Abstract
We present evidence to suggest the existence of a regulatory pathway for the Golgi apparatus to modulate the spatial positioning of otherwise distantly located lysosomes. Rab34, a new member of the Rab GTPase family, is associated primarily with the Golgi apparatus. Expression of wild-type or GTP-restricted but not GDP-restricted versions of Rab34 causes spatial redistribution of lysosomes from the periphery to the peri-Golgi region. The regulation of lysosomal positioning by Rab34 depends on its association with the membrane mediated by prenylation and its direct interaction with Rab-interacting lysosomal protein (RILP). This biological activity, mediated by Rab34-RILP interaction, is dependent on Lys82 in the switch I region. Our results have uncovered a novel mechanism for the Golgi apparatus to regulate the spatial distribution of another organelle.
INTRODUCTION
Rab proteins represent the largest family in the ras-like small GTPase superfamily and have been shown to participate in many trafficking events in the cells (Chavrier et al., 1999; Somsel et al., 2000; Pfeffer, 2001; Segev, 2001; Zerial and McBride, 2001). The roles of Rabs range from vesicle budding in ensuring that proper components are sorted or incorporated for subsequent events (Allan et al., 2000; Carroll et al., 2001; Valsdottir et al., 2001), movement of Rab-containing transport intermediates by interacting with components or cofactors of cytoskeleton networks (Chavrier et al., 1999; Nielsen et al., 1999; Marks and Seabra, 2001), and vesicle targeting via interacting with effectors that function as tethering factors to ensure specific tethering of transport intermediates with the target compartment so that subsequent docking and fusion mediated by soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) could take place faithfully (Moyer et al., 2001; Zerial and McBride, 2001).
Despite the importance of Rabs in diverse trafficking events and the fact that close to 60 distinct members have been identified in mammalian cells, only a fraction of these proteins have been investigated extensively. Members found in the Golgi apparatus are Rab1, Rab2, Rab6, and Rab33b. Rab1 is involved in transport from the endoplasmic reticulum (ER) to the Golgi and has been shown to recruit p115, a tethering factor, onto transport intermediates for subsequent tethering events (Allan et al., 2000). In addition, Rab1 also interacts with GM130 (Moyer et al., 2001), a cis-Golgi matrix protein that interacts with p115 and GRASP65. Rab2 regulates maturation of transport intermediates mediating ER-to-Golgi transport (Tisdale and Balch, 1996) and has recently been shown to regulate Golgi structure and function through an effector complex containing Golgin-45 and GRASP55 (Short et al., 2001). Rab6 controls retrograde transport from the Golgi to the ER and interacts with Rabkinesin-6 and GAPCenA (Darchen and Goud, 2000). An alternatively spliced form of Rab6 (called Rab6A′) has been identified and is implicated in regulating other events associated with the Golgi apparatus (Echard et al., 2000). Rab33b is also implicated in retrograde transport from the Golgi back to the ER (Valsdottir et al., 2001). Rab4, Rab5, Rab7, Rab9, and Rab11 are among the best-studied Rabs in the endocytic pathway (Chavrier et al., 1999; Somsel et al., 2000; Pfeffer, 2001; Segev, 2001; Zerial and McBride, 2001). Rab4, Rab5, and Rab11 regulate functions in earlier structures (sorting and/or recycling endosomes) of the endocytic pathway, whereas Rab9 is enriched in late endosomes and regulates traffic from the late endosome to the TGN, and it also coordinates interaction of TIP47 with cargo proteins. Lysosomal Rab7 regulates the distribution and function of late endosomes and lysosomes. Rab-interacting lysosomal protein (RILP) was recently identified as a Rab7 effector and can shift dynein–dynactin complex onto the peri-Golgi region (Cantalupo et al., 2001; Jordens et al., 2001). Several other Rabs are implicated in regulating traffic in a tissue/cell-specific manner. Rab27a is intimately involved in the spatial distribution of melanosomes in melanocytes (Marks and Seabra, 2001), whereas various Rab3s function in regulated secretion mediated by secretory granules or synaptic vesicles (Darchen and Goud, 2000). The existence of many other uncharacterized Rab proteins suggests that cellular processes are likely under the regulation of other Rabs through similar and/or novel mechanisms.
In the present study, we have uncovered a novel mechanism for interorganellar trans-regulation of spatial distribution of lysosomes by Golgi-associated Rab34. We show here that a Rab associated predominantly with one organelle could potentially mediate a long-range regulation of the spatial distribution of another organelle.
MATERIALS AND METHODS
cDNA Cloning and Mutagenesis
The coding region of mouse Rab34 was retrieved by PCR with oligonucleotides 1 (5′-AACTCGAGTGAACATTCTGGCGCCCGTGCGGAGG-3′) and 2 (5′-GCGGATCCTCAGGGACAACATGTGGCCTTCTT-3′) from a mouse EST clone (GenBank accession number AW 742422, clone ID 2780131). The PCR products were digested with enzymes XhoI and BamHI and resolved by agarose gel electrophoresis. The desired DNA fragment was purified and subcloned into the pEGFP-C1 vector (Clontech, Cambridge, UK) in corresponding XhoI and BamHI sites. Rab34T66N, Rab34K82Q, Rab34Q111L, and Rab34F132Y mutants were generated by standard PCR mutagenesis (Lu et al., 2001) and similarly subcloned into pEGFP-C1. All constructs were confirmed by DNA sequencing. The pEGFP-Rab6A′Q72L and pEGFP-Rab33bQ92L were prepared in our laboratory by Lu Lei and T. T. T. Hoai, respectively. The Golgi targeting chimera Rab34Q111L-GS15, Rab34K82Q-GS15, and Rab34Q111L-GRIP as well as Rab34Q111L (del CCP) were constructed by PCR and subcloned into pEGFP-C1 vector. GS15 cDNA was retrieved from the human brain cDNA library and confirmed by sequencing. The cDNA for the GRIP domain (670–748) of Golgin-97 was kindly provided by Lu Lei.
Antibodies
The polyclonal antibody against Rab34 was prepared as described previously (Lowe et al., 1996). Because Rab34 has an N-terminal extension that is unique, this N-terminal region (residues 1–58) was retrieved by PCR and subcloned into pGEX-KG vector into the BamHI and XhoI sites. The recombinant DNA was transformed into DH5α cells to express GST-Rab34(1–58). The fusion protein was purified as described previously (Lowe et al., 1996). The purified GST-Rab34(1–58) was used to inject rabbits at 2-week intervals. Affinity purification of specific antibodies against Rab34 was carried out as described previously (Lu et al., 2001). The monoclonal antibody (mAb) against rat Lamp1 and polyclonal antibody against syntaxin 6 were produced in our laboratory (Wong et al., 1999). The mAbs against human Lamp1, Lamp2, and CD63 developed by J. T. August and J. E. K. Hildreth, respectively, were obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa (Department of Biological Science, Iowa City, IA). The mAb against Golgi mannosidase II was purchased from Babco (Berkeley, CA). mAb against TGN38 was a gift from G. Banting (University of Bristol, UK). Polyclonal antibody against ribophorin was a gift from D.I. Meyer (University of California, Los Angeles, USA). mAb against GFP was purchased from Clontech. mAb against human transferrin receptor was from Transduction Laboratories. mAb against cathepsin D was from Oncogene Research Products. The FITC- and Texas Red–conjugated secondary antibodies were from Jackson ImmunoResearch, West Grove, PA.
Cell Culture and Transient Transfection
Normal rat kidney (NRK) and Hela cells were grown in DMEM supplemented with 10% fetal bovine serum (Gibco, Ann Arbor, MI) in a 5% CO2 incubator at 37°C. Transfection was performed using either Lipofectamine (Life Technologies, Gaithersburg, MD) for immunofluorescence analysis or Effectence (Qiagen, Hilden, Germany) for immunoblot analysis.
Immunoblot Analysis and GTP Overlay
Cells were lysed in standard SDS sample buffer. Approximately 30 μg (for 15 wells of mini-gel) or 50 μg (for 10 wells of mini-gel) of cell lysates was resolved by SDS-PAGE. For immunoblotting, proteins were transferred to a Hybon-C extra nitrocellulose filter. The filter was blocked with 5% skim milk in PBS overnight at 4°C and then incubated with primary antibody for 1 h at room temperature. The filter was washed and incubated with horseradish peroxidase–conjugated secondary antibody (Pierce, Rockford, IL) for 30 min at room temperature. The blots were detected using a chemiluminescence detection kit (Pierce). For the GTP overlay, the recombinant GST-Rab34 fusion proteins were resolved by SDS-PAGE and transferred onto nitrocellulose filter. The filter was incubated with α-(32P)GTP as described previously (Bucci et al., 1992).
Indirect Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described previously (Lu et al., 2001). Cells (control or transfected) grown on coverslips were washed twice with PBSCM (PBS containing 1 mM CaCl2 and 1 mM MgCl2) and fixed for 20 min with 3% paraformaldehyde in PBSCM at 4°C. After sequential washing with 50 mM NH4Cl in PBSCM and then PBSCM, cells were permeabilized with 0.1% saponin (Sigma, St. Louis, MO) in PBSCM for 15 min at room temperature. The permeabilized cells were incubated with primary antibodies in fluorescence dilution buffer (PBSCM containing 5% normal goat serum, 5% fetal bovine serum, and 2% bovine serum albumin) for 1 h at room temperature. After extensive washing, cells were incubated with FITC- and/or Texas Red–conjugated secondary antibodies in fluorescence dilution buffer for 1 h at room temperature. Cells were washed and mounted with Vectashield (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed using a Zeiss Axioplan II microscope equipped with a Bio-Rad (Hercules, CA) MRC-1024 confocal scanning laser. For brefeldin A or nocodazole treatment, cells were incubated with 10 μg/ml brefeldin A (Epicentre Technology, Madison, WI) or nocodazole (Sigma) for 1 h at 37°C before fixation.
Yeast Two-Hybrid Screens and Analysis
cDNAs for the wild-type and mutants of Rab34 in pEGFP-C1 vector were subcloned into the NdeI/BamHI sites of pGBKT7 in the same reading frame as the Gal4 DNA binding domain (Gal-BD) (Clontech). The constructs were transformed into yeast strain AH109 cells. For screening, AH109 cells expressing pGBKT7-Rab34Q111L were mated with a pool (∼5.5 × 107 independent colonies) of Y187 yeast cells pretransformed with human kidney cDNA library fused to Gal4 activation domain (AD) in pACT2 vector (Clontech). The interaction-positive diploid cells were selected on SD/-Trp/-Leu/-His/-Ade (QDO) plates. The Gal4-AD-cDNA in pACT2 vector was recovered from the positive yeast clones on QDO plates and sequenced. To confirm the interaction, the recovered DNA was retransformed into Y187 to perform yeast mating with AH109 cells expressing various bait proteins, and the diploids were selected on QDO plates. The control constructs, pGBKT7-Rab6A′Q72L and pGBKT7-Arl1Q71L, were prepared in our laboratory by Lu Lei. The pGBKT7-Rab33bQ92L was prepared in our laboratory by T. T. T. Hoai.
GST-Pulldown Experiment
A partial cDNA fragment of RILP was recovered from the interaction-positive yeast diploid. For the production of GST-RILP fusion protein, the coding region for residues 103–394 of RILP was retrieved by PCR with oligonucleotide A (5′-AAGAATTCGCG-CGGGGCCACAGGAGGAGCGC-3′) and B (5′-AAGGGCG GC-CGCCCCCAGACAAAGGTGTTCGTGGAG-3′) and then cloned into pGEX-4T-1 vector (Amersham Biosciences, Arlington Heights, IL). The fusion protein was expressed in DH5α cells and purified. The purified GST-RILP (103–394) was coupled to the GST-Sepharose 4B resin for in vitro binding assay. For pulldown experiments, Hela cells were transfected with enhanced green fluorescent protein (EGFP)-tagged Rab34wt, Rab34T66N, Rab34K82Q, Rab34Q111L, and Rab34F132Y. Cells were lysed in binding buffer (containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1% TX-100, and EDTA-free proteinase inhibitor cocktail [Roche]) for 1 h at 4°C. The lysates were spanned down using a TLA-100 rotor at 55,000 rpm for 30 min. The supernatants were incubated with 25 μg GST-RILP bound to the resin in the presence of 100 μM GTP-γ-S (or GDP for Rab34T66N only). After overnight incubation, the resins were washed three times with 20 mM HEPES, pH 7.4, 100 mM NaCl, and 5 mM MgCl2, and then the samples were analyzed by SDS-PAGE and immunoblotting with mAb against GFP.
In Vitro Direct Interaction of 35S-Met–labeled RILP with GST-Rab34
To establish a direct interaction between RILP and Rab34, the full-length RILP cDNA was cloned into pD-MYC vector (Xu et al., 2001). The myc-RILP (35S-Met–labeled) was generated by the use of TNT quick-coupled transcription/translation systems (Promega, Madison, WI) and purified by immunoprecipitation with polyclonal anti-myc tag antibody (Upstate Biotechnology, Lake Placid, NY). After elution, purified myc-RILP was diluted in binding buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2), incubated with GST-Rab34 coupled to GST-Sepharose 4B resin, and loaded with 100 μM GTPγS for the GTP form or 100 μM GDP for the GDP form. After overnight incubation, the resin was washed sequentially with binding buffer containing 1, 0.5, and 0.1% Triton-100. The samples were then resolved by SDS-PAGE. The gel was dried and analyzed by a PhosphorImager.
RESULTS
Rab34 is a 29-kDa Protein Associated Primarily with the Golgi Apparatus
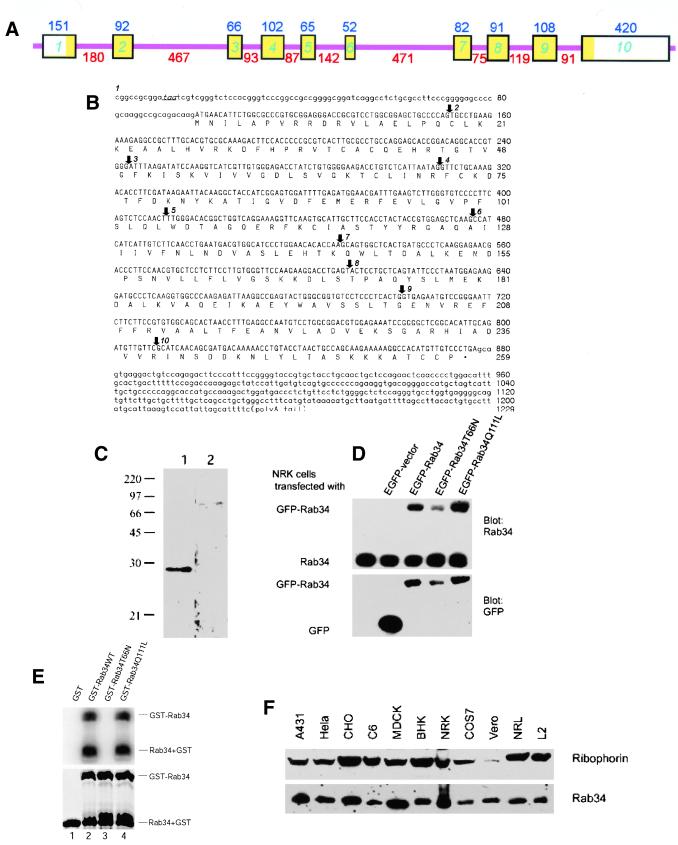
The partial cDNA for Rab34 was originally identified as Rah (ras-related homolog) (Morimoto et al., 1991). The amino acid sequence derived from the complete coding region clearly establishes Rab34/Rah as a member of the Rab family (GenBank accession numbers AF322068 and NM 033475) (Pereira-Leal and Seabra, 2000). The nucleotide sequence of a genomic clone from chromosome 11 (GenBank accession number AC004807) encompasses the entire cDNA sequence of Rab34. Sequence comparison of the cDNA with the genomic clone established that the Rab34 gene has at least 10 exons spanning a genomic region of ∼2954 base pairs with 9 intervening introns (Figure 1A). The complete cDNA and deduced amino acid sequences of Rab34 are shown in Figure 1B. The assigned initiation ATG codon is preceded by an in-frame stop codon (TAA), and its flanking sequence is in good agreement with the Kozak sequence. The predicted mouse Rab34 consists of 259 amino acids and is 94% identical to the human protein (GenBank accession number AAK09397). Rab34 is most homologous to Rab36 with 54% amino acid identity and is much less homologous (∼25% amino acid identity) to other Rab members (such as Rab6, Rab7, and Rab33b). Both Rab34 and Rab36 are relatively larger than other Rabs because of N-terminal extensions (Mori et al., 1999; Pereira-Leal and Seabra, 2000).
Figure 1.
Molecular and biochemical characterizations of Rab34. (A) Mouse Rab34 gene organization: the boxes indicate various exons interrupted by the introns depicted as thin pink lines. The sizes of the exons are indicated above each box, and those of introns are indicated below each intron. The exon regions corresponding to the open reading frame are indicated in yellow. (B) Nucleotide and deduced amino acid sequences of mouse Rab34: The in-frame stop codon preceding the initiation cordon is indicated in italics and underlined. Arrows indicate the starting points of exon 2–10 as indicated. (C) Antibodies raised against GST-Rab34 specially recognize a 29-kDa protein. Total lysates of NRK cells were resolved by SDS-PAGE and transferred to filters. The filters were incubated with anti-Rab34 antibodies in the presence of GST (lane 1) or GST-Rab34 (lane 2). As shown, detection of the 29-kDa polypeptide was abolished by preincubation with the antigen but not GST. (D) Characterization of EGFP-Rab34 fusion proteins. Control NRK cells and cells transfected with EGFP vector, EGFP-Rab34, GDP-restricted EGFP-Rab34T66N, and GTP-restricted EGFP-Rab34Q111L as indicated were analyzed by immunoblot with anti-Rab34 antibodies (top) or anti-GFP antibodies (bottom). In addition to the endogenous 29-kDa Rab34 polypeptide, anti-Rab34 also detected the fusion proteins of ∼59 kDa. (E) Wild-type and Rab34Q111L but not Rab34T66N bind GTP as revealed by GTP overlay assay. The recombinant GST proteins resolved by SDS-PAGE were either stained with Coomassie blue (bottom) or transferred to a filter and incubated with 32P-GTP (top). In addition to the GST-Rab34, autocleavage of fusion proteins into GST and Rab34 was observed. (F) Rab34 is widely expressed in mammalian cells. Approximately 30 μg of total lysates derived from the indicated cells were analyzed by immunoblot using anti-Rab34 antibodies or antibodies against ribophorin.
To study the properties of endogenous Rab34, a recombinant N-terminal unique region (residue 1–58) of Rab34 fused to GST (GST-Rab34) was produced in bacteria and used to raise polyclonal antibodies in rabbit. The affinity-purified antibodies detected a polypeptide of 29 kDa by immunoblot analysis (Figure 1C), and detection of this polypeptide was abolished by preincubation of antibodies with GST-Rab34 (lane 2) but not with GST (lane 1). The apparent size of Rab34 is in excellent agreement with the predicted molecular weight of 29.1 kDa. When cells were transfected with DNA constructs for expressing wild-type Rab34 (EGFP-Rab34), GDP-restricted form (EGFP-Rab34T66N), or GTP-restricted form (EGFP-Rab34Q111L), all N-terminally tagged with EGFP, a polypeptide of ∼59 kDa corresponding to the EGPF fused version was detected by Rab34 antibodies in addition to the endogenous 29-kDa polypeptide (Figure 1D, top). Consistent with this, antibodies against EGFP detected only the 59-kDa fusion proteins (Figure 1D, bottom). Together, these results establish that the antibodies are specific for Rab34. Recombinant full-length Rab34 (wild-type, GDP-restricted T66N, and GTP-restricted Q111L) were also produced in bacteria as fusion proteins with GST. The resulting purified fusion proteins (GST-Rab34WT, GST-Rab34T66N, and GST-Rab34Q111L) were resolved by SDS-PAGE and found to consist of both the fused version and the autocleaved form of GST as well as the various forms of Rab34 (Figure 1E, bottom). When these recombinant proteins were analyzed for GTP binding by overlay assay after being transferred to a filter (Bucci et al., 1992), only the wild-type and GTP-restricted Q111L were capable of binding GTP, whereas the GDP-restricted form failed to interact with GTP (Figure 1E, top). This is true for both the fused (upper form) and the autocleaved forms of Rab34 (lower form). These results suggest that these mutant Rab34 proteins do possess the expected biological properties. When total cell lysates were analyzed by immunoblot, Rab34 was detected in a variety of cell lines from various species (Figure 1F), suggesting that Rab34 is widely expressed in these cell lines and that epitopes recognized by these antibodies are well conserved.
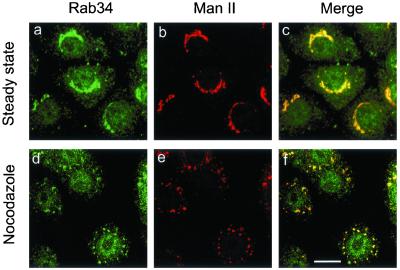
Using the Rab34-specific antibodies described above, we have examined the subcellular localization of endogenous Rab34 by indirect immunofluorescence microscopy (Figure 2). In addition to some diffuse cytosolic labeling, Rab34 was found to be concentrated in perinuclear structures (Figure 2a) marked by Golgi mannosidase II (Man II) (Figure 2b). Nocodazole is known to fragment perinuclear Golgi apparatus into numerous small dot-like mini-Golgi structures (Wong et al., 1999). In nocodazole-treated cells, the Golgi apparatus marked by Man II was fragmented into several small dotted structures spreading throughout the entire cell (Figure 2e). Importantly, Rab34 was found to be enriched in these fragmented Golgi structures (Figure 2d). These results suggest that the majority of Rab34 is associated primarily with the Golgi apparatus. When cells were transfected with a construct for expressing untagged Rab34 and then analyzed using a limiting amount of anti-Rab34 antibodies, it was revealed that the exogenous Rab34 was also preferentially delivered to the Golgi apparatus (data not shown). These results establish Rab34 as another member of the Rab family that is associated predominantly with the Golgi apparatus.
Figure 2.
Rab34 is associated primarily with Golgi apparatus. NRK cells at steady state (a–c) or treated with nocodazole (d–f) were fixed, permeabilized, and incubated with anti-Rab34 antibodies (a, d) and mAb against Golgi marker Man II (b, e). The merged images are also shown (c, f). As shown, Rab34 colocalizes with Man II in both intact and fragmented Golgi apparatus. Bar, 10 μm.
Alternation of Spatial Positioning of Lysosomes on Expression of Wild-Type or GTP-restricted Version of Rab34
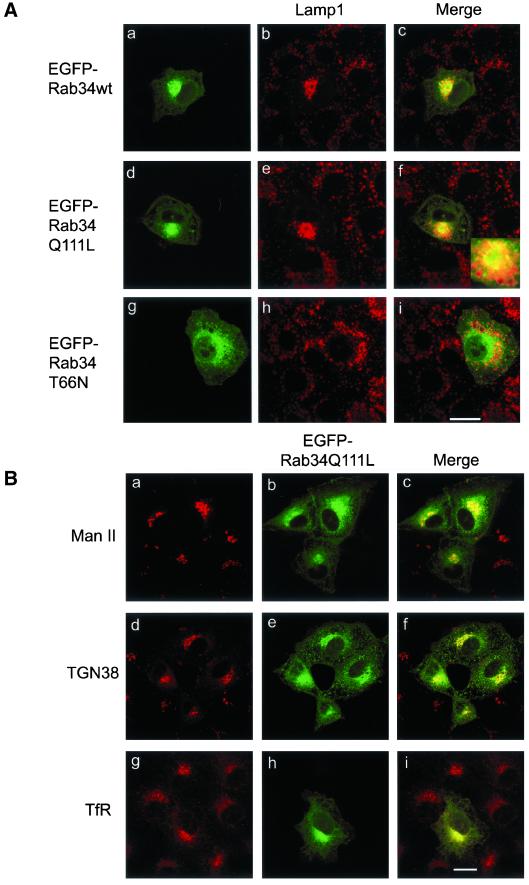
To unravel the biological property and potential physiological function of Rab34, we have examined the effect of expressing EGFP-Rab34, EGFP-Rab34Q111L, or EGFP-Rab34T66N on protein traffic through the secretory and the endocytic pathways. Intriguingly, it was found that the spatial distribution of lysosomes marked by Lamp1 in NRK cells was specifically altered by the expression of EGFP-Rab34 (Figure 3A, a–c) and EGFP-Rab34Q111L (Figure 3A, d–f) but not by EGFP-Rab34T66N (Figure 3A, g–i). Rather than being distributed throughout the entire cytoplasm (cells surrounding the transfected EGFP-positive cells, Figure 3A, b, e, and h), lysosomes were essentially shifted to a compact region that is marked by the expressed EGFP-Rab34 and EGFP-Rab34Q111L. When observing the labeling of lamp1 in several independent experiments, it was found that almost all cells (several hundreds) with compact lamp1 labeling had expression of EGFP-Rab34 or EGFP-Rab34Q111L. Conversely, >90% of cells (>100 randomly chosen cells ranging from low to high levels of expression) expressing EGFP-Rab34 or EGFP-Rab34Q111L had a significant amount of lamp1 being shifted to the perinuclear region. In most cells, the relocated lysosomes were seen not to overlie but rather to surround the Golgi-associated EGFP-Rab34 or EGFP-Rab34Q111L, suggesting that lysosomes are relocated to the peri-Golgi area (Figure 3A, f and Figure 5A, a–c). The expression of EGFP-Rab34T66N, conversely, did not significantly affect the distribution of lysosomes. Examination of >100 randomly chosen EGFP-Rab34T66N–positive cells ranging from low to high levels of expression revealed that 90% of EGFP-Rab34T66N–expressing cells had normal distribution of lysosomes.
Figure 3.
Expression of wild-type, GTP-restricted but not GDP-restricted forms of Rab34 specifically shifted lysosomal positioning from the periphery to the peri-Golgi region. (A) NRK cells were transfected with EGFP-Rab34wt (a–c), GTP-restricted EGFP-Rab34Q111L (d–f), and GDP-restricted EGFP-Rab34T66N (g–i). Transfected cells were fixed, permeabilized, and then labeled with mAb against lysosomal marker Lamp1. As shown, lysosomes were shifted from the periphery to the peri-Golgi region in cells expressing EGFP-Rab34wt and EGFP-Rab34Q111L but not EGFP-Rab34T66N. (B) NRK cells transfected with EGFP-Rab34Q111L were processed for indirect immunofluorescence microscopy with monoclonal antibodies against Man II (a–c), TGN marker TGN38 (d–f), or early endosomal transferrin receptor (TfR) (g–i). As shown, the spatial distribution of the Golgi apparatus and early endosomes was not significantly affected by the expression of EGFP-Rab34Q111L. Bars, 10 μm.
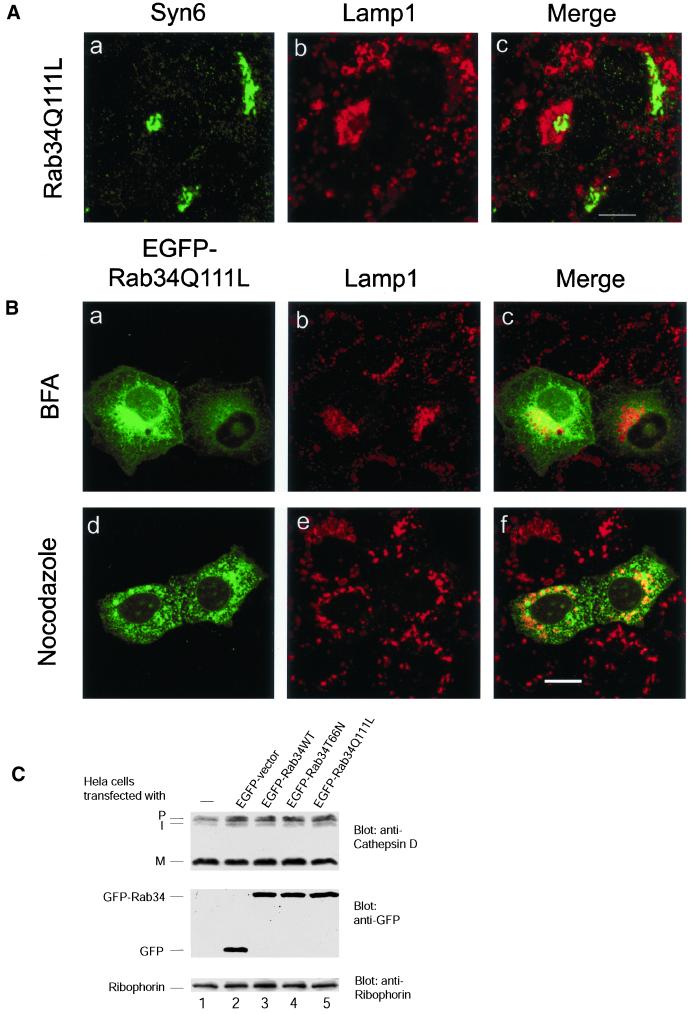
Figure 5.
Relocated lysosomes maintain some structural and functional identities. (A) NRK cells were transfected with Rab34Q111L and then processed to detect the Golgi apparatus marked by syntaxin 6 (a) and lysosomes marked by Lamp1 (b). As shown, redistributed lysosomes seem to embrace the Golgi apparatus. Bar, 10 μm. (B) NRK cells transfected with EGFP-Rab34Q111L was treated with brefeldin A (a–c) or nocodazole (d–f) (both at 10 μg/ml for 60 min at 37°C) and then processed to detect lysosomes by anti-Lamp1 labeling. As shown, lysosomal repositioning induced by EGFP-Rab34Q111L was not affected by brefeldin A but was randomized by nocodazole treatment. Bar, 10 μm. (C) Control or Hela cells transfected using Effectence (efficiency of transfection was estimated to be ∼60–70% by viewing EGPF positive cells) with constructs for expressing indicated proteins were processed for immunoblot analysis to detect various forms of cathepsin D (P for Golgi-associated form, I for endosomal intermediate, and M for mature lysosomal form) (top). As shown, the appearance of mature lysosomal form of cathepsin D was not affected by expression of any form of Rab34, suggesting that transport of cathepsin D from the Golgi to the lysosome was not significantly affected. The expressed EGFP and EGFP-Rab34 were revealed to be present at comparable levels in these transfected cells (middle), whereas the loading was normalized to the amount of ribophorin (bottom).
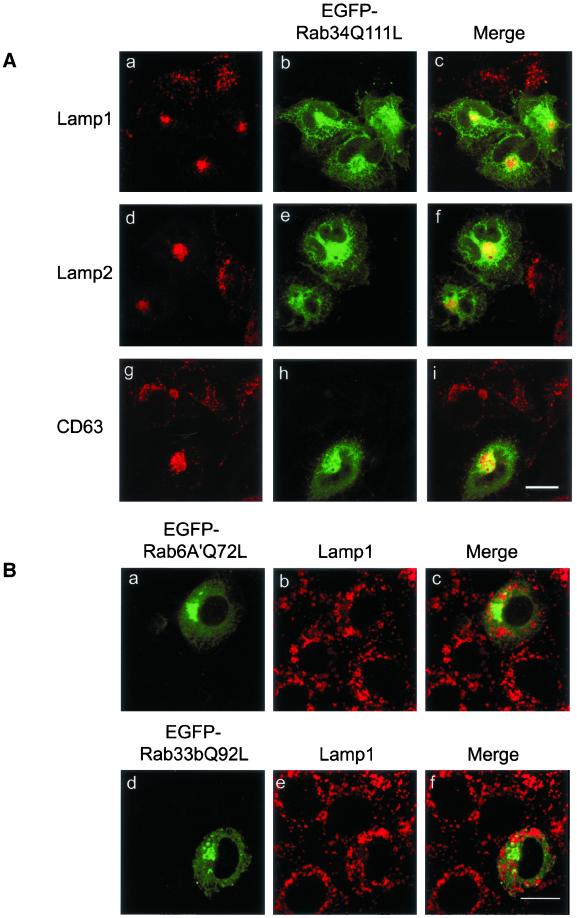
The observed effect of EGFP-Rab34 and EGFP-Rab34Q111L on lysosomes is specific, because the distributions of the Golgi cisternae marked by Golgi mannosidase II (Figure 3B, a–c), the TGN marked by TGN38 (Figure 3B, d–f) or syntaxin 6 (data not shown), and early endosomes marked by transferrin receptor (Figure 3B, g–i) were essentially not affected. Furthermore, transport to the surface of the envelope protein of vesicular stomatitis virus was also not affected by the expression of any form of Rab34 (data not shown). Similar to the Golgi association of endogenous Rab34, the majority of expressed EGFP-Rab34Q111L colocalized well with Man II and TGN38 (Figure 3B, c and f). Similar colocalization of EGFP-Rab34 with Man II and TGN38 was also observed (data not shown), further supporting our conclusion that Rab34 is preferentially associated with the Golgi apparatus. To complement our observation made in NRK cells, we have also examined the effect of EGFP-Rab34Q111L on the distribution of lysosomes in Hela cells using three different lysosomal makers (Figure 4A). As shown, lysosomes marked by Lamp1 (Figure 4, a–c), Lamp2 (Figure 4, d–f), and CD63 (Figure 4, g–i) were all shifted to the compact peri-Golgi region marked by the expressed EGFP-Rab34Q111L, which is more diffusely distributed than the more compact Golgi distribution in NRK cells shown in Figure 3.
Figure 4.
GTP-restricted Rab34 but not Rab6A′ or Rab33b shifted lysosomes from peripheral to peri-Golgi positioning. (A) Hela cells were transfected with EGFP-Rab34Q111L and then labeled with lysosomal markers Lamp1 (a–c), Lamp2 (d–f), and CD63 (g–i). As shown, lysosomes marked by all three markers were shifted to the peri-Golgi region in Hela cells. Bar, 10 μm. (B) NRK cells were transfected with GTP-restricted form of Rab6A′ (EGFP-Rab6A′Q72L) (a–c) or Rab33b (EGFP-Rab33bQ92L) (d–f) and then processed to detect lysosomes with anti-Lamp1 antibody. These two other Golgi-associated Rabs did not affect the distribution of lysosomes. Bars, 10 μm.
Because Rab36 is highly homologous to Rab34, we also checked its effect on lysosomal distribution. The results suggest that Rab36 also possesses the ability to regulate the spatial distribution of lysosomes (data not shown). In marked contrast, overexpression of GTP-restricted Rab6A′ or Rab33b, two other Rabs associated with the Golgi apparatus (Chavrier and Goud, 1999; Darchen and Goud, 2000; Zerial and McBride, 2001), had no noticeable effect on the distribution of lysosomes (Figure 4B), suggesting that the observed regulation of lysosome distribution by Rab34 and Rab36 is specific for these Rabs in the Golgi apparatus.
Redistributed Lysosomes Might Maintain Their Structural and Functional Identity
Because the redistributed lysosomes maintained their unique vacuole-like structure with a clear ring labeling for lysosomal membrane proteins (particularly in NRK cells) and are located in the peri-Golgi region, it seems that lysosomes retain some of their structural identity. As shown in cells transfected with GTP-restricted Rab34, the relocated lysosomes could clearly be seen to segregate from the Golgi apparatus marked by syntaxin 6 in that lysosomes seem to embrace the Golgi apparatus (Figure 5A). This interpretation was strengthened by the additional observation that the distribution of relocated lysosomes was not affected by treatment with brefeldin A (Figure 5B, a–c), which is known to redistribute Golgi proteins back to the ER and to cause TGN and early endosomes to fuse into a compact structure located near the microtubule-organizing center while having no significant effect on lysosomal distribution (Klausner et al., 1992). Furthermore, processing of procathepsin D to the mature lysosomal form in cells overexpressing various Rab34 constructs was not significantly affected (Figure 5C), suggesting that the function of lysosomes (at least the processing of cathepsin D) was also maintained regardless of their spatial distribution. These results indicate that Rab34 may selectively regulate the spatial distribution of lysosomes.
Although the distribution of relocated lysosomes was not affected by brefeldin A treatment, it was randomized by treatment with nocodazole (Figure 5B, d–f), suggesting that the effect of Rab34 on the spatial redistribution of lysosomes depends on an intact microtubule network.
Interaction of Rab34 with RILP
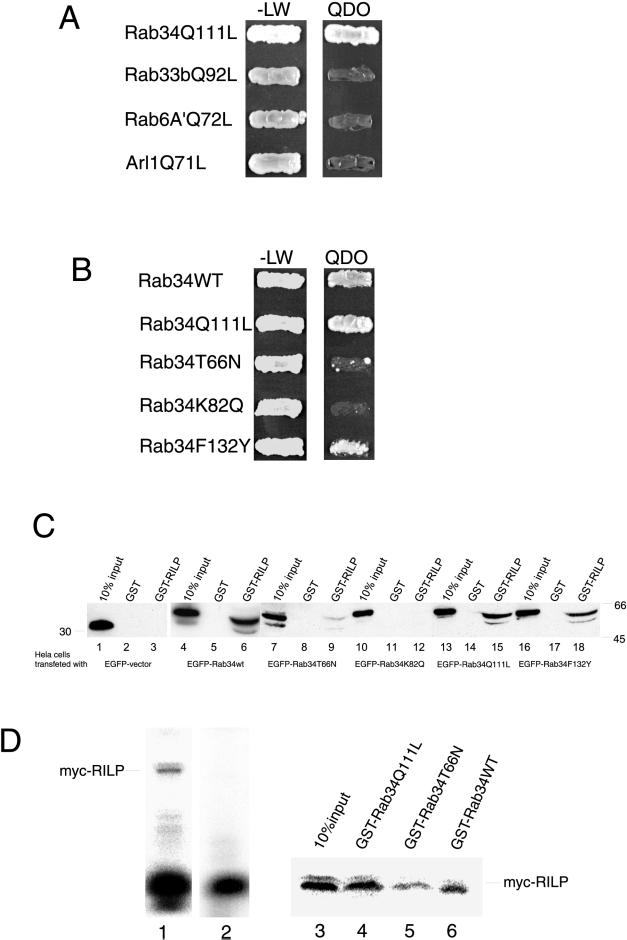
To determine the molecular mechanism underlying the role of Rab34 in regulating lysosomal positioning, we searched for its interacting partners. Because pulldown experiments and coimmunoprecipitations have not led to identification of specific interacting proteins (data not shown), we have exploited the more direct approach of yeast two-hybrid screens. The yeast strain AH109 harboring the pGBKT7 vector containing Rab34(Q111L) fused to the Gal4 DNA-binding domain was mated with 5.5 × 107 independent yeast colonies (strain Y187) containing a human kidney cDNA library fused to the Gal4 DNA activation domain in the pACT2 vector. Screening at the highest stringency revealed four independent clones that are positive for interaction with Rab34. DNA sequencing revealed that three of the clones contain partial cDNA encoding RILP, which was recently identified as a partner for activated GTP-restricted Rab7 and implicated in regulating lysosome distribution (Cantalupo et al., 2001; Jordens et al., 2001). Because RILP could potentially regulate dynein-dynactin distribution, it was proposed that RILP could regulate lysosome distribution through the dynein-dynactin complex (Jordens et al., 2001). The interaction of Rab34 with RILP is shown in Figure 6. The GTP-restricted Rab34 but not Rab33b, Rab6A′ (Echard et al., 2000), or Arl1 (Lu et al., 2001) showed interaction with RILP (Figure 6A). Wild-type and GTP-restricted Rab34 but not GDP-restricted Rab34 interacted with RILP (Figure 6B). The interaction of Rab34 with RILP was further supported by GST-pulldown experiments (Figure 6C). Lysates derived from cells expressing various forms of Rab34 fused to EGFP were incubated with immobilized GST or GST-RILP (residues 103–394). After extensive washing, the amounts of EGFP-fused proteins that were retained on the beads were determined by immunoblot analysis using antibodies against EGFP. Approximately 5–10% of EGFP-Rab34Q111L was retained by immobilized GST-RILP (lane 15) but not by GST (lane 14). Similar results were observed for EGFP-Rab34 (lanes 4–6). The amount of EGFP-Rab34T66N retained by immobilized GST-RILP was routinely <2% (lane 9) of the input. EGFP was not retained by immobilized GST (lane 2) or by GST-RILP (lane 3). These results suggest that RILP can interact preferentially with wild-type and GTP-restricted forms of Rab34. Because the yeast two-hybrid assay most likely measures a direct interaction between Rab34 (the bait) and RILP (the prey), it seems that Rab34 can interact directly with RILP. We have also investigated a direct interaction between Rab34 and RILP (Figure 6D). 35S-Met–labeled myc-RILP was generated by in vitro translation (lane 1). After immunoprecipitation using immobilized anti-myc antibodies, the labeled RILP was depleted from the translation reaction (lane 2). After being eluted from the beads, the affinity-purified RILP was incubated with various immobilized forms of Rab34 fused to GST, and the amounts of 35S-met–labeled RILP retained by the respective beads (lanes 4–6), along with 10% of input (lane 3), were analyzed by SDS-PAGE and PhosphoImager. As shown, RILP was most efficiently retained by immobilized GTP-restricted GST-Rab34Q111L (lane 4) and to a lesser extent by GST-Rab34WT (lane 6). An even smaller amount of RILP was retained by immobilized GST-Rab34T66N (lane 5). These results further suggest that Rab34 and RILP can interact directly, and this interaction is preferentially for the GTP-restricted form of Rab34. Together, these results suggest that RILP may serve as a possible effector protein for Rab34.
Figure 6.
RILP is an effector of Rab34. (A) Using Rab34Q111L as bait in a yeast two-hybrid system to screen a human kidney cDNA library, a partial coding region of RILP (residues 103–401) was identified. AH109 yeast cells expressing pGBKT7-Rab34Q111L, pGBKT7-Rab33bQ92L, pGBKT7-Rab6A′Q72L, or pGBKT7-Arl1Q71L were mated with Y187 yeast cells expressing pACT2-RILP(103–401). The diploids were grown on SD/-Leu/-Trp (-LW) or SD/-Leu/-Trp/-His/-Ade (QDO) plates. As shown, RILP specifically interacts with GTP-restricted form of Rab34 but not with Rab33b, Rab6A′, or Arl1, three other small GTPases associated with Golgi apparatus. (B) The interaction between Rab34 and RILP is preferential for the wild-type and GTP-restricted form and is dependent on K82 but not F132. AH109 yeast cells expressing pGBKT7-Rab34WT, pGBKT7-Rab34Q111L, pGBKT7-Rab34T66N, pGBKT7-Rab34K82Q, or pGBKT7-Rab34F132Y were mated with Y187 yeast cells expressing pACT2-RILP(103–401). The diploids were grown on either -LW or QDO plates. (C) In vitro binding assay showing K82-dependent preferential interaction of RILP with wild-type and GTP-restricted form of Rab34. Hela cells were transfected with EGFP-vector (lanes 1–3), EGFP-Rab34wt (lanes 4–6), EGFP-Rab34T66N (lanes 7–9), EGFP-Rab34K82Q (lanes 10–12), EGFP-Rab34Q111L (lanes 13–15), and EGFP-Rab34F132Y (lanes 16–18). The resulting cell lysates were incubated with GST-RILP(103–394) or GST, as indicated, coupled to GST-Sepharose 4B resin in the presence of 100 μM GTP-γ-S (or GDP for Rab34T66N) during the binding experiments. Proteins retained on the beads were resolved by SDS-PAGE (along with 10% of starting materials) and then processed for immunoblotting analysis using an mAb against GFP. As shown, EGFP-Rab34wt, EGFP-Rab39Q111L, and EGFP-Rab34F132Y but not others were efficiently retained (∼5–10% of total input) by immobilized RILP but not GST. Less than 1% of the GDP-restricted form of Rab34 (lane 7–9) was retained by the GST-RILP beads. (D) Direct interaction between RILP and Rab34. 35S-Met–labeled myc-RILP was generated by in vitro translation (lane 1, 10 μl of in vitro translation reaction). After immunoprecipitation with myc antibodies, labeled RILP was efficiently depleted (lane 2; 10 μl of in vitro translation reaction after immunoprecipitation). The immunopurified RILP was incubated with immobilized GST-Rab34Q111L (lane 4), GST-Rab34T66N (lane 5), or GST-Rab34wt (lane 6). The amounts of 35S-Met–labeled myc-RILP retained by these beads, together with 10% input (lane 3), were resolved by SDS-PAGE and revealed by PhosphorImager.
K82 in Switch I Region of Rab34 Is Important for Interaction with RILP
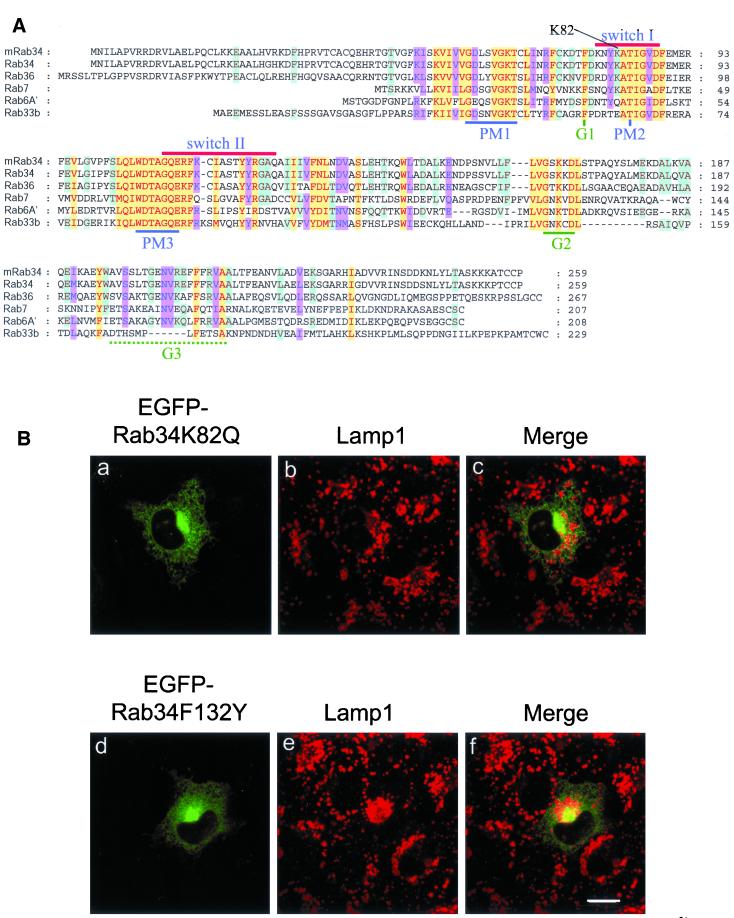
Because Rab34 shares similar levels of amino acid identities with Rab7 and other Rabs (such as Rab6A′ and Rab33b) and the steady-state distribution of the majority of Rab34 (the Golgi) and Rab7 (lysosomes and late endosomes) is different, it is intriguing to find that RILP is potentially a downstream effector shared by Rab34 and Rab7. To determine the structural basis underlying the interaction between Rab34 and RILP, we examined the amino acid residues that are selectively conserved in Rab34, Rab36, and Rab7 but not in Rab6A′ or Rab33b. In doing so, we found that the Lys residue at position 82 (K82) in the switch I region of Rab34 is also conserved in Rab36 and Rab7 (Figure 7A). In addition, a few other residues in the Rab34 switch II region (G124 and A125) and C-terminal flanking region (F132, L151, F162) are also selectively conserved in Rab36 and Rab7. A chimeric protein consisting of the N-terminal 142-residue of Rab34 and the C-terminal 104-residue of Rab6A′ remains capable of redistributing lysosomes (data not shown). This suggests that the structural information in Rab34 required for redistributing lysosomes lies within the N-terminal 142-residue region. We then focused our attention on K82, G124, A125, and F132. Mutations of each of these residues were performed by substituting them with the corresponding residues in Rab6A′, and it was found that K82 plays an essential role in interaction with RILP. When K82 of Rab34 was mutated to the corresponding residue (Q) in Rab6A′, the resulting mutant (Rab34K82Q) failed to interact with RILP as assessed by the yeast two-hybrid assay (Figure 6B) and by GST-pulldown experiments (Figure 6C, lanes 10–12). As a control, another mutant (Rab34F132Y), in which F132 was replaced by the corresponding residue (Y) in Rab6A′, remains capable of interacting with RILP, as analyzed by both yeast two-hybrid assay (Figure 6B) and GST-pulldown (Figure 6C, lanes 16–18). These results clearly establish that K82 in the switch region I of Rab34 is essential for interaction of Rab34 with RILP.
Figure 7.
K82, which is important for interaction of Rab34 with RILP, is also essential for Rab34 to effect lysosomal positioning. (A) Alignment of amino acid sequences of mouse Rab34 (mRab34) and other human Rabs as indicated. The characteristic regions of small GTPases and K82 conserved among Rab34, Rab36, and Rab7 but not Rab6A′ or Rab33b are indicated. Residues highly conserved in all Rabs are indicated in red under yellow background, residues conserved in five of these six Rabs are indicated in blue under pink background, and those conserved in four Rabs are shown in brown under light blue background. (B) NRK cells transfected with EGFP-Rab34K82Q (a–c) and EGFP-Rab34F132Y (d–f) were processed for indirect immunofluorescence microscopy using anti-Lamp1 antibody. As shown, although both forms of Rab34 were enriched primarily in the Golgi apparatus, lysosomal redistribution was observed in cells expressing EGFP-Rab34F132Y but not EGFP-Rab34K82Q. Bar, 10 μm.
Interaction of Rab34 with RILP Is Essential for Its Effect on Lysosome Positioning
Because wild-type and GTP-restricted but not GDP-restricted forms of Rab34 could effectively redistribute lysosomes to the peri-Golgi region and are capable of interacting with RILP, in conjunction with the known effect of RILP on the spatial distribution of lysosomes, it seems likely that the observed biological effect of Rab34 on lysosome distribution is mediated, at least in part, by its direct interaction with RILP. This possibility was established by the observation that expression of EGFP-Rab34K82Q failed to redistribute lysosomes to the peri-Golgi region, although the majority of it was faithfully targeted to the Golgi apparatus (Figure 7B, a–c). Examination of >100 randomly chosen EGFP-Rab34K82Q–positive cells ranging from low to high levels of expression revealed that 80% of EGFP-Rab34K82Q–expressing cells had normal distribution of lysosomes. Furthermore, expression of EGFP-Rab34F132Y, which remains capable of interaction with RILP, resulted in the redistribution of lysosomes to the peri-Golgi region (Figure 7B, d–f). These results suggest that K82 is essential not only for Rab34 to interact with RILP but also to regulate the spatial distribution of lysosomes, although this residue is not important for Golgi association. Taken together, these results suggest that interaction with RILP is most likely the underlying molecular mechanism for Rab34 in regulating the spatial distribution of lysosomes and that K82 is a key residue that couples the molecular interaction with the biological effect.
Interorganellar trans-Regulation of Lysosomal Positioning by the Golgi Apparatus
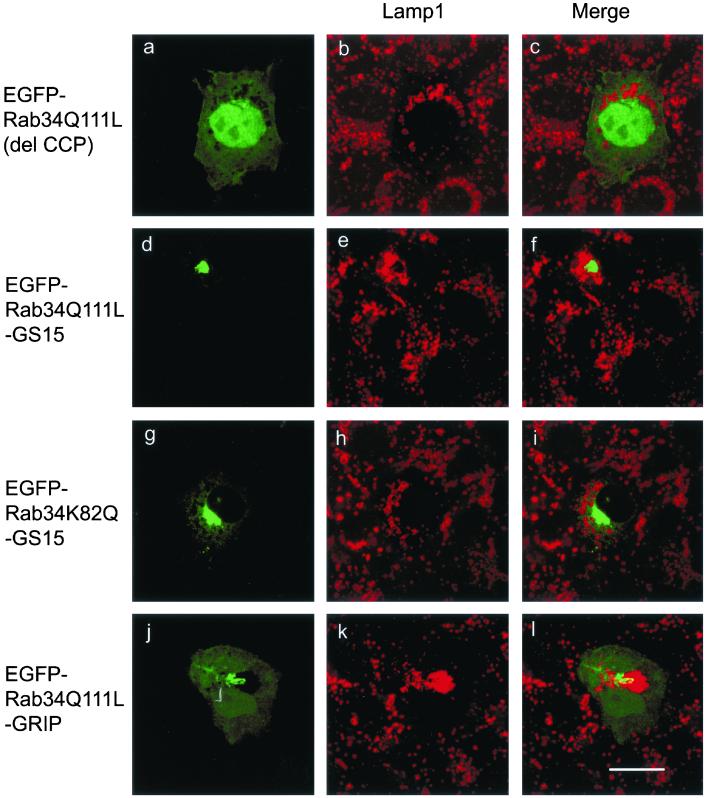
Because endogenous Rab34 as well as EGFP-Rab34wt and EGFP-Rab34Q111L are preferentially targeted to the Golgi apparatus, our results suggest that a regulatory pathway exists for the Golgi apparatus to modulate the lysosomal positioning. To substantiate this concept of interorganellar trans-regulation, we looked for additional evidence for Golgi-localized Rab34 to influence lysosomal distribution (Figure 8). To rule out the possibility that the observed effects of Rab34 may be a result of some mistargeting of Rab34 to the lysosome, we created two other versions of Rab34Q111L that are targeted to the Golgi apparatus. The first (Rab34Q111L-GS15) is the replacement of the C-terminal prenylation signal (CCP motif) of Rab34Q111L by GS15, a SNARE of the Golgi apparatus (Xu et al., 1997), so that Rab34Q111L is stably associated with Golgi membranes as an integral membrane protein. As a control, EGFP-Rab34K82Q-GS15 was similarly constructed, in which the K82 was replaced with Q so that it is not able to interact with RILP. Another chimera (Rab34Q111L-GRIP) was constructed by replacing the C-terminal CCP motif of Rab34Q111L by the GRIP domain (residue 670–748) of Golgin-97. Because the GRIP domain of Golgin-97 was shown to be sufficient for targeting to the TGN (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999), Rab34Q111L-GRIP will be delivered specifically to the TGN. As a negative control, a mutant form, EGFP-Rab34Q111L(del CCP), was created by deleting the C-terminal CCP motif, which is important for prenylation and membrane association. As shown in Figure 8 (a–c), EGFP-Rab34Q111L(del CCP) is no longer associated with the Golgi apparatus but rather was detected in the cytosol and the nucleus (Figure 8a). The distribution of lysosomes (Figure 8b) was essentially not affected by the expression of EGFP-Rab34Q111L(del CCP), suggesting that the observed effect of Rab34 on lysosomal positioning is dependent on its association with the Golgi membrane. Importantly, when stably anchored to the Golgi apparatus, Rab34Q111L-GS15 is still able to redistribute the lysosomes to the peri-Golgi region (Figure 8, d–f). This effect is dependent on the interaction of Rab34 with RILP, because Golgi-anchored EGFP-Rab34K82Q-GS15 did not affect lysosomal positioning (Figure 8, g–i). Alternatively, when Rab34 was targeted to the TGN via the GRIP domain, Rab34Q111L-GRIP was detected in the Golgi apparatus (Figure 8j), consistent with previous studies showing that Golgi targeting by GRIP domains is mediated by a saturable mechanism (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999). Rab34Q111L-GRIP was also detected in the cytosol and nucleus. Intriguingly, potent effects of Rab34Q111L-GRIP on lysosomal positioning were observed (Figure 8k). These results suggest that Rab34 could indeed mediate a long-range interorganellar trans-regulation of lysosomal positioning from the central Golgi apparatus, and this novel mechanism of Rab action depends on its Golgi membrane association and interaction with RILP.
Figure 8.
One of the Rab34 action sites is in the Golgi apparatus. NRK cells transfected with the indicated forms of Rab34 were labeled with anti-Lamp1 to detect lysosomes. EGFP-Rab34Q111L (del CCP) represents EGFP-Rab34Q111L whose C-terminal prenylation signal CCP was deleted and is distributed in the cytosol and nucleus. EGFP-Rab34Q111L-GS15 represents EGFP-Rab34Q111L whose C-terminal CCP was replaced by the entire coding region of GS15 and is localized to the Golgi apparatus as an integral membrane protein. EGFP-Rab34K82Q-GS15 represents EGFP-Rab34K82Q whose C-terminal CCP was replaced by the entire coding region of GS15 and is localized to the Golgi. EGFP-Rab34Q111L-GRIP represents EGFP-Rab34Q111L whose C-terminal CCP was replaced by the GRIP domain of Golgin-97 and is targeted to the Golgi (mainly TGN) by a saturable mechanism. Bar, 10 μm.
DISCUSSION
Molecular mechanisms governing the spatial distribution of intracellular organelles and/or protein complexes are receiving increasing attention because of their potentials in mediating/regulating diverse cellular events (Hayles and Nurse, 2001; Knoblich, 2001; Marks and Seabra, 2001). Each of the organelles of the secretory and endocytic pathways has its relatively unique spatial distribution in the cell (Farquhar and Palade, 1998; Gruenberg, 2001). The centrally localized Golgi apparatus not only serves to mediate membrane traffic in the secretory pathway but also functionally integrates the secretory with the endocytic pathway (Lippincott-Schwartz, 1998; Gu et al., 2001). The spatial distribution of the Golgi apparatus is mediated by an interlinked spectrum of protein interactions involving cytoskeletons, such as the microtubule network (Lippincott-Schwartz, 1998; Chabin-Brion et al., 2001) and intermediate filaments (Gao and Sztul, 2001), Golgi scaffold/matrix proteins, and integral membrane proteins of the organelle (Bock et al., 2001; Moyer et al., 2001).
Lysosomes and lysosome-related organelles not only mediated degradation of macromolecules such as proteins, lipids, and RNAs but also are intimately involved in many physiological processes of an organism, such as antigen presentation, pigmentation, and host defense (Mellman, 1996; Hirokawa, 1998; Dell'Angelica et al., 2000; Marks and Seabra, 2001). The most extensively studied example of lysosome-related organelles is melanosomes in melanocytes (Marks and Seabra, 2001). Possibly originating from the endosomal system, melanosomes are generated through several morphologically distinct intermediates and are transported from the central Golgi region to the cell periphery for subsequent transfer to neighboring keratinocytes. The migration of melanosomes from the Golgi region to the cell periphery is mediated by the microtubule network through the plus-end motor protein kinesin (Hirokawa, 1998; Dell'Angelica et al., 2000; Marks and Seabra, 2001; Karcher et al., 2002). Once at the cell periphery, melanosomes are actively retained there via the actin–myosin–based cytoskeleton before being transferred to the surrounding keratinocytes. The migration and positioning of melanosomes are important for the physiological function of melanocytes, because mutations in genes whose products participate in these processes are associated with diseases in humans and/or genetic defects in mice. Mutations in Rab27a, myosin 5a, and melanophilin are the molecular basis for the ashen, dilute, and leaden phenotypes in mice, respectively, and mutations in Rab27a and myosin5a have been shown to cause Griscelli syndrome in humans (Marks and Seabra, 2001). The cell biological defects caused by mutations of Rab27a, myosin 5a, and melanophilin probably involve the defective transport to and/or retention of melanosomes in the periphery by the actin–myosin cytoskeleton, which is coordinated by concerted action of Rab27a, myosin 5A, and melanophilin (Wu et al., 2002). This results in their repositioning in the Golgi region, probably mediated by microtubule network through the minus-end motor protein dynein. The migration of melanosomes from the periphery to center was exploited in fish and amphibians as a strategy to rapidly alter their skin color (Marks and Seabra, 2001). An example of genetic diseases with altered spatial distribution/migration of lysosomes and/or lysosome-related organelles is the Chediak-Higashi syndrome in human and the beige phenotype in mice. The defects arise from mutations in the same gene referred to as CHS1 in human and Lyst in mice (Marks and Seabra, 2001). CHS1 mutant cells have enlarged and centrally located lysosomes (and melanosomes in melanocytes), whereas overexpression of CHS1 leads to abnormally small and peripherally localized lysosomes, suggesting that CHS1/Lyst may regulate the spatial distribution of lysosomes and lysosome-related organelles (Perou et al., 1997).
Several other proteins have recently been shown to regulate the spatial distribution of lysosomes (Cantalupo et al., 2001; Caplan et al., 2001; Jordens et al., 2001). First, Rab7 and its interacting protein RILP have a positive role in shifting lysosomes toward the central Golgi region. Overexpression of the activated GTP-restricted but not GDP-restricted form of Rab7 results in redistribution of peripheral lysosomes to the peri-Golgi region. RILP is a cytosolic protein that interacts with the activated form of Rab7, and overexpression of RILP results in redistribution of lysosomes to the peri-Golgi region, even in the presence of the GDP-restricted dominant negative form of Rab7. Although it remains to be established that the effect of the activated form of Rab7 on lysosome distribution is dependent on its interaction with RILP, these studies indicate that RILP is a downstream effector of Rab7, because overexpression of RILP can bypass the requirement of Rab7. Overexpression of RILP results in redistribution of the dynein-dynactin complex and lysosomes to the peri-Golgi region (Jordens et al., 2001). Because dynein has been shown to associate with lysosomes (Lin and Collins, 1992) and is known to move organelles toward the minus-end of microtubules in the peri-Golgi region, one possibility is that overexpressed RILP activates the dynein-dynactin complex that drives movement of lysosomes toward the peri-Golgi region. Because direct interaction of RILP with subunits of the dynein-dynactin complex has not been shown, it could act either directly or indirectly with the dynein-dynactin complex. Another protein shown to regulate spatial distribution of lysosomes is human Vam6p, overexpression of which caused clustering and fusion of lysosomes in the peri-Golgi region (Jordens et al., 2001). Because a dominant-negative form of Rab7 did not interfere with the action of Vam6p, Vam6p could act either downstream of or in parallel with Rab7. It remains to be examined whether RILP and Vam6p act together or in parallel.
In our present study, we first established that Rab34 is associated with the Golgi apparatus. This conclusion is based on the observation that endogenous Rab34 was detected primarily in the Golgi apparatus, marked by Golgi mannosidase II, either in normal cells or when the Golgi was fragmented by treatment with the microtubule-disrupting agent nocodazole. Further support for this notion came from the demonstration that N-terminally EGFP-tagged wild-type and GTP-restricted forms of Rab34 are targeted to the Golgi apparatus. Because Rab34 is primarily Golgi-localized, it was intriguing that expression of the wild-type and GTP-restricted form of Rab34 could effectively redistribute the otherwise distantly located lysosomes to the peri-Golgi region. This interorganellar effect was observed in diverse cell lines and for several lysosomal markers, suggesting that Rab34 has a general role in regulating the distribution of lysosomes as intact organelles. Additional experiments were performed to further establish that the Golgi apparatus is most likely the site (or at least, one of the sites) of action of Rab34, because Rab34 could still modulate lysosomal positioning when targeted to the Golgi apparatus by fusion to either GS15 or the GRIP domain of Golgin-97. Because Golgi-localized Rab6A′ and Rab33b had no effect on lysosome distribution, the effect of Rab34 is specific rather than a result of some indirect secondary events. This is further strengthened by the observation that the GDP-restricted form had no significant effect and that the effect on lysosomal positioning can be abolished by replacement mutation of a single residue (K82). Because the relocated lysosomes maintain their vacuolar morphology in the peri-Golgi region (most obvious in NRK cells) and the location is not affected by treatment with brefeldin A, it seems that relocated lysosomes retain some properties of their structural identity. Furthermore, because intracellular transport and processing of cathepsin D were not significantly affected by overexpression of any form of Rab34, it was concluded that the relocated lysosomes also maintain some of their functional identity (at least, in terms of cathepsin D transport and processing). Hence, this report suggests that the Golgi-localized Rab34 regulates the spatial distribution of lysosomes without globally affecting their structural and functional identities, a property that is probably different from that of Rab7 and Vam6p (Cantalupo et al., 2001; Caplan et al., 2001).
The molecular mechanism and structural basis underlying the action of Rab34 was investigated by searching for its interacting partners by various approaches. The yeast two-hybrid screens proved to be the most fruitful, because three of the four clones identified from screens of >5.5 × 107 independent colonies encode RILP. RILP interacts preferentially with the wild-type and GTP-restricted forms of Rab34. The interaction of Rab34 with RILP was further confirmed by in vitro GST-pulldown experiments and by demonstration of a direct interaction of purified RILP generated by in vitro translation with recombinant GST-Rab34. Furthermore, no interaction of RILP with the activated forms of Rab6A′ or Rab33b was detected, and Rab6A′ and Rab33b did not affect the spatial distribution of lysosomes. This correlation between the interaction with RILP and effects on lysosome distribution indicates that the interaction with RILP could be the underlying mechanism for the regulation by Rab34 of the spatial distribution of lysosomes.
To prove that interaction with RILP is responsible for Rab34 to regulate lysosomal redistribution, we first investigated the structural basis for Rab34 to interact with RILP. Because the overall amino acid sequence homology of Rab34 with Rab7 is not significantly higher than those observed between Rab34 and other Rabs, we have carefully compared the amino acid sequences of Rab34, Rab36, Rab7, and other Rabs, such as Rab6A′ and Rab33b. We identified several residues (K82, G124, A125, F132, L151, and F162) that are selectively conserved in Rab34, Rab36, and Rab7 but not in Rab6 or Rab33b. Because a chimeric protein consisting of the N-terminal 142 residues of Rab34 and the C-terminal region of Rab6A′ possesses the ability to relocate lysosomes, it is likely that K82, G124, A125, and/or F132 may play a role in the interaction with RILP and regulation of lysosomal positioning. It was then found that K82 is crucial for Rab34 to interact with RILP, because a mutant version of Rab34 harboring a replacement of K82 by the corresponding residue (Q) in Rab6A′ failed to interact with RILP, as assessed by yeast two-hybrid or GST pulldown. Although this mutant was targeted to the Golgi, it failed to affect lysosome distribution. These results clearly establish that interaction with RILP is necessary for Rab34 to regulate lysosomal positioning. Because the role of RILP is likely to be mediated by dynein–dynactin complex, the effect of Rab34 on lysosome distribution could be mediated via downstream action of the dynein–dynactin complex through RILP as an adaptor to bridge these events. The dependence of Rab34 on an intact microtubular network to effect the spatial redistribution of lysosomes supports this possibility.
Another difference between Rab7- and Rab34-mediated regulation of lysosome distribution is that Rab7 acts in cis by affecting the host compartment, whereas Rab34 probably acts in trans by affecting another compartment, most likely through regulation of the microtubular cytoskeleton and its associated motor proteins. This interorganellar effect of Rab34 thus defines a novel mechanism of action of Rabs in regulating cellular processes. Although expression of Rab34 (either the wild-type or GTP form) results in the shift of lysosomal positioning to the peri-Golgi region, the sizes of the majority of the shifted lysosomes are similar to those found in control cells. Our preliminary comparison of Rab34 and Rab7 indicates that Rab34 is more potent in shifting peripheral lysosomes to the peri-Golgi region, whereas Rab7 has the additional property of inducing larger lysosomes (data not shown). This suggests that Rab34 may not have the capability to enhance the size of the lysosome. Our preliminary studies of RILP (data not shown) suggest that overexpression of RILP alone can lead to fewer but much larger lysosomes repositioned in the peri-Golgi region, suggesting that RILP may possess two properties, one to shift lysosomes from the periphery to the peri-Golgi region (more similar to Rab34 and, to a lesser extent, to Rab7) and the other to enhance the size of lysosomes (mainly similar to Rab7). One intriguing possibility is that RILP may have a feedback effect on both Rab34 and Rab7, and its overexpression may result in activation of both Rab34 and Rab7, which mediate the repositioning and enlargement of lysosomes, respectively. A feeding-back action of the effector on small GTPase activation was recently demonstrated for ARF1 (Zhu et al., 2000). In this regard, RILP may serve as a shared effector of Rab34 and Rab7 as well as a shared “activator” for these two Rabs by feedback loops. Strikingly, the cellular phenotype of fewer enlarged lysosomes in the peri-Golgi region caused by overexpression of RILP is similar to that reported in cells derived from patients suffering from Chediak-Higashi syndrome. One speculative possibility will be that RILP and beige/lyst may have opposing action on lysosomal positioning and sizes. Loss of beige/lyst function in Chediak-Higashi syndrome cells may lead to a net increased effect of RILP and result in effects similar to those observed after RILP overexpression. If this is true, we may expect that overexpression of beige/lyst will antagonize the effect of RILP. Whether beige/lyst could act by regulating activities of Rab34 and Rab7 may be worth future investigations. More studies are needed to test these speculations.
On the basis of the results presented here and in the context of the known properties of RILP, one working model for Rab34 to regulate spatial positioning of lysosomes could be envisioned. On or during activation via nucleotide exchange of GDP for GTP, Rab34 becomes associated with the Golgi apparatus, which in turn recruits cytosolic RILP to the Golgi region. This interaction either activates RILP or forms a functional Rab34-RILP complex, which then modulates the activity of dynein–dynactin to promote migration of lysosomes toward the minus-end of microtubule. Because Rab34 can exhibit a spatial regulation of otherwise distantly located lysosomes, most likely by affecting their migration, the relocated lysosomes maintain some of their structural and functional identity. Our studies thus have unveiled a novel mechanism for Golgi-associated Rab34 to regulate spatial distribution of lysosomes via direct interaction with RILP. This interaction could subsequently modulate dynein–dynactin–mediated organelle migration. Because the dynein–dynactin complex is involved in minus-end–driven movement of other intracellular organelles and/or protein complexes, particularly in retrograde transport in axons of neurons (Hirokawa, 1998; Karcher et al., 2002), it is conceivable that Rab34/RILP or similar interactions may exhibit a Golgi-originated regulation of this dynein–dynactin–mediated movement of organelles and/or protein complexes in peripheral regions, including axons. Evidence does exist that indicates the involvement of small GTPases in regulating organelle transport in the axoplasm (Hirokawa, 1998; Karcher et al., 2002). Further studies along these lines may yield additional understanding of not only spatial distribution/migration of organelles but also the mechanisms of action of Rab proteins as well as the cytoskeleton network.
ACKNOWLEDGMENTS
We thank Drs. Li-Fong Seet, Bor Luen Tang, Paren Singh, and Koh Pang Lim for careful reading of this manuscript; G. Banting for anti-TGN38 mAb and D. Meyer for rabbit anti-glycophorin antibodies; and Lu Lei and other members of Dr. Hong's laboratory for helpful discussions and assistance. This work was supported by A*Star (Agency for Science, Technology and Research), Singapore. W.H. is also a faculty member of the Department of Biochemistry, National University of Singapore.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0280. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0280.
REFERENCES
- Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase Rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan S, Hartnell LM, Aguilar RC, Naslavsky N, Bonifacino JS. Human Vam6p promotes lysosome clustering, and fusion in vivo. J Cell Biol. 2001;154:109–122. doi: 10.1083/jcb.200102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KS, Hanna J, Simon I, Krise J, Barbero P, Pfeffer SR. Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science. 2001;292:1373–1376. doi: 10.1126/science.1056791. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P, Goud B. The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol. 1999;11:466–475. doi: 10.1016/S0955-0674(99)80067-2. [DOI] [PubMed] [Google Scholar]
- Darchen F, Goud B. Multiple aspects of Rab protein action in the secretory pathway: focus on Rab3 and Rab6. Biochimie. 2000;82:375–384. doi: 10.1016/s0300-9084(00)00219-4. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Mullins C, Caplan S, Bonifacino JS. Lysosome-related organelles. FASEB J. 2000;14:1265–1278. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- Echard A, Opdam FJ, de Leeuw HJ, Jollivet F, Savelkoul P, Hendriks W, Voorberg J, Goud B, Fransen JA. Alternative splicing of the human Rab6A gene generates two close but functionally different isoforms. Mol Biol Cell. 2000;11:3819–3833. doi: 10.1091/mbc.11.11.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 1998;8:2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sztul E. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J Cell Biol. 2001;152:877–894. doi: 10.1083/jcb.152.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;2:721–730. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- Gu F, Crump CM, Thomas G. Trans-Golgi network sorting. Cell Mol Life Sci. 2001;58:1067–1084. doi: 10.1007/PL00000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Nurse P. A journey into space. Nat Rev Mol Cell Biol. 2001;2:647–656. doi: 10.1038/35089520. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Karcher RL, Deacon SW, Gelfand VI. Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 2002;12:21–27. doi: 10.1016/s0962-8924(01)02184-5. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA. A novel Golgi-localization domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol. 1999;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2:11–20. doi: 10.1038/35048085. [DOI] [PubMed] [Google Scholar]
- Lin SX, Collins CA. Immunolocalization of cytoplasmic dynein to lysosomes in cultured cells. J Cell Sci. 1992;101:125–137. doi: 10.1242/jcs.101.1.125. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J. Cytoskeletal proteins and Golgi dynamics. Curr Opin Cell Biol. 1998;10:52–59. doi: 10.1016/s0955-0674(98)80086-0. [DOI] [PubMed] [Google Scholar]
- Lowe SL, Wong SH, Hong W. The mammalian ARF-like protein (Arl1) is associated with the Golgi complex. J Cell Sci. 1996;109:209–220. doi: 10.1242/jcs.109.1.209. [DOI] [PubMed] [Google Scholar]
- Lu L, Horstmann H, Ng C, Hong W. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1) J Cell Sci. 2001;114:4543–4555. doi: 10.1242/jcs.114.24.4543. [DOI] [PubMed] [Google Scholar]
- Marks MS, Seabra MC. The melanosome: membrane dynamics in black and white. Nat Rev Mol Cell Biol. 2001;2:738–748. doi: 10.1038/35096009. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Mori T, Fukuda Y, Kuroda H, Matsumura T, Ota S, Sugimoto T, Nakamura Y, Inazawa J. Cloning and characterization of a novel Rab-family gene, Rab36, within the region at 22q11.2 that is homozygously deleted in malignant rhabdoid tumors. Biochem Biophys Res Commun. 1999;254:594–600. doi: 10.1006/bbrc.1998.9968. [DOI] [PubMed] [Google Scholar]
- Morimoto BH, Chuang CC, Koshland DE., Jr Molecular cloning of a member of a new class of low-molecular-weight GTP-binding proteins. Genes Dev. 1991;5:2386–2391. doi: 10.1101/gad.5.12b.2386. [DOI] [PubMed] [Google Scholar]
- Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- Munro S, Nichols BJ. The GRIP domain: a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2000;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Perou CM, Leslie JD, Green W, Li L, Ward DM, Kaplan J. The Beige/Chediak-Higashi syndrome gene encodes a widely expressed cytosolic protein. J Biol Chem. 1997;272:29790–29794. doi: 10.1074/jbc.272.47.29790. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- Short B, Preisinger C, Korner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somsel Rodman J, Wandinger-Ness A. Rab GTPases coordinate endocytosis. J Cell Sci. 2000;113:183–192. doi: 10.1242/jcs.113.2.183. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ, Balch WE. Rab2 is essential for the maturation of pre-Golgi intermediates. J Biol Chem. 1996;271:29372–29379. doi: 10.1074/jbc.271.46.29372. [DOI] [PubMed] [Google Scholar]
- Valsdottir R, Hashimoto H, Ashman K, Koda T, Storrie B, Nilsson T. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 2001;508:201–209. doi: 10.1016/s0014-5793(01)02993-3. [DOI] [PubMed] [Google Scholar]
- Wong SH, Xu Y, Zhang T, Griffiths G, Lowe SL, Subramaniam VN, Seow KT, Hong W. GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol Biol Cell. 1999;10:119–134. doi: 10.1091/mbc.10.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA., III Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- Xu Y, Wong SH, Zhang T, Subramaniam VN, Hong W. GS15, a 15-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) homologous to rbet1. J Biol Chem. 1997;272:20162–20166. doi: 10.1074/jbc.272.32.20162. [DOI] [PubMed] [Google Scholar]
- Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nature Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhu X, Boman AL, Kuai J, Cieplak W, Kahn RA. Effectors increase the affinity of ADP-ribosylation factor for GTP to increase binding. J Biol Chem. 2000;275:13465–13475. doi: 10.1074/jbc.275.18.13465. [DOI] [PubMed] [Google Scholar]