, and VP.C are not incorporated into the F-actin cytoskeleton and are therefore candidates for nonpolymerizable mutants.
Actins R62D, G13R, and VP.C Are Freely Soluble In Vivo
Correct folding of nascent actin requires its transient association with the CCT chaperonin particle (reviewed by Lewis et al., 1996). Actin folding mutants that irreversibly associate with CCT have been identified previously (McCormack et al., 2001a). We examined the behavior of one such mutant, actin G150P, by using the immunofluorescence assay; as with actin R62D, G13R, and VP.C, actin G150P failed to colocalize with cellular F-actin structures and was completely extractable by detergent before staining (Figures 1 and 2, bottom). Although actin R62D interacts normally with CCT in vitro (McCormack et al., 2001b), we were therefore concerned to demonstrate that actins R62D, G13R, and actin VP.C are freely soluble in vivo. To do this we examined their solubility in cell extracts.
Cells transfected with wild-type or mutant actin expression plasmids were extracted with 0.5% Triton X-100 and separated into 100,000-g supernatant and pellet fractions. Under these conditions, G-actin is found in the supernatant fraction, and polymerized actin in the pellet. Endogenous actin partitioned approximately equally between the supernatant and pellet fractions (Figure 3A, left; our unpublished data). In contrast, lamin B, a nuclear envelope component, was found only in the pellet fraction (our unpublished data). Transiently transfected wild-type FLAG-actin partitioned between the fractions in a similar manner, indicating that expression of wild-type actin does not alter the F:G-actin ratio (Figure 3A, left; see below). In this assay, mutant actins G13R, R62D, and VP.C were completely detergent extractable and soluble, remaining in the 100,000-g supernatant. In contrast, although actin G150P was readily extractable from cells by detergent treatment (Figure 2; our unpublished data), it was recovered quantitatively in 100,000-g pellet fraction, as expected from its association with the 700-kDa CCT particle (Figure 3A, left lanes). These data suggest that the failure of actins G13R, R62D, and VP.C to colocalize with cellular F-actin does not arise through their irreversible association with CCT. Consistent with this, actin G13R, like actin R62D, exhibits normal interaction with CCT in vitro (McCormack and Willison, personal communication).
Figure 3.
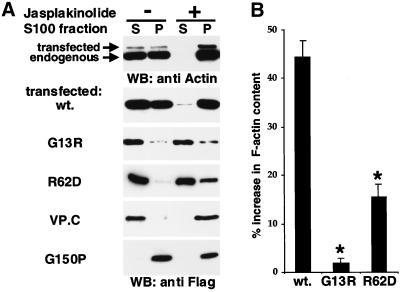
Actins G13R, R62D, and VP.C are soluble and do not enter the F-actin pool. (A) Solubility. Cells transfected with the indicated FLAG-tagged actin mutants were either left untreated (lanes 1 and 2) or treated with the actin-stabilizing drug jasplakinolide (0.3 μM; lanes 3 and 4) for 90 min before lysis and separation into 100,000-g supernatant (S) and pellet (P) fractions. Equal amounts were separated by SDS-PAGE, and proteins in each fraction were detected by immunoblotting by using anti-actin antibodies (top) or anti-FLAG antibodies (bottom). (B) G13R and R62D expression increase cellular F-actin substantially less than wild-type. Cells were transfected with the indicated FLAG-tagged actins; 36 h later they were fixed and stained for FLAG epitope and F-actin; F-actin levels in the transfected population were then quantitated relative to the untransfected population by using FACS, as described in MATERIALS AND METHODS. The relative increase in mean F-actin content is shown (mean of at least three independent experiments ± SEM). Asterisks indicate statistical significance at p < 0.01 according to the unpaired Student's t test.
To investigate further the interactions of the mutant actins with F-actin we tested the effect of treatment of the cells with jasplakinolide, which stabilizes F-actin (Bubb et al., 1994) and which might be expected to stabilize weakened subunit interactions in filaments containing mutant actin (cf. Kuang and Rubenstein, 1997). Jasplakinolide treatment resulted in quantitative recovery of transfected and endogenous cellular actin in the 100,000-g pellet fraction (Figure 3A, right lanes). In contrast, the majority of actin G13R remained soluble in jasplakinolide-treated cell extracts, whereas a small proportion of actin R62D moved to the pellet; only actin VP.C moved entirely to the insoluble fraction (Figure 3A, right lanes). These results suggest that unlike actin G13R, actin VP.C, and to a lesser extent actin R62D, retains a residual interaction with actin that can be enhanced by the drug.
Finally, we analyzed the ability of G13R and R62D to interact with wild-type actin by testing whether their overexpression resulted in an increase in total cellular F-actin. Transfected cell populations were fixed and stained for FLAG-actin expression and for F-actin with FITC-phalloidin, and the mean amount of F-actin present in transfected cells was compared with that in the untransfected population by using the FACS. Expression of wild-type actin increased the mean cellular F-actin content by ∼40%, presumably as a consequence of the increased total cellular actin (Figure 3B); we shall demonstrate below that FACS analysis of G-actin content in cells overexpressing wild-type actin exhibits a similar relative increase, showing that in this case the F:G-actin ratio remains unchanged (Figure 6D). In contrast, expression of actin G13R had no significant effect on mean cellular F-actin content, whereas actin R62D increased the phalloidin staining of transfected cells to some extent, but substantially less than wild-type actin (Figure 3B). Taken together, the data presented in this and the preceding section show that actins G13R, R62D, and VP.C exhibit substantially defects in their ability to polymerize in vivo, and interaction between actin G13R and wild-type actin is not detectable in any of the assays used.
Expression of Nonpolymerizable Actin Inhibits SRF Activation by Serum and Actin-binding Drugs
We next tested the ability of the various mutant actins to inhibit the activation of SRF after serum stimulation. Increasing amounts of FLAG-actin expression plasmids were contransfected with the SRF reporter 3D.ALuc, which contains a luciferase cDNA controlled by three SRF binding sites (Mohun et al., 1987; Hill et al., 1995). Forty-eight hours later reporter activity was measured before and after stimulation with 15% serum. Increasing amounts of wild-type actin expression effectively inhibited SRF activation (Figure 4A), as previously observed in microinjection experiments (Sotiropoulos et al., 1999). Because expression of wild-type actin does not alter the F:G-actin ratio, this result suggests that it is the increase in G-actin that inhibits SRF activity. Consistent with this view, expression of the nonpolymerizable actins G13R, R62D, or VP.C also strongly inhibited SRF activity (Figure 4, A and B). Neither wild-type actin nor the mutants affected direct activation of the reporter gene by the constitutively active SRF derivative SRF-VP16 (our unpublished data). Actins G13R and R62D seemed to act more effectively than wild-type actin, given their expression levels, but this was not the case for actin VP.C (compare insets, Figure 4, A and B). We also used actin containing an N-terminal nuclear localization signal (NLS) sequence to test whether forcing actin to accumulate in the nucleus affected its ability to inhibit SRF. NLS-actin showed strong but not exclusively nuclear staining, and inhibited SRF similarly to the wild-type protein (Figure 4B). All the other mutants tested effectively inhibited SRF activation (our unpublished data). Only expression of actin G150P failed to inhibit SRF activation (Figure 4C), suggesting that retention of actin G150P on the CCT particle is incompatible with SRF regulation. Taken together, these results show that actin does not need to be competent to enter the treadmilling cycle to inhibit SRF activity, and strongly support the notion that G-actin, or a subpopulation of it, participates directly in the regulation of SRF.
We previously showed that SRF activity is also strongly potentiated by cytochalasin D, and proposed that this occurs because the drug interferes with a presumptive regulatory function of G-actin (Sotiropoulos et al., 1999). Expression of wild-type actin, actin G13R, or actin R62D substantially inhibited SRF activation by cytochalasin D, although less effectively than they inhibited activation by serum (Figure 4D).
Physical Interaction between Actin and SRF Is Not Detectable In Vivo
The results presented above strongly support the view that unpolymerized actin somehow regulates SRF activity. We used two-hybrid approaches in mammalian cells and yeast to test for physical interaction between SRF and actin, exploiting the two fusions proteins actin VP.N and actin VP.C, which contain the transcriptional activation domain from the HSV VP16 protein. We showed above that expression of the nonpolymerizable actin VP.C does not potentiate SRF activity in serum-deprived cells, but instead inhibits SRF activation after serum stimulation (Figure 4B). Similar results were obtained with actin VP.N, in which the transcriptional activation domain is linked to the actin N terminus (Figure 4B). These results did not reflect failure of the VP16-tagged actin to reach the nucleus, because SRF activity was effectively inhibited by expression of a derivative of VP.N containing an N-terminal nuclear localization signal, which exhibited substantial nuclear accumulation (Figure 4B; our unpublished data). These data strongly suggest that physical interactions between actin and SRF do not occur on DNA, although they cannot exclude the possibility that the VP16 domain is not functional in this context. We also tested SRF–actin interactions in two-hybrid assays the yeast. No interaction between the two proteins was detectable, either by using actin tethered to DNA via the Gal4 DNA-binding domain or with SRF directly bound to DNA; no interaction between G13R and SRF was observed in this assay either (our unpublished data). These results suggest that direct physical interaction between SRF and actin does not occur.
Activation of SRF Is Independent of CRM1
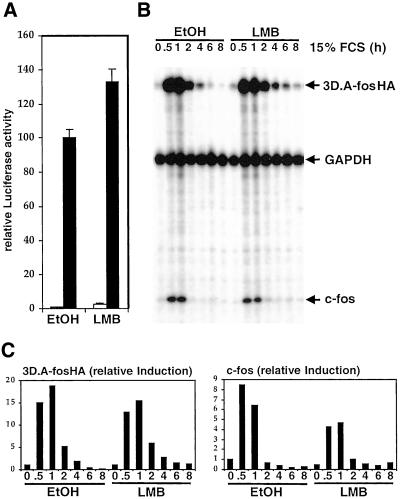
To address the possibility that SRF activation involves regulated nuclear-cytoplasmic shuttling of a large protein cofactor via the CRM1 nuclear export machinery we tested its sensitivity to leptomycin B, which inactivates CRM1. Because SRF induction occurs within minutes, if a CRM1-mediated process regulates its activity any effect of leptomycin B should also be seen rapidly. In control experiments, we found that RanBP1, a known leptomycin B-sensitive protein, became localized to the nucleus within 30 min (our unpublished data). Under these conditions leptomycin B treatment did not inhibit activation of SRF by serum stimulation, however, although a slight increase in basal reporter activity was observed in both starved and cycling cells (Figure 5A; our unpublished data). To monitor more precisely the time course with which serum-induced SRF activation and shutdown occurs we used RNase protection assays of the SRF reporter gene 3D.AFosHA. Serum induction and shutoff of both the SRF reporter and the endogenous c-fos gene were unaffected by leptomycin B pretreatment (Figure 5, B and C), and treatment with leptomycin B alone had no effect on either gene (our unpublished data). A previous report has suggested that CRM1 mediates nucleocytoplasmic shuttling of actin in mammalian cells (Wada et al., 1998). However, neither leptomycin B nor inactivating mutations of both the putative β-actin nuclear export sequences led to rapid nuclear accumulation of transiently expressed FLAG-actin (our unpublished data; see MATERIALS AND METHODS). Taken together, these results show that SRF regulation does not require CRM1-dependent protein export from the nucleus, although we cannot exclude the involvement of CRM1-independent nuclear shuttling or free diffusion of small factors through the nuclear pores.
Figure 5.
SRF activation does not require CRM1. (A) Leptomycin B does not inhibit serum activation of SRF. NIH3T3 cells transfected with 3D.ALuc were serum starved and restimulated in the presence or absence of 5 nM leptomycin B (LMB). (B) Leptomycin B does not inhibit SRF transcriptional shutdown after serum stimulation. Total cell RNA was prepared from 3D.A-fosHA cells, which contain a stably integrated SRF reporter gene, at various times after serum stimulation after pretreatment with 10 nM LMB as indicated. Transcripts of the 3D.A-fosHA reporter, endogenous c-fos, and control GAPDH were analyzed by RNase protection. An identical result was obtained using 50 nM LMB. (C) Quantification of RNase protection assay. The experiment in B was analyzed using the PhosphorImager. Charts show reporter and c-fos transcript levels relative to GAPDH control.
Actins V159N and S14C, which Favor F-Actin Formation, Activate SRF
We next sought to establish whether SRF activity is affected by expression of actin mutants that alter actin dynamics to favor F-actin accumulation, rather than inhibit it. The yeast actin mutant V159N is likely to represent such a protein: the mutation stabilizes F-actin by inhibiting the destabilizing conformational changes that occur after ATP hydrolysis and phosphate release (Belmont and Drubin, 1998; Belmont et al., 1999a). We used immunofluorescence and biochemical assays to characterize β-actin V159N and an additional mutant, actin S14C, identified during our investigation of nucleotide binding pocket mutants, which has similar properties. For comparison, we examined wild-type actin and actin S14A.
Actins V159N and S14C exhibited wild-type behavior in the yeast two-hybrid assay (Table 1). When expressed in NIH3T3 cells, both mutants substantially colocalized with phalloidin-stainable F-actin and were resistant to detergent extraction; S14C was indistinguishable from S14A (Figure 6A). In contrast to actin S14A or wild-type actin, however, actins V159N and S14C preferentially entered the detergent-insoluble F-actin fraction in the cell fractionation assay (Figure 6B). To test whether the mutants could copolymerize with wild-type actin, we examined their ability to alter the behavior of coexpressed HA-tagged wild-type actin in the detergent extraction assay. In a control experiment coexpression of LIMK1 substantially decreased extractability of transfected wild-type HA-actin (Figure 6C) consistent with its ability to stabilize F-actin (Arber et al., 1998; Yang et al., 1998). Coexpression of actins S14C and V159N also led to substantially decreased extractability of wild-type HA-actin, whereas coexpression of wild-type FLAG-actin or actin S14A had no effect (Figure 6C). These data suggest that actins S14C and V159N can copolymerize with wild-type actin to produce F-actin of increased stability.
To confirm directly that the F:G-actin ratio is altered by expression of actins S14C and V159N but not by expression of wild-type actin we used the FACS to quantitate phalloidin- and DNase I-stainable actin in cells expressing these actins. Expression of wild-type actin or actin S14A increased both mean cellular F-actin and G-actin contents to a similar extent, leaving the F:G-actin ratio essentially unaltered (Figure 6D). In contrast, expression of S14C and V159N caused a proportionately greater increase in mean F-actin content than G-actin content, indicating that expression of these mutants increases the F:G-actin ratio (Figure 6D). Thus, both the S14C and the V159N mutations alter actin dynamics in favor of F-actin.
Having demonstrated that expression of actins S14C and V159N can alter the dynamics of actin in vivo in favor of F-actin, we tested the effect of these mutants upon activity of the SRF reporter gene. Expression of actin V159N activated the reporter to >50% of the level observed after serum stimulation, whereas activation by actin S14C expression was more effective than by serum; in neither case was SRF activity further enhanced by serum treatment (Figure 6E). In contrast, actin S14A expression substantially inhibited the serum induction of SRF, although somewhat less efficiently than wild-type actin (Figure 6E). These data demonstrate that interference with the dynamic properties of actin itself can cause activation of SRF. Moreover, because cells expressing actin S14C or V159N do not exhibit an absolute decrease in DNase I-stainable G-actin, the results suggest that these mutants are defective in the repressive function of actin on SRF (see DISCUSSION).
DISCUSSION
Rho GTPases signal to SRF via alterations in actin dynamics: signals and agents that promote F-actin formation increase SRF activity, whereas inhibition of actin polymerization prevents SRF activation. Because expression of actin itself, which increases levels of both G- and F-actin, inhibits rather than promotes SRF activity, we previously proposed that SRF is regulated in response to G-actin (Sotiropoulos et al., 1999). Herein, we have demonstrated that expression of wild-type actin does not alter the F:G-actin ratio. We have also identified and characterized three actin mutants, actins R62D, G13R, and VP.C, which cannot polymerize in vivo, and shown that expression of these proteins is sufficient to inhibit SRF activity. Taken together, these results demonstrate that it is G-actin (or a G-actin subpopulation), rather than the F:G-actin ratio, that controls SRF activity. Our studies also identified two actin mutants, actins S14C and V159N, that alter actin dynamics in favor of F-actin formation. Expression of these mutants potentiates rather than inhibits SRF activity, but does not reduce the absolute level of G-actin as assessed by DNase I binding. It seems, therefore, that these mutants are unable to inhibit SRF activity. Our data provide strong evidence that actin itself participates directly in signaling to SRF. Study of the mechanism by which the actin mutants interfere with or promote activation of SRF will give useful insights into the nature of the link between actin and SRF.
Our data do not allow distinction between models in which RhoA signaling promotes SRF activation by reduction in the absolute levels of G-actin, or transient changes in the concentration of a G-actin subpopulation distinguished by conformation or nucleotide binding status. The disparity between actin and SRF levels, and the fact that most unpolymerized actin is bound to proteins such as β-thymosin and profilin, make it likely that only a small G-actin subpopulation is actually involved in SRF regulation. The experiments with actin G150P, which binds irreversibly to the actin chaperone CCT, suggest that this regulatory subpopulation is unlikely to involve nascent actin or the CCT itself. We were unable to detect direct physical interaction between actin and SRF in two-hybrid assays in yeast or tissue culture cells, making unlikely a simple model in which G-actin itself enters the nucleus and acts as an SRF corepressor. Although CRM1 has been reported to mediate nucleocytoplasmic shuttling of actin (Wada et al., 1998), our experiments with leptomycin B indicate that SRF regulation does not involve rapid CRM1-mediated redistribution of either actin itself or a large SRF cofactor. Rather, we favor the view that a G-actin subpopulation regulates the activity of an as-yet-unidentified SRF coactivator. Candidates for such a coactivator might be the BAF and TIP60 chromatin remodeling complexes, which are reported to contain actin itself (Zhao et al., 1998; Ikura et al., 2000; Shen et al., 2000; reviewed by Rando et al., 2000) or proteins of the myocardin/MAL family of SRF coactivators (Ma et al., 2001; Mercher et al., 2001; Wang et al., 2001). We are currently investigating the roles of coactivator and chromatin remodeling complexes in RhoA-actin signaling to SRF and its target genes.
Actins R62D, G13R, and the actin-VP16 fusion protein VP.C do not polymerize in vivo, as assessed by several biochemical and cell biological criteria. The mutant actins neither become stably incorporated into the F-actin cytoskeleton nor disrupt it, in contrast to the toxins and drugs previously demonstrated to inhibit SRF activation such as C3 transferase, C2 toxin, and latrunculins (Hill et al., 1995; Sotiropoulos et al., 1999). The inhibitory effects of nonpolymerizable actins must therefore arise from interference with actin treadmilling itself, rather than indirectly through the disruption of F-actin–dependent signaling complexes. Indeed, expression of actin G13R effectively inhibits SRF activation by constitutively active forms of the mDia1, which acts to promote F-actin accumulation, demonstrating that it must act downstream of this Rho effector (Copeland and Treisman, 2002). Nonpolymerizable actin also inhibits SRF activation by cytochalasin D, suggesting that the target of this drug involved in signaling to SRF is G-actin. Multiple signal pathways converge at SRF (Hill et al., 1994, 1995), and these nonpleiotropic inhibitors of RhoA-actin signaling will allow the contribution of this pathway to SRF function to be assessed.
Why should actin R62D, G13R, and VP.C fail to polymerize? These mutations must alter the conformation of actin monomer in such a way as to prevent its incorporation into the filament. The R62D mutation is likely to prevent formation of a salt bridge between actin subdomains 2 and 4 (Kabsch et al., 1990; Wertman et al., 1992), which may wedge open the nucleotide-binding cleft and/or lead to conformational changes of subdomain 2. The inefficient expression of actin G13R suggests that it may be defective in nucleotide binding. This is likely due to the large side chain, because actin G13A behaves similarly to wild-type actin in our assays (Posern, unpublished data). However, nucleotide binding is not required for actin polymerization per se (De La Cruz et al., 2000). We speculate that both the R62D and G13R mutations lock the protein into a conformational state similar to that of free ADP-actin, in which subdomain 2 is reorganized (Otterbein et al., 2001). This may also be the case for actin VP.C, because deletion or chemical modification of the actin C terminus can also bring about substantial conformational changes in subdomain 2 (Johannes and Gallwitz, 1991; Otterbein et al., 2001). Alternatively, the C-terminal VP16 domain in this fusion mutant might directly obstruct actin monomer addition to the filament-barbed end. It will be interesting to examine these mutants at the structural level.
We identified two mutant actins, V159N and S14C, that apparently increase the stability of F-actin. In yeast actin, the V159N mutation uncouples conformational change in F-actin from phosphate release (Belmont et al., 1999a), and our data suggest that in human β-actin the V159N and S14C mutations may have a similar effect. The mechanism by which S14C might affect phosphate release is not obvious. In ATP-actin, S14 is involved in both hydrogen bonding to the ATP gamma-phosphate and interactions with subdomain 2 (Schutt et al., 1993); in contrast, in free ADP-actin S14 adopts its alternative (and preferred) rotamer to interact with the nucleotide beta-phosphate (Otterbein et al., 2001). It is thus likely that S14 is involved in the structural reorganization of subdomain 2 after ATP hydrolysis; it may also transiently interact with the departing phosphate (Wriggers and Schulten, 1999). Because cysteine has both a lower hydrogen-bonding capacity and the opposite rotamer preference to serine (Ponder and Richards, 1987), the S14C mutation might inhibit the destabilizing structural changes that occur after ATP hydrolysis. In yeast, the actin S14C mutation is lethal, however, in contrast to actin V159N, suggesting that it may also affect other aspects of actin function.
Unlike expression of wild-type actin, expression of actins S14C or V159N, which alter actin dynamics in favor of F-actin, activates rather than represses SRF activity. Combination of the S14C or V159N mutations with the R62D mutation generated proteins that failed to colocalize with endogenous F-actin and that repressed SRF function (Posern, unpublished observations). However, it cannot be concluded from this observation that actins S14C and V159N must polymerize to affect SRF activity, because it remains unclear whether the structural changes induced by the S14C or V159N mutations remain intact upon introduction of the second mutation. Although actins S14C or V159N exhibit enhanced F-actin accumulation, our studies indicate that their overexpression still results in an increase in the total amount of DNase I-bindable actin, albeit to a lesser degree than that induced by expression of wild-type actin. It is thus unlikely that overexpression of these proteins activates the system merely by reducing bulk levels of G-actin below that found in their absence. Instead, our findings suggest that actins S14C or V159N must at least be defective in the repressive function of actin on SRF; indeed, it remains possible that they represent an “activated” conformation of actin that directly promotes transcriptional activation by SRF. These mutants will be useful tools with which to investigate the mechanism of signaling to SRF via actin.
ACKNOWLEDGMENTS
We thank John Copeland for FACS-based F-actin analysis protocol; Derek Davies and Ayad Eddaoudi from the Cancer Research UK FACS laboratory for assistance; and Eisuke Nishida for leptomycin B. We thank Elizabeth McCormack and Keith Willison for communication of unpublished data on actin mutants, stimulating discussions, the actin G150P mutant, and the CCT-binding analysis of actin G13R. Finally, we thank laboratory members, Caroline Hill and Michael Way for helpful discussions and comments on the manuscript. G.P. is the recipient of a European Molecular Biology Organization long-term fellowship.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–05–0068. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–05–0068.
REFERENCES
- Algrain M, Turunen O, Vaheri A, Louvard D, Arpin M. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J Cell Biol. 1993;120:129–139. doi: 10.1083/jcb.120.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Drubin DG. The yeast V159N actin mutant reveals roles for actin dynamics in vivo. J Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Orlova A, Drubin DG, Egelman EH. A change in actin conformation associated with filament instability after Pi release. Proc Natl Acad Sci USA. 1999a;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Patterson GM, Drubin DG. New actin mutants allow further characterization of the nucleotide binding cleft and drug binding sites. J Cell Sci. 1999b;112:1325–1336. doi: 10.1242/jcs.112.9.1325. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Senderowicz AM, Sausville EA, Duncan KL, Korn ED. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J Biol Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- Chen X, Cook RK, Rubenstein PA. Yeast actin with a mutation in the “hydrophobic plug” between subdomains 3 and 4 (L266D) displays a cold-sensitive polymerization defect. J Cell Biol. 1993;123:1185–1195. doi: 10.1083/jcb.123.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Peng J, Pedram M, Swenson CA, Rubenstein PA. The effect of the S14A mutation on the conformation and thermostability of Saccharomyces cerevisiae G-actin and its interaction with adenine nucleotides. J Biol Chem. 1995;270:11415–11423. doi: 10.1074/jbc.270.19.11415. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubenstein PA. A mutation in an ATP-binding loop of Saccharomyces cerevisiae actin (S14A) causes a temperature-sensitive phenotype in vivo and in vitro. J Biol Chem. 1995;270:11406–11414. doi: 10.1074/jbc.270.19.11406. [DOI] [PubMed] [Google Scholar]
- Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- Copeland, J., and Treisman, R. (2002). The diaphanous-related formin mDia1 controls serum response factor (SRF) activity through its effects on actin polymerization. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Mandinova A, Steinmetz MO, Stoffler D, Aebi U, Pollard TD. Polymerization and structure of nucleotide-free actin filaments. J Mol Biol. 2000;295:517–526. doi: 10.1006/jmbi.1999.3390. [DOI] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneste O, Copeland J, Treisman R. LIM kinase and diaphanous cooperate to regulate serum response factor and actin dynamics. J Cell Biol. 2002;157:831–838. doi: 10.1083/jcb.200203126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of SRF target gene. J Biol Chem. 2001;276:24531–24539. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Johannes FJ, Gallwitz D. Site-directed mutagenesis of the yeast actin gene: a test for actin function in vivo. EMBO J. 1991;10:3951–3958. doi: 10.1002/j.1460-2075.1991.tb04965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kuang B, Rubenstein PA. Beryllium fluoride and phalloidin restore polymerizability of a mutant yeast actin (V266G,L267G) with severely decreased hydrophobicity in a subdomain 3/4 loop. J Biol Chem. 1997;272:1237–1247. doi: 10.1074/jbc.272.2.1237. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Vainberg IE, Cowan NJ. Chaperonin-mediated folding of actin and tubulin. J Cell Biol. 1996;132:1–4. doi: 10.1083/jcb.132.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28:220–221. doi: 10.1038/90054. [DOI] [PubMed] [Google Scholar]
- Mack CP, Somlyo AV, Hautmann M, Somlyo AP, Owens GK. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J Biol Chem. 2001;276:341–347. doi: 10.1074/jbc.M005505200. [DOI] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- McCormack EA, Llorca O, Carrascosa JL, Valpuesta JM, Willison KR. Point mutations in a hinge linking the small, and large domains of beta-actin result in trapped folding intermediates bound to cytosolic chaperonin CCT. J Struct Biol. 2001a;135:198–204. doi: 10.1006/jsbi.2001.4385. [DOI] [PubMed] [Google Scholar]
- McCormack EA, Rohman MJ, Willison KR. Mutational screen identifies critical amino acid residues of beta-actin mediating interaction between its folding intermediates and eukaryotic cytosolic chaperonin CCT. J Struct Biol. 2001b;135:185–197. doi: 10.1006/jsbi.2001.4389. [DOI] [PubMed] [Google Scholar]
- Mercher T, et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci USA. 2001;98:5776–5779. doi: 10.1073/pnas.101001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohun T, Garrett N, Treisman R. Xenopus cytoskeletal actin and human c-fos gene promoters share a conserved protein-binding site. EMBO J. 1987;6:667–673. doi: 10.1002/j.1460-2075.1987.tb04806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak KJ, et al. Mutations in the skeletal muscle alpha-actin gene in patients with actin myopathy and nemaline myopathy. Nat Genet. 1999;23:208–212. doi: 10.1038/13837. [DOI] [PubMed] [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Zhao K, Crabtree GR. Searching for a function for nuclear actin. Trends Cell Biol. 2000;10:92–97. doi: 10.1016/s0962-8924(99)01713-4. [DOI] [PubMed] [Google Scholar]
- Sahai E, Alberts AS, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 1998;17:1350–1361. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler H, Korenbaum E, Schutt CE, Lindberg U, Karlsson R. Mutational analysis of Ser14 and Asp157 in the nucleotide-binding site of beta-actin. Eur J Biochem. 1999;265:210–220. doi: 10.1046/j.1432-1327.1999.00716.x. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Tominaga T, Sahai E, Chardin P, McCormick F, Courtneidge SA, Alberts AS. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 2000;5:13–25. doi: 10.1016/s1097-2765(00)80399-8. [DOI] [PubMed] [Google Scholar]
- Tran Quang C, Gautreau A, Arpin M, Treisman R. Ezrin function is required for ROCK-mediated fibroblast transformation by the Net and Dbl oncogenes. EMBO J. 2000;19:4565–4576. doi: 10.1093/emboj/19.17.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Weinhold B, Schratt G, Arsenian S, Berger J, Kamino K, Schwarz H, Ruther U, Nordheim A. Srf(−/−) ES cells display non-cell-autonomous impairment in mesodermal differentiation. EMBO J. 2000;19:5835–5844. doi: 10.1093/emboj/19.21.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Schulten K. Investigating a back door mechanism of actin phosphate release by steered molecular dynamics. Proteins. 1999;35:262–273. [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]