Abstract
The molecular mechanisms of peroxisome biogenesis have begun to emerge; in contrast, relatively little is known about how the organelle functions as cells age. In this report, we characterize age-related changes in peroxisomes of human cells. We show that aging compromises peroxisomal targeting signal 1 (PTS1) protein import, affecting in particular the critical antioxidant enzyme catalase. The number and appearance of peroxisomes are altered in these cells, and the organelles accumulate the PTS1-import receptor, Pex5p, on their membranes. Concomitantly, cells produce increasing amounts of the toxic metabolite hydrogen peroxide, and we present evidence that this increased load of reactive oxygen species may further reduce peroxisomal protein import and exacerbate the effects of aging.
INTRODUCTION
Peroxisomes are essential subcellular organelles of eukaryotic cells. These multifunctional structures arise through the carefully orchestrated reactions of some two dozen proteins, called peroxins (Terlecky and Fransen, 2000). These are critical processes; defects leave cells either devoid of peroxisomes or with organelles rendered unable to carry out the myriad biochemical and metabolic functions ascribed to them. Often, such failings result in disease (Gould and Valle, 2000).
Despite major recent advances in an understanding of how the peroxisome arises and functions, only scant information is available regarding the relationship of the organelle and cellular aging. It is unclear, for example, how the organelle functions as cells age and what role, if any, the peroxisome plays in the aging process. This work attempts to initiate such an examination.
The model system used in these studies is human diploid fibroblasts (HDFs)—cells with a finite replicative lifespan. These somatic cells will divide (or double) in culture until they reach a limit referred to as the “Hayflick number” (Hayflick, 1965). At this point, they cell-cycle arrest and are called “senescent.” There is accumulating evidence that this process of cellular senescence occurs in aged whole organisms (Dimri et al., 1995).
Among the contributing factors to cellular senescence are telomere shortening, DNA damage and related genomic instability, modified expression of genes, and the accumulation of reactive oxygen species (ROS) (reviewed in Johnson et al., 1999). With respect to the latter, mitochondria are widely regarded as the chief cellular generators of ROS and ironically, a major focus of free radical assault (Beckman and Ames, 1998; Lee and Wei, 2001). However, mitochondria are not the only source of cellular ROS.
Among their constituent enzymes, peroxisomes house a variety of hydrogen peroxide–generating oxidases. The organelle also contains catalase, which decomposes hydrogen peroxide and presumably prevents accumulation of this toxic compound. Thus, the peroxisome maintains a delicate balance with respect to the relative concentrations or activities of these enzymes to ensure no net production of ROS. How the organelle maintains this equilibrium is unclear. It is also not known what happens to these regulatory mechanisms as cells (and organisms) age.
Proteins are directed to the peroxisome by specific peptide sequences called peroxisomal targeting signals (PTSs), which are recognized by receptor molecules. All but a select few human peroxisomal proteins contain PTS1, a carboxy-terminal sequence (Subramani, 1998). PTS1 is identified and shuttled to the peroxisome by the soluble peroxin Pex5p (Dammai and Subramani, 2001). For the majority of peroxisomal enzymes, PTS1 is a tripeptide consisting of serine-lysine-leucine or a closely related variant (Subramani, 1998). In contrast, the PTS1 of catalase is a noncanonical PTS1 consisting of the four amino acids lysine-alanine-asparagine-leucine (Purdue and Lazarow, 1996). As we document in this report, it is these distinct PTS1s that lead to dissimilar recognition by Pex5p and, in aging cells, to significantly different import efficiencies. We also show that aging fibroblasts produce increasing amounts of ROS, an apparent consequence of this uncoupling of peroxisomal pro-oxidants and antioxidants. Finally, our characterization of peroxisomes in aging cells reveals changes in the size and number of organelles, as well as their ability to cycle Pex5p from their surfaces.
MATERIALS AND METHODS
Cell Culture
Early-passage IMR90 and Hs27 HDFs, obtained from the National Institute of Aging, Aging Cell Repository/Coriell Institute for Medical Research (Camden, NJ), and ATCC (Manassas, VA), respectively, were cultured in DMEM supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY), penicillin, and streptomycin. The cells were maintained at 37°C in humidified incubators supplemented with 5% CO2. To achieve higher passage levels, the cells were expanded through subcultivation. Late-passage cells were confirmed to be at or near replicative senescence by staining for senescence-associated β-galactosidase as described (Dimri et al., 1995).
Where indicated, cells were grown on glass coverslips pretreated with ProNectin F (Biosource International, Camarillo, CA).
In Vitro Import Assays
Peroxisomal protein import was examined in semipermeabilized cells by use of enzyme-linked immunosorbent assay (ELISA)- and immunofluorescence-based in vitro assays. Both approaches used the PTS1(-SKL)-containing substrate protein luciferase. For the ELISA system, luciferase was biotinylated and import quantified either directly in cells or after isolation of cellular organelles/peroxisomes as described (Terlecky, 2002). To ensure that comparisons were being made from equivalent numbers of cells, DNA content was measured and appropriate corrections were made in all experiments. The fluorometric method of DNA quantification was as described by Downs and Wilfinger (1983), except that SYBR Green (Molecular Probes, Eugene, OR) was used as the DNA-binding dye.
The immunofluorescence-based import assay was carried out as specifically detailed for IMR90 fibroblasts in Legakis and Terlecky (2001).
To examine the effects of H2O2 on import, cells were pretreated overnight with 125 μM H2O2 in serum-containing medium and for 2 h before harvest/permeabilization with 250 μM H2O2 in serum-free medium.
Immunocytochemistry and Microscopy
Cells grown on glass coverslips were fixed for 10 min in 4% (wt/vol) paraformaldehyde, treated for 10 min with 10 mM NH4Cl, and permeabilized for 5 min with 1% (vol/vol) Triton X-100. Cells were blocked for 1 h with 4% (wt/vol) bovine serum albumin (BSA) and incubated with primary antibody for 1 h and secondary antibody for 30–45 min. Rabbit anti-PMP70 (peroxisomal membrane protein of 70 kDa) antibodies were used at a 1/250 dilution, rabbit anti-catalase antibodies were used at a 1/500 dilution, rabbit anti-Pex5p antibodies were used at a 1/500 dilution, and CY3-conjugated goat anti-rabbit antibodies were used at a 1/300 dilution. All reactions were conducted in PBS. Coverslips were mounted using Slowfade antifade (Molecular Probes). A Zeiss LSM-310 confocal microscope was used to obtain all fluorescent images.
For the detection of cellular hydrogen peroxide, the method used was modified from Ohba et al. (1994) and Bass et al. (1983). Here, cells were washed three times with PBS and treated for 10 min at 37°C with 25 μM 2′,7′-dichlorofluorescin diacetate. The cells were washed again, and cellular fluorescence was examined by confocal microscopy using an excitation wavelength of 488 nm. Where indicated, early-passage Hs27 HDFs were labeled by allowing them to endocytose red FluoSphere microspheres (Molecular Probes). After an overnight incubation, these cells were washed and seeded onto coverslips containing (unlabeled) late-passage cells. These mixed populations of cells were then examined for the generation of hydrogen peroxide.
Enzyme Latency
Latency experiments were as modified from Wanders et al. (1984). Briefly, a confluent 15-cm dish was washed two times with Hanks' balanced salt solution, and the cells were removed by trypsinization and resuspended in ∼10 ml of 10 mM HEPES (pH 7.4), 0.25 M sucrose, and 0.1% (vol/vol) EtOH (buffer A). The cells were then pelleted in a clinical centrifuge, washed once with buffer A, resuspended in buffer A, and divided into aliquots into appropriate digitonin- and Triton X-100–containing buffer A reaction solutions. Permeabilization was carried out for 5 min at 4°C, after which the cells were microfuged (2 min) and the resultant supernatants assayed for lactate dehydrogenase or catalase as described (Storrie and Madden, 1990).
Preparation of plasmids/proteins
The pGFP-KANL and pDsRed2-SKL mammalian expression vectors were created by adding a 15-nucleotide sequence to the 3′ end of green fluorescent protein (GFP) in the pEGFP-C3 vector (Clontech, Palo Alto, CA) and a 12-nucleotide sequence to the 3′ end of DsRed2 in the pDsRed2-C1 vector (Clontech) by PCR amplification. For GFP-KANL, the forward primer, 5′-GTGAACCGTCAGATCCGCT-3′, complemented the nucleotide sequence upstream of the GFP ATG start site, which contained an Eco47III site. The reverse primer, 5′-CGTctcgagTTATAGATCAGCTTTCAGCTCGTCCATGCCGAGAGTGATCC-3′, complemented the last 22 nucleotides of GFP and created an in-frame 3′ end that included nucleotides coding for the peroxisomal targeting signal of catalase, −KANL (italic letters), a stop codon, and an XhoI site (lower-case letters). For DsRed2-SKL, the forward primer, 5′-CCGCTAGCGCTACCGGTCGCCACCATGGCC-3′, complemented the nucleotide sequence upstream of the DsRed2 ATG start site, which contained an Eco47III site. The reverse primer, 5′-CGTctcgagTTATAATTTGGACAGGAACAGG-TGGTGGCGGCC-3′, complemented the last 21 nucleotides of DsRed2 and created an in-frame 3′ end that included nucleotides coding for the peroxisomal targeting signal −SKL (italic letters), a stop codon, and an XhoI site (lower-case letters). PCR was performed on a Perkin Elmer-Cetus GeneAmp PCR System 2400, using Pwo polymerase (Roche, Laval, Canada), yielding fragments that encoded either GFP-KANL or DsRed2-SKL flanked by Eco47III and XhoI sites. The pEGFP-C3 and pDsRed2-C1 vectors were digested using XhoI and Eco47III, resulting in release of the GFP- and DsRed2-containing fragments, respectively. The linearized vectors were then ligated overnight with the appropriate digested PCR fragment, either GFP-KANL or DsRed2-SKL, using T4 DNA Ligase (Roche). The results were pGFP-KANL and pDsRed2-SKL, mammalian expression plasmids with GFP-KANL and DsRed2-SKL, respectively, under the control of the cytomegalovirus promoter. Ligation products were transformed into JM109 bacterial host and plated on Luria-Bertani plates containing 50 μg/ml kanamycin. One transformant of each was selected and amplified, and the (pGFP-KANL and pDsRed2-SKL) plasmids were isolated and sequenced (Robarts Research Institute Sequencing Facility) to confirm proper construct sequence. pGFP-SKL was similarly constructed, except that nucleotides coding for the peroxisomal targeting signal −SKL were used instead of those for −KANL.
For use in the Pex5p-binding assays, three (His)6-tagged human catalase proteins differing solely by the identity of their carboxy-terminal residues were expressed in bacteria and purified by use of Ni-NTA agarose. The recombinant proteins were designed to contain at their carboxy-terminus either the naturally occurring KANL sequence (KANL), an SKL sequence (SKL), or no PTS1 sequence at all (−). In the latter case, the KANL sequence was simply deleted. To generate these molecules, the human catalase gene was PCR-amplified from a full-length cDNA clone (Invitrogen, Paisley, UK). The same forward primer was used to amplify each of the three constructs. This nucleotide primer, 5′-ACGCaggcctGCTGACACGCGGGATCCCGCC-3′ complemented the amino-terminal sequence of human catalase along with an StuI restriction site (lower-case letters). Three reverse primers, 5′-GGGCGCaagcttTCACAGATTTGCCTTCTCCCT-3′, 5′-GGGCGCaagcttTCACA-G TTTCGATTTCTCCCTTGCCGCCAAGT-3′, and 5′-GGCGCaagctt-TCACTCCCTTGCCGCCAAGTG-3′, were designed to produce the KANL, SKL, and “−” versions of catalase, respectively. These primers contained nucleotide changes that coded for the appropriate amino acid substitutions and/or deletions. A HindIII restriction site (lower case) was also incorporated downstream of the stop codon. Each of the “catalase” genes were amplified by PCR (Eppendorf Mastercycler), digested appropriately, and ligated into pQE30-Xa (Qiagen, Hilden, Germany). Ligation products were transformed into the Escherichia coli strain DH5α, and recovered plasmids were confirmed to be correct by restriction analysis and DNA sequencing. The sequence-verified (His)6-tagged human catalase constructs were then expressed and purified according to the manufacturer's instructions (Qiagen).
Nuclear Microinjection and Imaging
Early- and late-passage Hs27 cells grown on glass coverslips were microinjected on a Leitz Labovert FS equipped with a microinjector. Glass capillary needles (World Precision Instruments, Sarasota, FL) were prepared with a Kopf vertical pipette puller. Plasmids were diluted to 15 μg/ml in an injection buffer consisting of 100 mM KCl and 20 mM KH2PO4 (pH 7.4). Cells were nuclear-injected with either the pGFP-SKL or the pGFP-KANL and incubated for 18 or 45 h. Live fluorescence images of microinjected cells were collected on a Zeiss Axiovert S100 inverted microscope equipped with a fluorescein isothiocyanate filter set and a charge-coupled device camera. Images were processed using SensiCam imaging software (PCO CCD Imaging).
When pGFP-KANL and DsRed2-SKL were nuclear-microinjected simultaneously into late-passage HDFs, they were added at concentrations of 20 and 15 μg/ml, respectively. These cells were grown on glass coverslips, microinjected, and fixed 42 h later. After mounting on glass slides, the cells were imaged on a Zeiss Axioplan2 microscope.
Pex5p Binding Assays
Human Pex5p was isolated as a glutathione S-transferase fusion protein from E. coli as described (Amery et al., 2001). Proteins (obtained from Sigma, St. Louis, MO) were coated onto microtiter well strips (Maxisorp Immunomodule; Nunc, Naperville, IL) overnight in 50 mM sodium carbonate (pH 9.0). (Equivalent coating of proteins in microwells was confirmed by the Bio-Rad protein assay performed in situ.) The wells were washed twice with PBS and blocked for 4 h at 30°C with 10 mg/ml nonfat milk in PBS plus 0.05% (vol/vol) Tween-20. The wells were washed again and incubated overnight with 1.6 μg GST-HsPex5p in PBS. To determine the amount of GST-Pex5p bound, the wells were washed and incubated with rabbit anti-GST antibodies (dilution 1:2500), followed by peroxidase-labeled goat anti-rabbit antibodies (dilution 1:2500). After washing, the wells were developed and stopped as described (Smythe et al., 1992; Terlecky, 2002). A microplate reader was used to determine the absorbance at 490 nm.
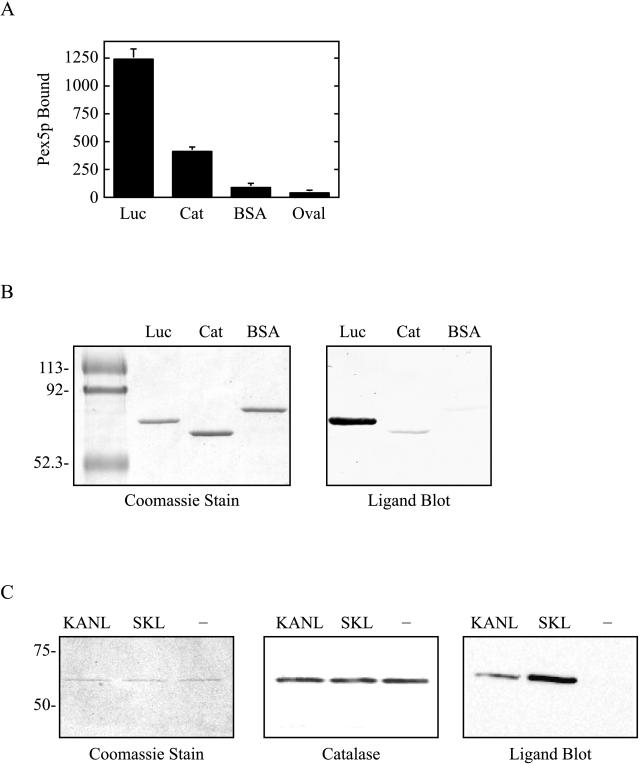
(Pex5p) ligand blots were carried out as described in Fransen et al. (1998), with the following changes. Here, no methionine was used in the reaction buffer, and the ligand was GST-Pex5p. Also, after the binding and washing steps, GST-Pex5p was detected with rabbit anti-GST antibodies (1:2500) and peroxidase-labeled goat anti-rabbit secondary antibodies (1:2500).
Immunoprecipitation and Protease Protection
Immunoprecipitation and protease protection experiments were performed on organelles from IMR90 fibroblasts. To prepare them, equivalent numbers of cells (confirmed by DNA content measurements, as above) were washed with Hanks' balanced salt solution, harvested in homogenization buffer (10 mM ethanolamine [pH 7.8], 10 mM acetic acid, 1 mM EDTA, 0.1% EtOH, 0.25 M sucrose) with a rubber policeman, and disrupted by passage through a narrow-gauge needle followed by Dounce homogenization. Nuclei and unbroken cells were removed by centrifugation at 1000 × g for 10 min at 4°C, and organelles were isolated by centrifugation at 10,000 × g for 20 min at 4°C. (The latter step quantitatively pellets PMP70/peroxisomes from these cells.) For immunoprecipitation, the organelles were lysed with a modified RIPA buffer (50 mM Tris/HCl [pH 7.4], 150 mM NaCl, 1% [vol/vol] NP40, 0.5% [vol/vol] deoxycholate, 0.1% [wt/vol] SDS) plus protease inhibitors (complete cocktail; Sigma), and anti-Pex5p (or preimmune) antibodies were added. After 2 h at 4°C on a nutator, protein A Sepharose (Sigma) was added for 30 min at 4°C. The immunoprecipitate was collected by centrifugation, washed, and run on a 10% SDS-PAGE gel. After transfer to nitrocellulose, the blots were probed with anti-Pex5p antibodies followed by chemiluminescent secondary antibodies (KPL, Gaithersburg, MD).
To protease treat, the organelles were incubated with 50 μg/ml proteinase K (Sigma) for 30 min on ice. The reaction was terminated by the addition of 2 mg/ml phenylmethylsulfonylfluoride. SDS-PAGE sample buffer was then added to the samples, and the proteins were separated on a 10% gel. After transfer to nitrocellulose, immunoblots were performed with anti-Pex5p or anti-catalase antibodies as above. Where indicated, organelles were disrupted with 1% Triton X-100 before protease treatment.
RESULTS
Age-Related Decline in PTS1-Import Efficiency
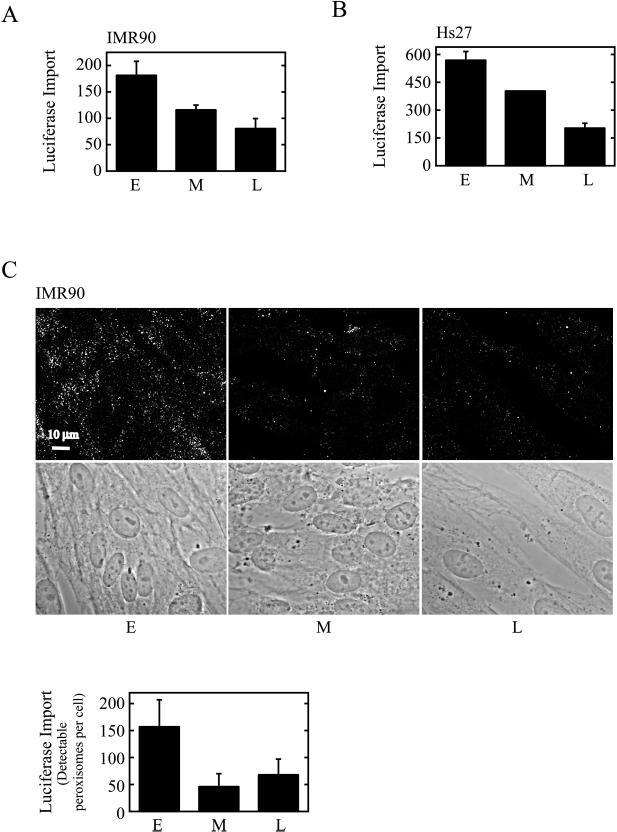
Biochemically defined in vitro assays were used to show that peroxisomal PTS1-protein import is reduced in aging cells (Figure 1). The cells used in this analysis, either IMR90 or Hs27 HDFs, were serially passaged to achieve appropriate population-doubling levels (PDLs). The PDL of a cell may be considered akin to its age (Dice, 1993; for review, see Beckman and Ames, 1998), and for our purposes here, we define (IMR90) early-passage cells as PDL 1–35, middle-passage cells as PDL 36–45, and late-passage cells as PDL 46–60. IMR90 cells reach replicative senescence at ∼PDL60. Hs27 cells, which senesce at comparable passage numbers, were similarly analyzed at early, middle, and late passage. Interestingly, both cell types showed import deficits beginning in middle passage (Figure 1).
Figure 1.
PTS1(-SKL)-protein import in early-, middle-, and late-passage HDFs. (A) Semi-intact IMR90 HDFs were incubated at 37°C with biotinylated luciferase in an in vitro import reaction. After 45 min, the cells were centrifuged and homogenized, and an organelle pellet/peroxisome fraction was prepared. The level of import in equivalent portions of the organelle pellets was determined by ELISA. Values presented (means and ranges of duplicate samples) are absorbance units (at 490 nm) with time zero values subtracted. E, Early-passage cells; M, middle-passage cells; L, late-passage cells. (B) Semi-intact Hs27 HDFs were incubated with biotinylated luciferase as in (A) except that import was assayed directly in cells. (C) Streptolysin-O–permeabilized IMR90 cells were incubated with luciferase for 1 h at 37°C. The cells were then fixed, treated with detergent, and examined by indirect immunofluorescence with anti-luciferase antibodies and CY3-labeled secondary antibodies. Bottom, phase-contrast images of the cells at top. The graph represents quantification of the import results. The number of detectable peroxisomes was determined by counting immunoreactive structures in each of the cells. For this analysis, a greater number of cells were evaluated than are shown in the immunofluorescence image. Values presented are the means ± SD.
The import substrate in these assays was luciferase, a PTS1 protein containing the carboxy-terminal sequence serine-lysine-leucine (Gould et al., 1987). In Figure 1, A and B, we used a biotinylated version of this substrate and an ELISA-based quantitative assay to evaluate import. This assay uses semipermeabilized cells and measures the accumulation of biotinylated-luciferase inside peroxisomes (Terlecky et al., 2001; Terlecky, 2002). After the transport reaction, biotin groups on unimported substrates are blocked, and import is assessed either in organelles prepared by cellular homogenization and fractionation (Figure 1A) or in (lysed) cells (Figure 1B). Irrespective of the cell type or assay variation used, PTS1-protein import was reduced by up to 60% in late-passage HDFs. Qualitatively similar results were obtained using an immunofluorescence-based import assay (Rapp et al., 1993; Wendland and Subramani, 1993) in which cells are semipermeabilized with streptolysin-O and the peroxisomal accumulation of luciferase is determined (Figure 1C). Note that with this system, import seems to be even more dramatically affected in middle-passage cells, perhaps reflecting the threshold nature of the assay. That is, the immunofluorescence signal obtained is largely all-or-none; import reduced below a certain critical level will simply not be detected.
Characteristics of Peroxisomes in Aging Cells
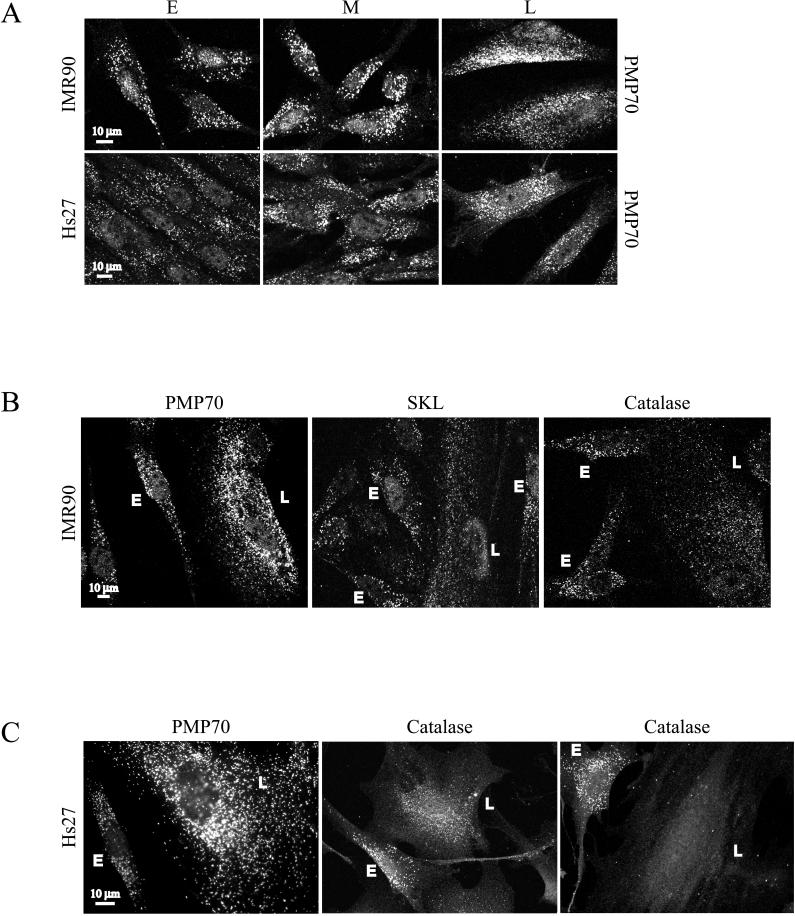
Peroxisomes of early-, middle-, and late-passage HDFs were examined by indirect immunofluorescence microscopy (Figure 2A). The organelles, identified by their reactivity with antibodies to the peroxisomal membrane protein of 70 kDa (PMP70), appeared as randomly scattered punctate structures in early-passage cells. In middle- and late-passage cells, the number of these structures increased. To more carefully document this point, we counted the number of immunoreactive structures per unit area in early-, middle-, and late-passage IMR90 cells. We found that for every one such structure in early-passage cells, there were 1.6 in middle-passage cells and 2.2 in late-passage cells. Similar results were obtained with Hs27 cells (Figure 2A). Furthermore, this increase in peroxisome abundance was also observed with antibodies to the membrane peroxin, Pex14p (data not shown).
Figure 2.
Peroxisomes in early-, middle-, and late-passage HDFs. (A) The peroxisomal membrane marker PMP70 was examined by indirect immunofluorescence in early (E)-, middle (M)-, and late (L)-passage HDFs. (B and C) PMP70, serine-lysine-leucine (SKL)-containing proteins, and catalase were examined by indirect immunofluorescence in cocultured early- (E) and late- (L) passage IMR90 (B) or Hs27 (C) HDFs.
To compare early- and late-passage cells more directly, we analyzed peroxisomal markers in cocultured cells (Figure 2, B and C). For these experiments, early- and late-passage HDFs were subcultivated onto the same culture dishes and coverslips before immunostaining. (The identity of late-passage cells was confirmed by staining with the histochemical biomarker, senescence-associated β-galactosidase [data not shown].) Once again, after staining with antibodies to PMP70, differences in peroxisome number and form were manifest in cells of distinct ages (Figure 2, B and C).
Peroxisomal matrix proteins were also examined by immunocytochemistry in cocultured cells (Figure 2, B and C). Two antibodies were used for this purpose: those generated to catalase and those specific for a peptide containing the carboxy-terminal PTS1 sequence serine-lysine-leucine. Both antibodies recognized punctate structures in early-passage cells (Figure 2, B and C). In late-passage cells, however, the staining was noticeably different; in IMR90s, both matrix markers appeared less intense, with a considerable amount of diffuse, cytosolic staining (Figure 2B). The behavior of peroxisomal matrix markers in late-passage cells was also more variable. Consider Hs27s, for example, in which in some old cells catalase appeared in distinct, peroxisomal structures but also in the cytosol. In others, the staining was more completely cytosolic (compare the two catalase images shown in Figure 2C). These results suggest that at least a portion of cellular catalase and other PTS1-containing enzymes are mislocalized in late-passage cells.
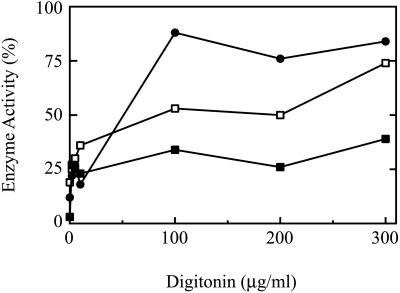
To investigate this point further, we performed latency analysis (Figure 3). In this assay, early- and late-passage (IMR90) cells were treated with increasing concentrations of digitonin, and the release of (cytosolic) lactate dehydrogenase and (peroxisomal) catalase was measured enzymatically. At 100 μg/ml digitonin, lactate dehydrogenase was almost completely released in early-passage cells (Figure 3). (A similar profile was obtained with late-passage cells, but is not shown for clarity.) This concentration establishes the point at which the plasma membrane was compromised and access to the cytosolic compartment was afforded. At this and greater concentrations of digitonin, the relative amount of detectable catalase was significantly higher in the late-passage cells (Figure 3), confirming the mislocalization suggested by immunofluorescence. Note that complete release of catalase was realized only in buffers supplemented with Triton X-100.
Figure 3.
Catalase latency. IMR90 HDFs were treated with increasing concentrations of digitonin, and the levels of catalase (▪) and lactate dehydrogenase (●) were determined. Data are presented as percentage of total cellular activity (set at 100), which was determined in the presence of 1% Triton X-100. Solid symbols, early-passage cells; open symbols, late-passage cells.
Catalase Contains a Weak PTS1 That Interacts Poorly with Pex5p
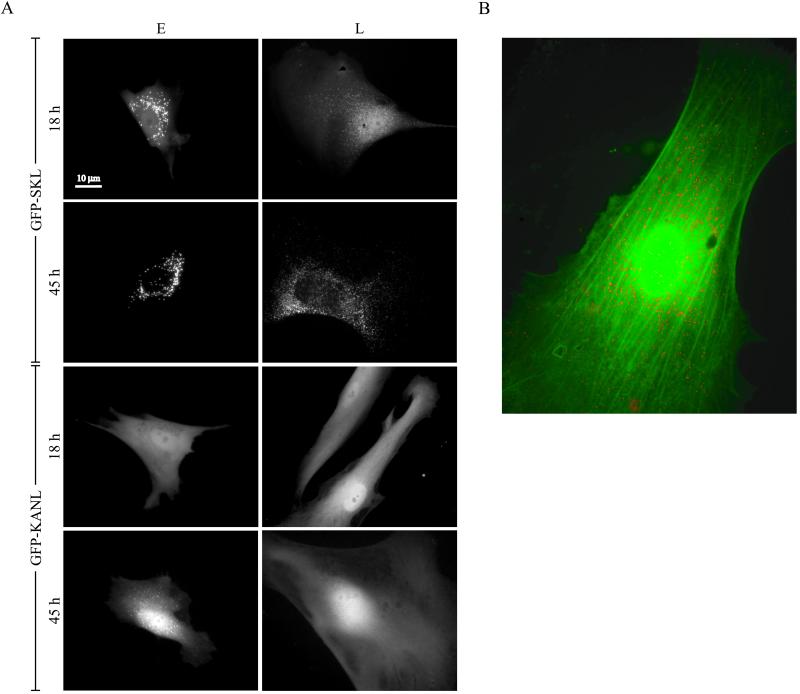
The import of PTS1 proteins containing the prototypical serine-lysine-leucine carboxy-terminus is clearly compromised in aged cells (Figures 1 and 2). Catalase, which contains a divergent PTS1, specifically, lysine-alanine-asparagine-leucine, also shows age-related declines in its import efficiency (Figures 2 and 3). To investigate whether one of these signals is more significantly affected than the other, we nuclear-microinjected plasmids encoding the GFP coupled to either serine-lysine-leucine (GFP-SKL) or lysine-alanine-asparagine-leucine (GFP-KANL) into early- and late-passage HDFs. Live cells were then examined for the expression of the hybrid proteins 18 and 45 h later under a fluorescence microscope (Figure 4A).
Figure 4.
Peroxisomal import of PTS1(−SKL and −KANL)-tagged constructs in early- and late-passage HDFs. (A) Early (E)- and late (L)-passage Hs27 HDFs were nuclear-microinjected with a plasmid encoding GFP-SKL or GFP-KANL as indicated. Live images were taken 18 or 45 h after injection on an inverted microscope. (B) Late-passage Hs27 HDFs were nuclear-microinjected with plasmids encoding DsRed2-SKL and GFP-KANL, incubated for 42 h, fixed, and imaged.
GFP-SKL was efficiently imported in early-passage cells, accumulating in peroxisomes by 18 h. In late-passage cells, import was delayed, with only faint fluorescent structures appearing at 18 h. Only by 45 h did GFP-SKL seem to have been imported to a significant extent. GFP tagged with the catalase PTS1 GFP-KANL did not appear in peroxisomes of early-passage cells until 45 h after microinjection and did not accumulate at all at 45 h in peroxisomes of late-passage cells. In old cells, it took some 115 h before import of GFP-KANL was finally detected (data not shown).
We also examined the import of reporters containing the two PTS1 sequences in the same (senescent) cell (Figure 4B). In this experiment, DsRed2 was coupled to serine-lysine-leucine (DsRed2-SKL), and GFP was coupled to lysine-alanine-asparagine-leucine (GFP-KANL). Note that at 42 h after microinjection, DsRed2-SKL appeared in peroxisomes, whereas GFP-KANL remained largely in the cytosol. Clearly, aging compromises the peroxisomal protein import apparatus, with the PTS of catalase particularly affected.
Import of PTS1-containing proteins is mediated by Pex5p, a soluble receptor molecule that shuttles between the cytosol and the organelle (Dodt and Gould, 1996; Dammai and Subramani, 2001). The functional cycle of Pex5p commences with the binding of its cargo in the cytosol. A potential explanation for differences in the import efficiencies of two PTS1-containing proteins is dissimilar recognition by Pex5p at this step. To determine whether Pex5p displayed preferential interaction with proteins containing a serine-lysine-leucine PTS1, e.g., luciferase, versus those containing a lysine-alanine-asparagine-leucine PTS1, e.g., catalase, we performed solid-phase and ligand-blot binding assays (Figure 5, A and B). In the former, luciferase, catalase, and two control proteins (BSA and ovalbumin) were coated onto the wells of microplates and the binding of GST-tagged human Pex5p was examined. Our results indicate that binding of Pex5p to luciferase was consistently three to four times higher than to catalase (see representative experiment shown in Figure 5A). Only little binding was observed to the control proteins (Figure 5A), and no binding was detected in experiments conducted with no Pex5p added, no proteins coated, or heat-denatured Pex5p (data not shown). Similar results were obtained with ligand blots, in which luciferase, catalase, and BSA were separated by SDS-PAGE, transferred to nitrocellulose, and blotted with Pex5p. Once again, the binding of Pex5p to luciferase was dramatically higher than that to the other proteins tested (Figure 5B).
Figure 5.
Analysis of Pex5p binding. Solid phase assay. (A) Here, 2 μg of luciferase (Luc), catalase (Cat), bovine serum albumin (BSA), or ovalbumin (Oval) were coated onto microtiter wells, and the binding of GST-Pex5p was examined. Values shown are absorbance units (at 490 nm) and represent the mean ± SD for n = 5. Ligand blot assays. (B) Here, 800 ng of Luc, Cat, or BSA were separated by SDS-PAGE and either stained with Coomassie blue (left) or transferred to nitrocellulose membranes and blotted with GST-Pex5p (right). (C) Here, 200 ng of recombinant human catalase containing its own PTS1 (KANL), an altered PTS1 (SKL), or no PTS1 (−), were separated by SDS-PAGE and either stained with Coomassie blue (left) or transferred to nitrocellulose membranes and immunoblotted with anti-catalase antisera (center) or blotted with GST-Pex5p (right).
We also addressed this point by examining binding of Pex5p to purified recombinant human catalase molecules differing only by the identity of their carboxy-terminal residues. In this experiment, catalase molecules were engineered to contain a polyhistidine tag (for purification) and either their own PTS1 (KANL), an altered PTS1 (SKL), or no PTS1 (−). After expression, purification, and characterization (Figure 5C, left and center), the three species were blotted with Pex5p. As shown in Figure 5C, right, Pex5p preferentially binds catalase with the “SKL” PTS1.
Pex5p Cycling
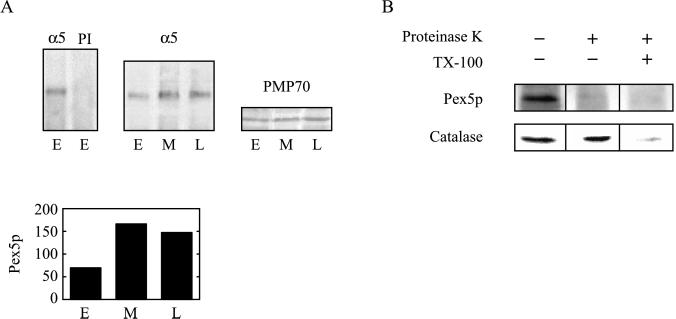
As part of its “extended shuttle” to the peroxisome, Pex5p and bound cargo interact with docking proteins on the organelle membrane. This interaction is transient; accumulation of the receptor at the peroxisome membrane is associated with errors in the cycling mechanism and a resultant reduction in protein import (Dodt and Gould, 1996). To examine whether aberrant Pex5p cycling was associated with aging cells, we analyzed the level of peroxisome-associated Pex5p. To do this, we isolated organelles, which were normalized to equal amounts of PMP70, from HDFs at different ages and immunoprecipitated Pex5p (Figure 6A). Importantly, the level of membrane-associated Pex5p was consistently higher in middle- and late-passage cells. Control experiments revealed that the total amount of cellular Pex5p did not change in these cells, but only the amount associated with organelle membranes was altered (data not shown). Immunostaining of Pex5p in cells confirmed this age-related increase in peroxisome association (data not shown).
Figure 6.
Association of Pex5p with organelle membranes. (A) Pex5p accumulates on organelles of aging HDFs. Pex5p was immunoprecipitated from detergent-solubilized organelles of early-passage (E), middle-passage (M), or late-passage (L) IMR90 HDFs using anti-Pex5p antisera (α5) or preimmune sera (PI), as indicated, and immunoblotted with anti-Pex5p antisera. Organellar PMP70 was also examined by immunoblotting. The graph represents quantification of the α5 immunoblot with a Fujifilm LAS100plus luminescent image analyzer. The units on the ordinate are arbitrary. Equivalent results were obtained in three different experiments. (B) Pex5p accumulates on the outside of organelles from late-passage cells. Organelles from late-passage IMR90 HDFs were treated with proteinase K and Triton X-100 or untreated, as indicated, and the resultant organelles were immunoblotted with anti-Pex5p or anti-catalase antisera.
Recently, Pex5p was shown to actually enter the peroxisome as part of its reaction cycle (Dammai and Subramani, 2001). To determine whether the peroxisome-associated Pex5p observed in late-passage cells was on the membrane or inside the organelle, we performed a protease-protection assay (Figure 6B). In this experiment, organelles from late-passage cells were treated with proteinase-K and immunoblotted. Our results indicate that Pex5p was completely degraded by the protease under conditions in which the luminal enzyme catalase was largely insensitive. Importantly, Pex5p and catalase were both completely degraded by the protease when the organelles were pretreated with detergent. Together, these results suggest that aging cells accumulate Pex5p on the surface of their peroxisomes. It should be noted that Pex5p also appears on the surface of peroxisomes (and is largely protease sensitive) in early-passage cells (data not shown), presumably reflecting normal trafficking of the PTS1 import receptor.
Role of Hydrogen Peroxide in Peroxisome Senescence
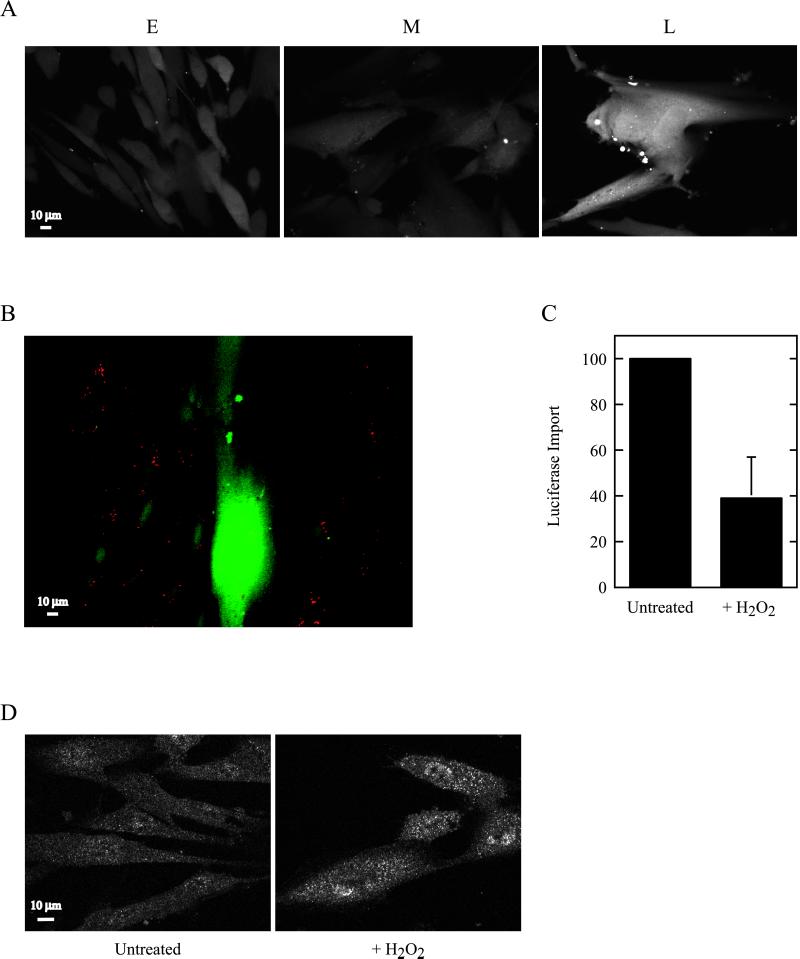
A potential consequence of peroxisomes exhibiting a reduced capacity to import enzymes is a loss of homeostatic regulation. That is, perhaps there is an alteration in the balance between those peroxisomal enzymes that generate hydrogen peroxide and other ROS and those, like catalase, that degrade the toxic metabolites. One manifestation of such a disequilibrium would be a buildup of hydrogen peroxide in cells. To analyze this, we treated HDFs of various ages with the oxidation-sensitive dye 2′,7′-dichlorofluorescin diacetate (Bass et al., 1983; Ohba et al., 1994). This compound enters cells and is converted to a nonfluorescent, cell-impermeant derivative. Exposure to hydrogen peroxide converts the compound to the fluorescent version, 2′,7′-dichlorofluorescein, which is readily visualized by confocal microscopy. As shown in Figure 7A, little hydrogen peroxide was seen in early- and middle-passage cells. However, in late-passage cells, a dramatic increase of the ROS appeared. Similar results were obtained when this assay was performed with cocultured early- and late-passage HDFs (Figure 7B). Also, treatment of early-passage HDFs with the catalase inhibitor aminotriazole resulted in an induction of hydrogen peroxide very similar to that seen in late-passage cells (data not shown). Thus, although our studies certainly support the idea that peroxisomes may contribute to the production of hydrogen peroxide in late-passage cells, the extent of this contribution remains an important open question. Also not entirely clear is why hydrogen peroxide accumulates largely in late-passage cells, although peroxisomal protein import is already impaired in middle-passage cells (Figure 1). Perhaps this reflects the involvement of glutathione peroxidase or other cytosolic hydrogen peroxide–degrading activities whose capacity to process the ROS is eventually overwhelmed in late-passage cells.
Figure 7.
Hydrogen peroxide accumulates in aging HDFs and inhibits peroxisomal protein import. (A) IMR90 HDFs at early (E), middle (M), and late (L) passage were examined for the generation of hydrogen peroxide using the fluorescent dye 2′,7′-dichlorofluorescin diacetate. (B) Hydrogen peroxide in cocultured early- and late-passage Hs27 HDFs. Early-passage cells are readily identified as those having endocytosed red FluoSphere microspheres. (C) Effect of hydrogen peroxide on PTS1 (−SKL)-protein import. Early-passage IMR90 HDFs were pretreated with hydrogen peroxide or untreated as indicated, and peroxisomal protein import was examined as in Figure 1A. Results presented (mean ± one SD) are pooled from five experiments and normalized to the untreated control cell value (arbitrarily set at 100) to permit comparison. (D) Effect of hydrogen peroxide on localization of the PTS1 receptor. Pex5p was examined by indirect immunofluorescence in early-passage HDFs pretreated with hydrogen peroxide or untreated as indicated.
The relationship between cellular accumulation of hydrogen peroxide and the induction of a senescent phenotype has already been established. Specifically, Chen and Ames (1994) showed that HDFs treated with sublethal doses of hydrogen peroxide displayed many characteristics of senescent cells, including growth arrest, reduced activity of critical cellular enzymes, and an “aged” morphology. We addressed a slightly different question in Figure 7C. Specifically, we tested whether exposing early-passage cells to hydrogen peroxide would induce “peroxisome senescence” and effect a reduced ability of the organelle to import its constituent enzymes. Our results suggest that this is the case, because hydrogen peroxide treatment of cells significantly reduced PTS1-protein import (Figure 7C). Furthermore, these cells accumulated Pex5p on their peroxisomes, as determined by immunofluorescence (Figure 7D) and immunoprecipitation (data not shown). In sum, these results suggest that hydrogen peroxide amasses in aging cells and that such accumulation may contribute to a reduction in the functional integrity of peroxisomes. These phenomena presumably contribute not only to the “aging” of peroxisomes but to cellular aging as well.
DISCUSSION
The peroxisome is a ubiquitous organelle of nucleated cells. Its role in various physiological processes, including lipid metabolism and specific steps of cholesterol, bile acid, and plasmalogen biosynthesis, make it indispensable for human health. The organelle carries out a form of respiration, with its oxidases producing hydrogen peroxide as an end product. This highly poisonous ROS is rapidly converted to water through the action of peroxisomal catalase, at least under most circumstances. This work raises the possibility that as human cells age, the ability of the peroxisome to maintain this balance of hydrogen peroxide–generating and –degrading activities and to prevent oxidative stress is compromised. Indeed, it is conceivable from our data that peroxisomal dysfunction actually contributes to the cellular aging process. We have characterized peroxisomes in aging HDFs and offer a preliminary explanation for how this state of lost equilibrium and reduced organelle function arises.
Peroxisomes import enzymes posttranslationally from the cytosol (Lazarow and Fujiki, 1985). We examined age-related changes in the import efficiency of the organelle using several in vitro and in vivo approaches and in two human cell types. Our results indicate that human peroxisomes import a serine-lysine-leucine–containing PTS1 reporter less efficiently with advancing age. Importantly, this import deficit appears in middle-passage cells, long before cells become truly “senescent.”
PTS1 is a name given to a class of peptide sequences that direct proteins to the peroxisome. Although all PTS1-containing enzymes are thought to engage the cycling receptor Pex5p as part of their transport mechanism, other factors are certain to play a role in the import efficiency of one protein versus another. For example, the strength of the interaction with Pex5p may be an important determinant, as may the structure of the protein and its concentration and the presence or absence of requisite accessory factors. Catalase contains a PTS1, but one that is considerably different from all others. Appending its PTS1 to GFP resulted in a fusion protein that was less efficiently imported than a serine-lysine-leucine-tagged GFP reporter in early-passage cells and considerably less well imported than the other reporter in late-passage cells (Figure 4). These results are perhaps not wholly unexpected; catalase has already been described as a relatively poor substrate for the peroxisomal protein import apparatus (Lazarow et al., 1982). In more recent studies, two fibroblast cell lines have been isolated from patients with Zellweger-like disorders, in which the import of catalase is selectively compromised (Sheikh et al., 1998). “Catalase-less peroxisomes” have also been described in a patient with infantile Refsum's disease (IRD) (Fujiwara et al., 2000). These same investigators also characterized a temperature-sensitive Chinese hamster ovary (CHO) cell mutant that, at the nonpermissive temperature, compartmentalized all PTS1-proteins tested except catalase. Interestingly, both the IRD patient fibroblasts and the temperature-sensitive CHO cells were compromised for the membrane peroxin Pex2p. An IRD patient fibroblast has been identified that harbors a mutant form of Pex5p, a defect that renders the cell selectively deficient in catalase import (Shimozawa et al., 1999). Finally, a temperature-sensitive CHO cell exists that, at the nonpermissive temperature, expresses a form of Pex5p that renders the cell able to import all PTS1 proteins tested except catalase (Ito et al., 2001).
Two questions arise: Why is catalase imported less efficiently even in early-passage cells, and why is the effect exacerbated in late-passage cells? The answer to the first question appears to be partly because Pex5p only poorly recognizes catalase (Figure 5). These binding results are supported by recent experiments performed in the yeast Candida boidinii. In this organism, catalase is transported into the peroxisome with an efficiency that is described as “suboptimal” (Horiguchi et al., 2001). These investigators used the two-hybrid system to show that CbPex5p bound to the catalase PTS1, specifically asparagine-lysine-phenylalanine, some 60% less well than it did to another PTS1 sequence from an enzyme in the same organism. Perhaps it is not a coincidence that this is much the same difference we observe in the relative binding affinity of Pex5p for catalase versus luciferase.
With respect to why catalase and other PTS1-containing enzymes are imported with reduced efficiency in aging cells, our work identifies at least one critical mechanistic step that is affected: that of Pex5p cycling. We show that peroxisomes in middle- and late-passage cells accumulate more than twice as much Pex5p on their membranes (Figure 6). Similar aberrant Pex5p trafficking is seen in the cells of patients with defects in certain peroxins (Dodt and Gould, 1996). One such peroxin is Pex2p, a zinc-finger peroxisomal membrane protein directly implicated in PTS1-protein import (Terlecky et al., 2001). Pex2p is a particularly interesting molecule in that it is, to the best of our knowledge, the only peroxin whose (mRNA) expression is dramatically reduced with age (Lee et al., 1999). Although these were mouse expression studies, an intriguing possibility is that Pex2p levels are indeed reduced in aging human cells and that this phenomenon is related to the observed accumulation of membrane-associated Pex5p and defective PTS1-protein import. We are currently examining these points and investigating the role of Pex2p in human peroxisome senescence.
One additional point regarding Pex5p: our work does not address whether its ability to bind PTS1 is compromised with age. It is known that the molecular chaperone hsp70 regulates binding of Pex5p to PTS1 (Harano et al., 2001) and is required for PTS1 protein import into the peroxisome (Walton et al., 1994). Furthermore, the expression of hsp70, like that of Pex2p, decreases significantly with age (Lee et al., 1999). However, the extent to which decreased cellular hsp70 levels affect Pex5p function and contribute to the reduced import capacity of peroxisomes in older cells remains to be determined.
Although absolutely essential, matrix protein import is but one facet of peroxisome biogenesis. Peroxisomes are thought to arise by growth and division (Purdue and Lazarow, 2001). In a simplified view, the organelle membrane assembles appropriate lipids and proteins, matrix enzymes are imported, and the organelle divides. Presumably, these processes are interconnected and carefully regulated. Our observation that peroxisomes are more abundant in older cells (Figure 2), yet display a reduced capacity to import proteins (Figure 1), suggests an age-related disconnect of at least two of the steps. Perhaps aging somehow alters regulation of peroxisome growth and division, leading to organelle proliferation in the absence of normally required cellular cues. PEX11 proteins are directly involved in peroxisome division (Li and Gould, 2002), and it would be interesting to know whether their activity is altered in aging cells.
It seems certain that the accumulation of oxidatively damaged macromolecules plays a role in cellular senescence and is an important determinant of organismal longevity (Beckman and Ames, 1998; Johnson et al., 1999; Lee and Wei, 2001). A number of degenerative diseases may also be linked to ROS-induced alterations in cellular functions (Masters and Crane, 1995). Our results suggest that the peroxisome, an organelle vital to lipid and membrane biosynthesis and functioning, may be a contributor to the oxidative load experienced by aging cells. The organelle converts nearly all of the molecular oxygen it consumes to hydrogen peroxide (Singh, 1996). Coupled with estimates of hepatic peroxisomes consuming 10% or more of total cellular oxygen, it is clear that this is a significant amount of ROS under consideration. The reduced capacity of the peroxisome to import PTS1-containing enzymes, especially catalase, creates functionally compromised organelles in aging cells. These structures do not efficiently metabolize hydrogen peroxide, with serious potential downstream consequences. For example, the activities of peroxisomal enzymes in catalase-deficient aged organelles are probably compromised, as has been demonstrated previously in cells with mislocalized catalase (Sheikh et al., 1998). Also, accumulated hydrogen peroxide will add to oxidative stress and damage cellular constituents. Finally, the effects of hydrogen peroxide actually may further decrease the efficiency of peroxisomal matrix protein import and result in a self-perpetuating negative spiral. Importantly, this spiral may be acting early, before any obvious characteristics of aging are observed and may contribute to the initial stages of peroxisome dysfunction and cellular senescence.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant DK-56299 to S.R.T. and a Canadian Institutes of Health Research operating grant to P.A.W. The authors also acknowledge the Center for Molecular and Cellular Toxicology with Human Applications in Michigan (funded by National Institute of Environmental Health Sciences grant 1-P30-ES-06639).
Footnotes
DOI: 10.1091/mbc.E02–06–0322.
REFERENCES
- Amery L, Sano H, Mannaerts GP, Snider J, Van Looy J, Fransen M, Van Veldhoven PP. Identification of Pex5p-related novel peroxisome targeting signal 1 (PTS1)-binding proteins in mammals. Biochem J. 2001;357:635–646. doi: 10.1042/0264-6021:3570635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds MC, Thomas M. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J Immunol. 1983;130:1910–1917. [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ames BN. Senescent-like growth arrest induced by hydrogen peroxide in human diploid fibroblasts. Proc Natl Acad Sci USA. 1994;91:4130–4134. doi: 10.1073/pnas.91.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- Dice JF. Cellular and molecular mechanisms of aging. Physiol Rev. 1993;73:149–159. doi: 10.1152/physrev.1993.73.1.149. [DOI] [PubMed] [Google Scholar]
- Dimri G, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 import receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs TR, Wilfinger WW. Fluorometric quantification of DNA in cells and tissue. Anal Biochem. 1983;2:538–547. doi: 10.1016/0003-2697(83)90212-9. [DOI] [PubMed] [Google Scholar]
- Fransen M, Terlecky SR, Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc Natl Acad Sci USA. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara C, Imamura A, Hashiguchi N, Shimozawa N, Suzuki Y, Kondo N, Imanaka T, Tsukamoto T, Osumi T. Catalase-less peroxisomes: implication in the milder forms of peroxisome biogenesis disorder. J Biol Chem. 2000;275:37271–37277. doi: 10.1074/jbc.M006347200. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller G, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Valle D. Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 2000;16:340–345. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- Harano T, Nose S, Uezu R, Shimizu N, Fujiki Y. Hsp70 regulates the interaction between the peroxisome targeting signal type 1 (PTS1)-receptor Pex5p and PTS1. Biochem J. 2001;357:157–165. doi: 10.1042/0264-6021:3570157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Horiguchi H, Yurimoto H, Goh T-K, Nakagawa T, Kato N, Sakai Y. Peroxisomal catalase in the methylotrophic yeast Candida boidinii: transport efficiency and metabolic significance. J Bacteriol. 2001;183:6372–6383. doi: 10.1128/JB.183.21.6372-6383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito R, Huang Y, Yao C, Shimozawa N, Suzuki Y, Kondo N, Imanaka T, Usuda N, Ito M. Temperature-sensitive phenotype of Chinese hamster ovary cells defective in Pex5 gene. Biochem Biophys Res Commun. 2001;288:321–327. doi: 10.1006/bbrc.2001.5773. [DOI] [PubMed] [Google Scholar]
- Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell. 1999;96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Robbi M, Fujiki Y, Wong L. Biogenesis of peroxisomal proteins in vivo and in vitro. In: Kindl H, Lazarow PB, editors. Peroxisomes and Glyoxysomes. New York: Annals of the New York Academy of Sciences, Volume; 1982. p. 386. 285–300. [DOI] [PubMed] [Google Scholar]
- Lee C-K, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- Lee H-C, Wei Y-H. Mitochondrial alterations, cellular response to oxidative stress and defective degradation of proteins in aging. Biogerontology. 2001;2:231–244. doi: 10.1023/a:1013270512172. [DOI] [PubMed] [Google Scholar]
- Legakis JE, Terlecky SR. PTS2 protein import into mammalian peroxisomes. Traffic. 2001;2:252–260. doi: 10.1034/j.1600-0854.2001.90165.x. [DOI] [PubMed] [Google Scholar]
- Li X, Gould SJ. PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol. 2002;156:643–651. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CJ, Crane DI. On the role of the peroxisome in ontogeny, ageing and degenerative disease. Mech Ageing Dev. 1995;80:69–83. doi: 10.1016/0047-6374(94)01563-2. [DOI] [PubMed] [Google Scholar]
- Ohba M, Shibanuma M, Kuroki T, Nose K. Production of hydrogen peroxide by transforming growth factor-β1 and its involvement in induction of egr-1 in mouse osteoblastic cells. J Cell Biol. 1994;126:1079–1088. doi: 10.1083/jcb.126.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-targeting sequence. J Cell Biol. 1996;134:849–862. doi: 10.1083/jcb.134.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- Rapp S, Soto U, Just WW. Import of firefly luciferase into peroxisomes of permeabilized Chinese hamster ovary cells: a model system to study peroxisomal protein import in vitro. Exp Cell Res. 1993;205:59–65. doi: 10.1006/excr.1993.1058. [DOI] [PubMed] [Google Scholar]
- Sheikh FG, Pahan K, Khan M, Barbosa E, Singh I. Abnormality in catalase import into peroxisomes leads to a severe neurological disorder. Proc Natl Acad Sci USA. 1998;95:2961–2966. doi: 10.1073/pnas.95.6.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa N, et al. Functional heterogeneity of C-terminal peroxisome targeting signal 1 in Pex5p-defective patients. Biochem Biophys Res Commun. 1999;262:504–508. doi: 10.1006/bbrc.1999.1232. [DOI] [PubMed] [Google Scholar]
- Singh I. Mammalian peroxisomes: metabolism of oxygen and reactive oxygen species. In: Reddy JK, Suga T, Mannaerts GP, Lazarow PB, Subramani S, editors. Peroxisomes: Biology and Role in Toxicology and Disease. New York: Annals of the New York Academy of Sciences, Volume; 1996. p. 804. 612–627. [DOI] [PubMed] [Google Scholar]
- Smythe E, Redelmeir TE, Schmid SL. Receptor-mediated endocytosis in semiintact cells. Methods Enzymol. 1992;219:223–234. doi: 10.1016/0076-6879(92)19024-z. [DOI] [PubMed] [Google Scholar]
- Storrie B, Madden EA. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–180. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Terlecky SR. In vitro analysis of peroxisomal protein import. In: Bonifacino JS, Dasso M, Lippincott-Schwartz J, Hartford JB, Yamada KM, editors. Current Protocols in Cell Biology. New York: John Wiley & Sons; 2002. pp. 11.15.1–11.15.10. [DOI] [PubMed] [Google Scholar]
- Terlecky SR, Fransen M. How peroxisomes arise. Traffic. 2000;1:465–473. doi: 10.1034/j.1600-0854.2000.010604.x. [DOI] [PubMed] [Google Scholar]
- Terlecky SR, Legakis JE, Hueni SE, Subramani S. Quantitative analysis of peroxisomal protein import in vitro. Exp Cell Res. 2001;263:98–106. doi: 10.1006/excr.2000.5111. [DOI] [PubMed] [Google Scholar]
- Walton PA, Wendland M, Subramani S, Rachubinski RA, Welch WJ. Involvement of 70-kD heat-shock proteins in peroxisomal import. J Cell Biol. 1994;125:1037–1046. doi: 10.1083/jcb.125.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJA, Kos M, Roest B, Meijer AJ, Schrakamp G, Heymans HSA, Tegelaers WHH, van den Bosch H, Schutgens RBH, Tager JM. Activity of peroxisomal enzymes and intracellular distribution of catalase in Zellweger syndrome. Biochem Biophys Res Commun. 1984;123:1054–1061. doi: 10.1016/s0006-291x(84)80240-5. [DOI] [PubMed] [Google Scholar]
- Wendland M, Subramani S. Cytosol-dependent peroxisomal protein import in a permeabilized cell system. J Cell Biol. 1993;120:675–685. doi: 10.1083/jcb.120.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]