Abstract
We have cloned a full-length cDNA encoding a novel myosin II heavy chain kinase (mhckC) from Dictyostelium. Like other members of the myosin heavy chain kinase family, the mhckC gene product, MHCK C, has a kinase domain in its N-terminal half and six WD repeats in the C-terminal half. GFP-MHCK C fusion protein localized to the cortex of interphase cells, to the cleavage furrow of mitotic cells, and to the posterior of migrating cells. These distributions of GFP-MHCK C always corresponded with that of myosin II filaments and were not observed in myosin II-null cells, where GFP-MHCK C was diffusely distributed in the cytoplasm. Thus, localization of MHCK C seems to be myosin II-dependent. Cells lacking the mhckC gene exhibited excessive aggregation of myosin II filaments in the cleavage furrows and in the posteriors of the daughter cells once cleavage was complete. The cleavage process of these cells took longer than that of wild-type cells. Taken together, these findings suggest MHCK C drives the disassembly of myosin II filaments for efficient cytokinesis and recycling of myosin II that occurs during cytokinesis.
INTRODUCTION
During cytokinesis, cells are pinched into two parts by constriction of contractile rings. The contractile rings contain parallel filaments of actin and myosin II, a configuration suitable for constriction of the ring, in animal cells (Mabuchi and Okuno, 1977; Mabuchi, 1986; Glotzer, 1997; Robinson and Spudich, 2000) and in the amoeba cells of the cellular slime mold Dictyostelium discoideum (Yumura and Fukui, 1985; Fukui and Inoue, 1991). It is known that disassembly of the contractile ring components, including the myosin II filaments, accompanies the progression of cytokinesis, ultimately leading to fusion of the opposing plasma membranes and separation of the two daughter cells (Yumura et al., 1984). Little is known, however, about how disassembly of myosin II filaments is regulated during this process.
D. discoideum is a powerful experimental system that enables functional analysis of various myosin II mutants in the absence of the wild-type form (De Lozanne and Spudich, 1987; Manstein et al., 1989; Uyeda and Yumura, 2000). Through the use of such myosin II mutants, for instance, the functional significance of the phosphorylation of the three threonine residues at positions 1823, 1833, and 2029 in the tail region of Dictyostelium myosin II was demonstrated: their phosphorylation state regulates the assembly and disassembly of myosin II filaments (Kuczmarski and Spudich, 1980; Egelhoff et al., 1991, 1993). One mutant in which alanine residues were substituted for the three threonines (3XALA myosin) mimics the dephosphorylated state and accumulates excessively in the equatorial region of mitotic cells (Egelhoff et al., 1993). In contrast, 3XASP myosin, in which the threonine residues are substituted with aspartate residues, mimics the phosphorylated state, cannot form bipolar filaments, and shows markedly reduced accumulation in the cleavage furrows (Sabry et al., 1997). More recently, systematic mutational analysis revealed that a single negative charge at position 1823 is able to perturb filament assembly (Nock et al., 2000), which suggests that phosphorylation of threonine 1823 primarily mediates disassembly of myosin II filaments and their translocation out of the cleavage furrow and that it must be both temporally and spatially regulated.
Three myosin heavy chain kinases, MHCK A (Futey et al., 1995), MHCK B (Clancy et al., 1997), and MHC-PKC (Ravid and Spudich, 1992), have been cloned from Dictyostelium; however, none of these have been shown to play a role in regulating localization of myosin II within contractile rings. We therefore searched the database of the D. discoideum cDNA Project and found a fragmentary sequence with a high homology to both MHCK A and B. We then noticed that its partial genomic sequence had been deposited under the name mhckC and that its catalytic domain had been expressed and shown to phosphorylate a peptide modeled on the MHCK A target site in the Dictyostelium myosin II tail in vitro, although the specificity of the reaction was not shown (Luo et al., 2001). In this report, we describe our cloning of both the full-length genomic DNA and the cDNA of mhckC, generation of knockout strains, characterization of the intracellular distribution of MHCK C, and examination of the effects of MHCK C on the distribution of myosin II. The results clearly show that MHCK C is required for efficient disassembly of myosin II filaments in cleavage furrows during cytokinesis, and consequently, efficient cytokinesis, in Dictyostelium cells.
MATERIALS AND METHODS
Cell Culture
Parental D. discoideum wild-type AX2 cells and myosin II− HS1 cells (Ruppel et al., 1994) were grown axenically in HL-5 medium (Sussman, 1987) supplemented with penicillin and streptomycin at 22°C. mhckC− cells were cultured in HL-5 in the presence of penicillin, streptomycin, and 10 μg/ml blasticidin-S. Cells carrying the Dictyostelium expression vector pBIG (Ruppel et al., 1994), or its derivatives, were grown in medium supplemented with 10 μg/ml G418. Cells expressing FLAG-tagged MHCK A were described previously (Steimle et al., 2001b).
Gene Disruption in Dictyostelium
To construct a vector targeting the mhckC gene, the blasticidin resistance gene cassette was inserted at the MscI site of the mhckC gene. As BamHI and SacI sites were engineered flanking the termini of the gene, the construct was excised using these enzymes, after which 10 μg of the linearized fragment was introduced into Dictyostelium cells by electroporation. The transformant clones were then cultured in axenic medium containing 10 μg/ml blasticidin S. Selective disruption of mhckC gene by the targeting vector was confirmed by genomic polymerase chain reaction (PCR).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis
AX2 cells developed on agar containing 16.7 mM phosphate buffer, pH 6.2 (Fukui et al., 1990). Total RNA was purified by ISOGEN reagent (Nippon Gene, Tokyo, Japan) from AX2 cells at the indicated times following the manufacturer's recommendations, after which reverse transcription with MMLV reverse transcriptase (Toyobo Engineering, Osaka, Japan) and PCR were performed for 28 cycles with primers specific to mhckC, i.e., 5′-ATCAAAATTCCCAGTTGCCGATGT-3′ and 5′-TCGCTTCATTGAATTTCTCTGCCCA-3′. These oligonucleotide primers were designed to amplify a segment encompassing exon 1 and 2 of the mhckC gene, so that the potential amplification of contaminated genomic DNA can be easily detected. We used the IG7 message as an internal control for RT-PCR (Chang et al., 1996). A pair of oligonucleotide primers for IG7 was 5′-TTACATTTATTAGACCCGAAACCAAGCG-3′ and 5′-TTCCCTTTAGACCTATGGACCTTAGCG-3′. The mixture contained 5 μl of 10× ExTaq buffer, 250 μM each dNTP, 1.0 mM MgCl2, and 0.5 U of ExTaq polymerase (Takara, Kyoto, Japan).
Fluorescence Microscopy
Dictyostelium cells were transfected with green fluorescent protein (GFP)-myosin II/pTIKL (Liu et al., 2000) or GFP-mhckC/pBIG by electroporation, after which the resultant transfectants were transferred to plastic Petri dishes with thin glass bottoms (Iwaki, Funabashi, Japan), and the culture medium was replaced with medium modified to decrease the background fluorescence (Nagasaki et al., 2002). Cells expressing GFP fusion proteins were observed under a fluorescence microscope (IX50; Olympus, Tokyo, Japan) equipped with an UPlan Apo 100× oil immersion objective lens (Olympus). Time-lapse pictures were acquired with a charge-couple device camera (C5985; Hamamatsu Photonics, Hamamatsu, Japan) at intervals of 30 s by using a time-lapse recording system (ARGAS-20; Hamamatsu Photonics). For montage sequences, video images were digitized using NIH Image software, version 1.61.
Immunofluorescence staining of cells was carried out as described previously (Steimle et al., 2001b). Monoclonal antibody against the FLAG epitope was purchased from Sigma-Aldrich (St. Louis, MO).
RESULTS
Identification of a Novel Myosin Heavy Chain Kinase Gene (mhckC)
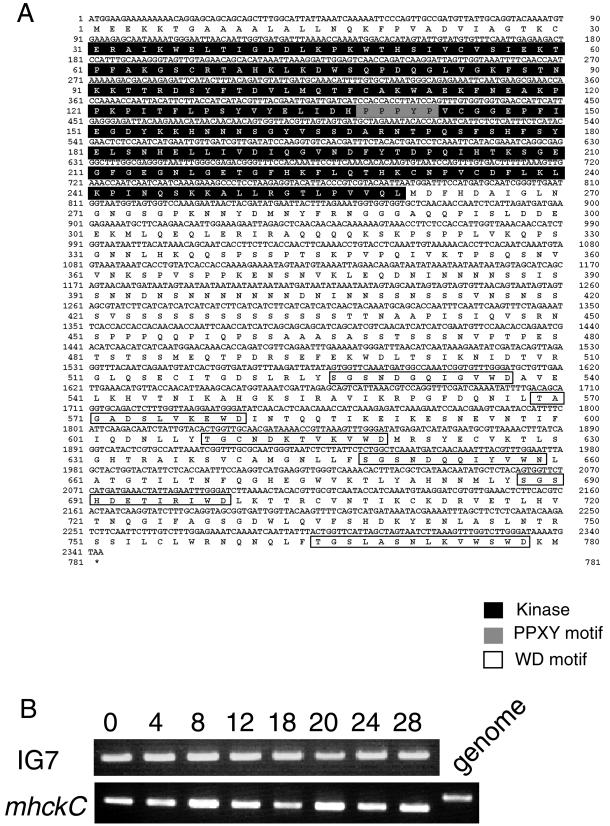
To clone the mhckC gene, we first assembled the full-length sequence of the cDNA on a computer by using fragmentary sequences found in the GenBank database (no. AF079447) and the database of the D. discoideum cDNA Project. This enabled us to design a pair of oligonucleotides that would amplify the full-length sequence from genomic DNA or a cDNA library from Dictyostelium cells. Sequence analysis of the complete cDNA revealed that it encodes a 780-amino acid polypeptide (MHCK C) having a calculated molecular mass of 86 kDa (Figure 1A).
Figure 1.
(A) Nucleotide and predicted amino acid sequences of MHCK C, which consists of 780 amino acid residues derived from mhckC cDNA. The N-terminal catalytic domain is shown in white letters on a black background. The putative binding site for a WW domain (PPXY) is shown in white letters on a gray background within the catalytic domain. The six C-terminal repeats of the WD domain are boxed. The GenBank accession number is AB079663; a partial sequence of mhckC has been deposited by N. Iranfar and W.F. Loomis under the accession number AF079447. (B) RT-PCR analysis of the expression of mhckC during vegetative and developmental stages. The products of RT-PCR are smaller than that from genomic DNA, due to the absence of an intron. IG7 is expressed at a constant level during development and is used as an internal control.
The primary structure of MHCK C is similar to those of MHKC A and B from Dictyostelium. The members of this kinase family have two domains in common: a kinase catalytic domain in the N-terminal half of the polypeptide and a WD repeat domain in the C-terminal half. The catalytic domain of MHCK C spans amino acid residues 31–259 and, unlike other members of the family, contains a PPXY sequence, a putative WW domain-binding motif (Einbond and Sudol, 1996; Sudol and Hunter, 2001; Ilsley et al., 2002), whereas six WD repeat motifs extend from residues 526–776 (Figure 1A).
To determine when during the life cycle of Dictyostelium the mhckC gene is expressed, samples of total RNA were isolated at various stages of the life cycle and analyzed by RT-PCR. As shown in Figure 1B, the mhckC transcript is expressed continuously throughout the growth and developmental phases.
MHCK C Is Enriched in Cell Cortex and Cleavage Furrow
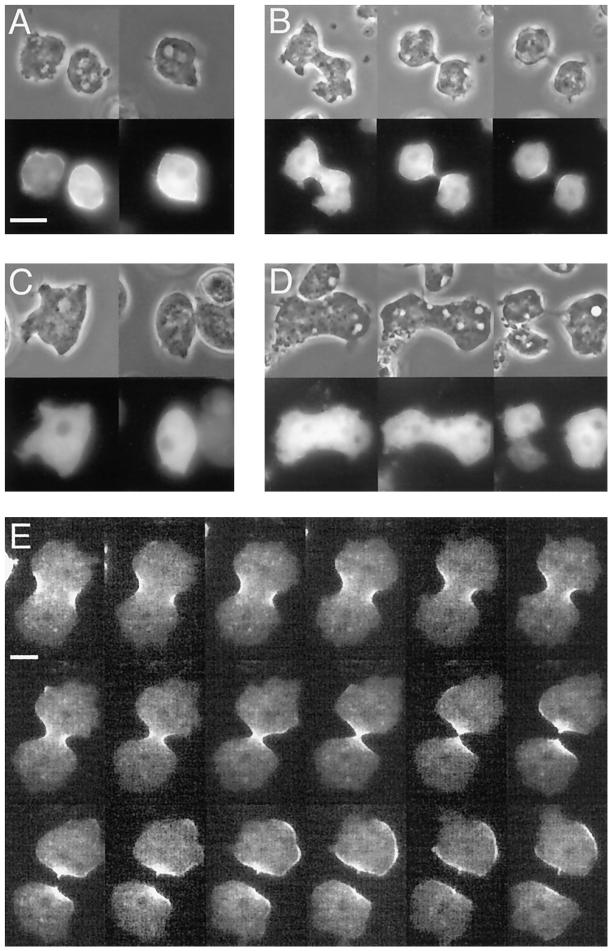
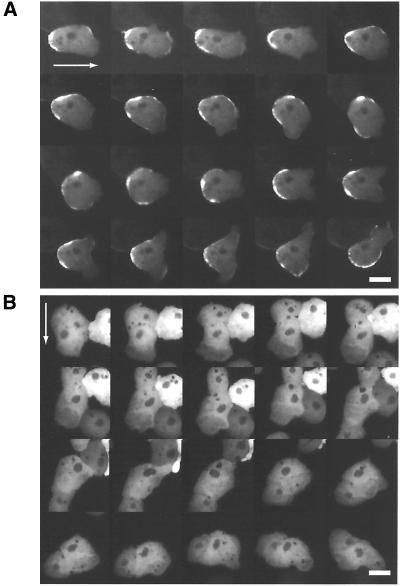
To observe MHCK C dynamics in living Dictyostelium cells, wild-type and myosin II− cells were transformed with an extrachromosomal vector harboring a gfp-mhckC fusion gene, the expression of which was driven by the constitutive actin 15 promoter. In wild-type cells, GFP-MHCK C was localized in the cell cortex during interphase (Figure 2A) and seemed to be slightly enriched in the cleavage furrow during mitosis (Figure 2B), but due to the thickness of the cells, it was not possible to resolve spatial details in that region. To improve our resolution, we cultured wild-type cells expressing GFP-MHCK C under a thin sheet of agarose. Under those conditions, GFP-MHCK C could be clearly identified in the cleavage furrows of mitotic cells (Figure 2E). Interphase cells under agarose sheets spread out pseudopodia and moved vigorously. In the wild-type cells, under these conditions, GFP-MHCK C was localized in the posterior (Figure 3A), where myosin II was also enriched (Yumura et al., 1984; our unpublished data).
Figure 2.
Localization of GFP-tagged MHCK C in wild-type and myosin II− cells during interphase and mitosis. Wild-type (A and B) and myosin II− cells (C and D) that expressed GFP-MHCK C were placed on a dish with a glass bottom. In wild-type cells, GFP-MHCK C was localized in the cell cortex during interphase (A) and accumulated in the cleavage furrow during mitosis (B). In myosin II− cells, in contrast, GFP-MHCK C was distributed diffusely in the cytoplasm during both interphase (C) and mitosis (D). Localization of GFP-MHCK C in mitotic cells under an agarose sheet (E). Cells expressing GFP-MHCK C were placed in a dish with a glass bottom and overlaid with a thin agarose sheet. GFP-MHCK C accumulated in the cleavage furrow during cytokinesis. The fluorescent images were recorded with 30-s intervals between frames. Bar, 10 μm.
Figure 3.
Localization of GFP-MHCK C in motile cells under an agarose sheet. Under agarose sheets, cells formed large pseudopodia and moved actively. Wild-type (A) and myosin II− (B) cells expressing GFP-MHCK C. The arrows show their original direction of migration. In wild-type cells, GFP-MHCK C localized to the posterior of the cells, whereas GFP-MHCK C was diffusely distributed in the cytoplasm of myosin II− cells. The fluorescent images were recorded with 30-s intervals between frames. Bar, 10 μm.
In contrast, GFP-MHCK C exhibited no specific localization in myosin II− cells during either interphase or mitosis without agarose sheets; instead, it was distributed diffusely in the cytoplasm (Figure 2, C and D). Under agarose sheets, GFP-MHCK C in myosin II− cells again exhibited no specific localization (Figure 3B). Behavior of GFP-MHCKC in mitotic myosin II− cells under agarose sheets was difficult to follow because they often failed to divide under those conditions (Yumura and Uyeda, 1997; our unpublished data). However, we were unable to detect any sign of its accumulation in the equatorial regions in these cells.
Disruption of mhckC Slows Down Cleavage Process, but Does Not Significantly Affect Development
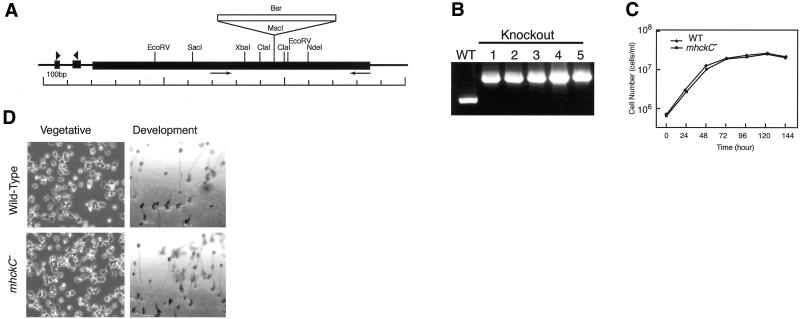
To investigate its function in vivo, we eliminated MHCK C from Dictyostelium wild-type strain AX2 by gene disruption with the blasticidin-S resistance cassette as a marker for selection (Figure 4A). Mutants were identified by a 1.3-kilobase pair shift in the size of the mhckC PCR products, which corresponds to the size of the inserted marker gene (Figure 4B)
Figure 4.
(A) Genomic organization of the mhckC locus and the knockout construct. The mhckC gene consists of three exons represented as black boxes. The targeting vector used to disrupt the mhckC gene was constructed by insertion of the blasticidin-S resistant gene at the MscI site. The arrows show the positions of the primer used for genomic PCR. The arrowheads indicate the positions of the primer pair for RT-PCR. (B) Genomic PCR using cell lines in which the mhckC gene was disrupted by homologous recombination. Mutant cells were identified by the shift in the size of the PCR products. (C) Growth of Dictyostelium cells in axenic suspension. Wild-type AX2 and mhckC− cells were diluted in fresh medium at 105 cells/ml and then shaken at 22°C. (D) Phenotypic analysis of wild-type and mhckC− cells. The cells were placed on plastic dishes containing HL-5 (vegetative) or were incubated for 24 h on agar containing 16.7 mM phosphate buffer (development).
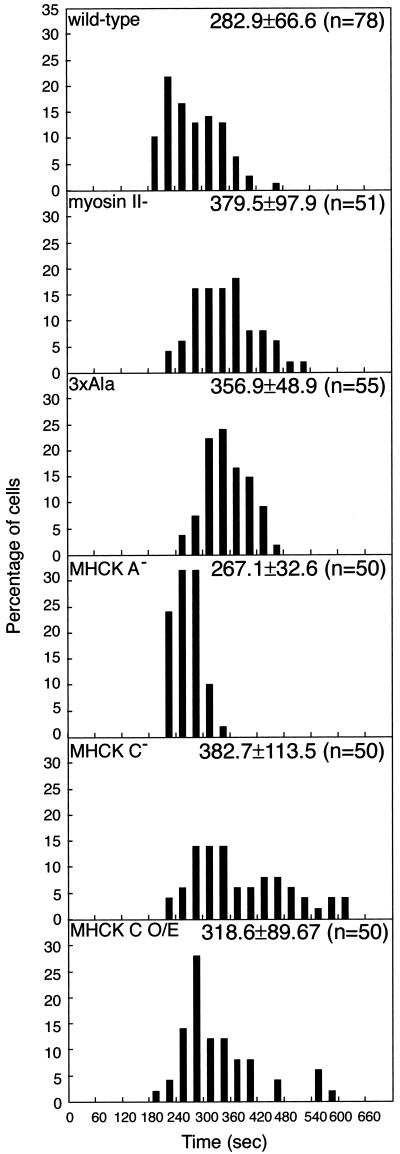
There were no noticeable morphological differences between mhckC− cells and the parental wild-type cells during the growth phase on plastic plates (Figure 4D). However, mhckC− cells took ∼35% longer time to complete cytokinesis compared with the wild-type cells (Figure 5). Similarly, cells expressing 3XALA myosin II and myosin II− cells also needed 26 and 35% longer, respectively, than the wild-type cells to divide. That myosin II− cells on substrates are able to divide in a cell cycle-coupled manner has been reported previously (Neujahr et al., 1997b; Zang et al., 1997; Nagasaki et al., 2002). In contrast, mhckA− cells completed division at a speed comparable with that of wild-type cells (Figure 5). The time required for cell division of mhckA− cells sharply peaked between 200 and 300 s under our conditions, whereas some wild-type cells took >300 s to divide. The significance of this subtle difference between mhckA− and wild-type cells, however, is not clear. When expression vector carrying gfp-mhckC was introduced into mhckC− cells (MHCK C O/E), the time required for cytokinesis decreased compared with mhckC− cells.
Figure 5.
Time required to complete cleavage process in wild-type and mutant cells. The time each cell spends for the cleavage process, that is, beginning from the mataphase, as judged by the spherical appearance of the cell and the reduction of intracellular vesicle movements, until the complete scission of the cytoplasmic strand connecting the two daughter cells, was measured using recorded video sequences. The histogram shows the distribution of the time for each strain. The numbers in the upper right corner of each box shows the average and the number of cells.
MhckC− cells did not exhibit noticeable phenotypic defects in our other assays. Growth on bacterial lawns was also normal (our unpublished data). In suspension culture, mhckC− cells grew at a rate similar to that of wild-type cells (Figure 4C), and they formed normal fruiting bodies on agar plates (Figure 4D).
Localization of Myosin II in mhckC-Null Cells
To study the distribution of myosin II in Dictyostelium cells in more detail, a GFP-myosin II fusion protein was expressed in wild-type and mhckC− cells. During interphase, GFP-myosin II localized to the cell cortex in both cell types (our unpublished data).
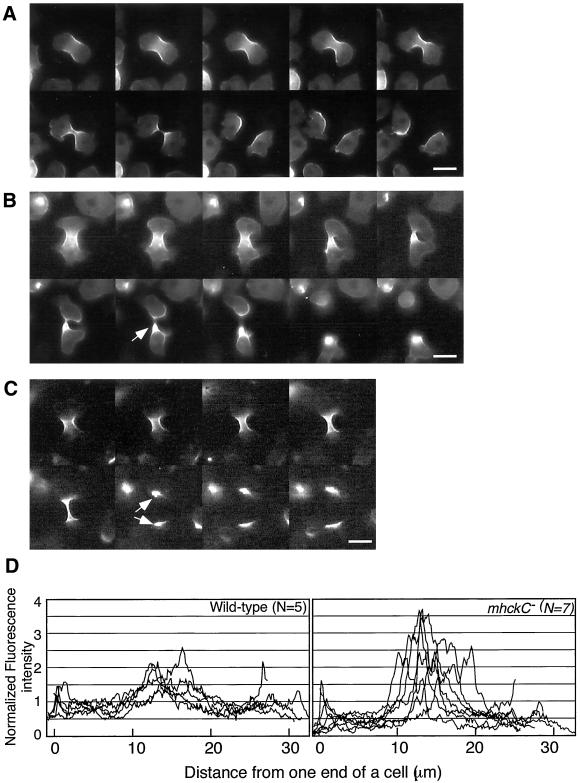
Early in the mitotic phase, at a time when cleavage furrows were apparent, GFP-myosin II accumulated in the equatorial region of wild-type cells. Immediately after cell division, the accumulated GFP-myosin II filaments were present in the posterior of both daughter cells, which migrated away from each other. Shortly thereafter, however, the high concentration of myosin II was smoothly redistributed to the posterior cortex, where it is normally found in migrating interphase cells (Figure 6A). In mitotic mhckC− cells, GFP-myosin II accumulated excessively in the cleavage furrow (Figure 6B), and after separation, the aggregated GFP-myosin II remained in the posterior of one or both of the daughter cells for a prolonged period (Figure 6B, arrow). For comparison, Figure 6C shows a mitotic cell expressing GFP-3XALA myosin II, which mimics the dephosphorylated state. Note that it, too, accumulated excessively in the equatorial region of mitotic cells, and large aggregates were often observed in postmitotic cells. Figure 6D shows fluorescence intensity profiles of GFP-myosin II along the long axis of mitotic wild-type and mhckC− cells. In mhckC− cells, fluorescence intensity of GFP-myosin that accumulated in cleavage furrows was 1.5–2 times as high as that of wild-type cells.
Figure 6.
Distribution of GFP-myosin II in mitotic wild-type (A) and mhckC− (B) cells under agarose sheets. In mhckC− cells, GFP-myosin II accumulated excessively in the cleavage furrow and remained aggregated (arrow) after the cells were separated. (C) Mitotic wild-type cell expressing 3XALA myosin II. Arrows indicate excessive accumulation of GFP-myosin II. Bar, 10 μm. (D) Profiles of relative fluorescent intensity of GFP-myosin in mitotic wild-type and mhckC− cells. Fluorescence intensity was measured by scanning a line along the long axis of a cell by using the NIH Image software. Subsequently, these intensities were normalized with the average of the total intensity in each cell.
MHCK A Localizes to Actin-rich Regions in Mitotic Cells
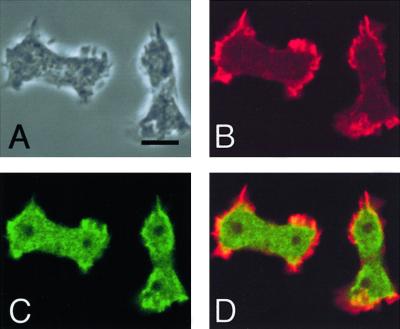
Steimle et al. (2001b) investigated the cellular distribution of MHCK A in Dictyostelium cells during interphase and the developmental phase, but its localization in mitotic cells was not described. We therefore expressed FLAG-tagged MHCK A in Dictyostelium cells, and after fixation with 3.7% formalin and ethanol, the distributions of MHCK A and actin filaments were investigated using an anti-FLAG antibody and rhodamine-phalloidin, respectively (Figure 7). In mitotic cells, actin filaments were abundant at the poles of the cells (Figure 7B). MHCK A was also localized at the poles of the cells (Figure 7C), its distribution corresponding to that of the actin filaments (Figure 7D).
Figure 7.
Immunofluorescence localization of MHCK A in mitotic cells. (A) Phase contrast image of fixed mitotic cells expressing FLAG-MHCK A. (B) Fluorescence images of actin filaments visualized by staining with rhodamine-phalloidin. (C) MHCK A visualized by staining with anti-FLAG antibody. (D) Merged image of actin filaments and MHCK A. Control staining of wild-type cells with the anti-FLAG antibody yielded a negligible signal (our unpublished data). Bar, 10 μm.
DISCUSSION
Contractile rings contain two major components, actin filaments and nonmuscle myosin II filaments, and the active interaction between these two filament systems is believed to power the constriction process. It is not known, however, how myosin II filaments accumulate in the equatorial regions of cells when contractile rings are assembled, or how they diffuse back to the cytoplasm when the cleavage is complete. In Dictyostelium, formation of myosin II filaments is regulated by phosphorylation of three threonine residues in the tail region of the myosin heavy chain (Vaillancourt et al., 1988; Egelhoff et al., 1993; Neujahr et al., 1997a; Sabry et al., 1997; Nock et al., 2000; Redowicz, 2001). It has therefore been speculated that myosin heavy chain kinase plays a key role in regulating myosin II function in mitotic cells (Sabry et al., 1997; Yumura and Uyeda, 1997). In that regard, three myosin II kinases have been cloned from Dictyostelium. Among them, MHC-PKC, a myosin II heavy chain-specific protein kinase C, is not expressed in the vegetative phase; it is specifically expressed during the developmental stage and is implicated in the regulation of myosin II translocation in response to chemoattractant cAMP when the cells aggregate to form fruiting bodies (Ravid and Spudich, 1992; Dembinsky et al., 1996, 1997). MHCK A (Futey et al., 1995) and MHCK B (Clancy et al., 1997) belong to a novel class of protein kinases and are unrelated to conventional eukaryotic protein kinases. MHCK A has been demonstrated to phosphorylate and drive disassembly of myosin II filaments both in vitro and in vivo (Futey et al., 1995; Kolman et al., 1996; Kolman and Egelhoff, 1997; Steimle et al., 2001a), whereas MHCK B has been shown to phosphorylate myosin II heavy chains in vitro (Clancy et al., 1997; Luo et al., 2001). There is no evidence, however, that either of these kinases is involved in the phosphorylation of myosin II in cleavage furrows.
We have identified a novel kinase, MHCK C, involved in the disassembly and reorganization of myosin II filaments present in cleavage furrows. Possessing specific kinase activity against myosin II heavy chain in vitro (Egelhoff, personal communication), MHCK C is normally localized in the cleavage furrows of mitotic cells (Figure 2). Knockout of the mhckC gene results in excessive accumulation of myosin II filaments in the cleavage furrow (Figure 6, B and D) and the slower cleavage process (Figure 5). The excessive accumulation of myosin II in the cleavage furrows of mhckC− cells, as well as the slower cleavage process, is reminiscent of the phenotype of cells expressing 3XALA, a myosin II mutant that mimics the dephosphorylated state (Figure 6C). These results suggest that MHCK C catalyzes the phosphorylation of myosin II required for disassembly of myosin II filaments in contractile rings and that the excessive accumulation of myosin II in the contractile rings and the slower cleavage process of mhckC− cells are due to the inefficient phosphorylation of the three threonine residues in the distal tail region of myosin II. Nonetheless, mhckC− cells grew at normal rates (Figure 4C), both in suspension cultures and on plastic dishes (our unpublished data), indicating that the inability to remove myosin II filaments from cleavage furrows is not particularly deleterious. This is not surprising because the 3XALA cells exhibited only modest reduction in growth rates compared with the wild-type cells (Egelhoff et al., 1991). We speculate that mhckC− cells are less sicker than cells expressing 3XALA myosin, because perhaps other kinases are able to inefficiently phosphorylate filamentous myosin II in mhckC− cells. In contrast, 3XALA myosin filaments cannot be phosphorylated at all at these three positions and should be even slower to disassemble than wild-type myosin II in mhckC− cells. On the other hand, overexpression of MHCK C diminished the amount of myosin II present in the cytoskeletal fraction (our unpublished data). In addition, when cells overexpressing GFP-MHCK C were grown in suspension, the expression of the kinase became undetectable within a week, although it persisted in cells maintained on plates (our unpublished data). It may be that, in suspension, cells expressing GFP-MHCK C at higher levels grow comparatively slowly, enabling them to be outgrown by cells that have somehow turned off the MHCK C expression. Consistent with that idea, the 3XASP mutant is unable to support cell division in suspension at all (Egelhoff et al., 1991).
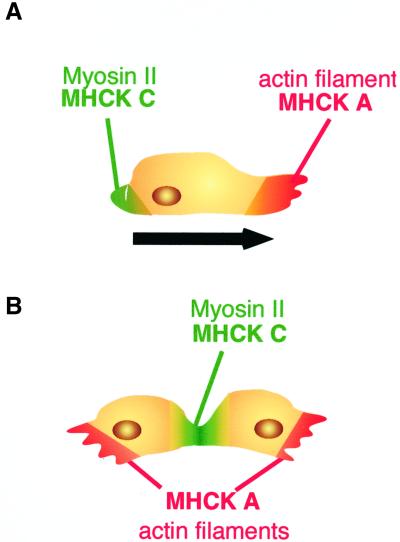
GFP-tagged MHCK C localized to the cortex of interphase cells (Figure 2) and to the posterior of migrating cells (Figure 3), i.e., the intracellular distribution of MHCK C always corresponded to that of myosin II filaments. That this intracellular localization of MHCK C was abolished in myosin II− cells (Figure 2, C and D) is indicative of the myosin II dependence of the cellular distribution of MHCK C. Recently, localization of MHCK A was studied in living Dictyostelium cells by using a GFP fusion protein (Steimle et al., 2001b). Unlike MHCK C, MHCK A is localized in the actin-rich protrusions at the anterior of migrating cells. Moreover, MHCK A translocated from the cytoplasm to the cell cortex in response to cAMP, even in myosin II− cells. Thus, the intracellular distribution of MHCK A is complementary to that of MHCK C and is independent of myosin II in interphase cells. This suggests that MHCK A is involved in preventing myosin II assembly in the actin rich regions of the cell anterior, whereas MHCK C mediates myosin II turnover at the posterior (Steimle et al., 2001b). We have extended this analysis on MHCK A localization to mitotic cells, and found that MHCK A is enriched in the polar regions during cytokinesis (Figure 7). This is consistent with the fact that mhckA− cells carried out the cleavage process at a rate comparable with that of wild-type cells (Figure 5). We suggest that the two kinases have complementary functions in vivo in both mitotic and interphase cells (Figure 8).
Figure 8.
Schematic diagram illustrating the differences in the distributions of MHCK A and MHCK C in migrating (A) and mitotic (B) Dictyostelium cells. In migrating cells, myosin II filaments are localized at the posterior cortices, whereas actin filaments are localized at the leading edges as well as at the posterior cortices (Yumura et al., 1984; Westphal et al., 1997). The distribution of MHCK A corresponds to that of the anterior actin filaments. It has been speculated that MHCK A prevents formation of myosin II filaments within the actin rich protrusions (Steimle et al., 2001b). In contrast, MHCK C is localized at the posterior in migrating cells. We propose that by phosphorylating the myosin II tail, MHCK C performs a key function in the disassembly of myosin II filaments, facilitating turnover of myosin II. In mitotic cells, myosin II filaments are localized in cleavage furrows (Neujahr et al., 1997a), whereas actin filaments concentrate in the polar regions, with a small accumulation in the furrow (Yumura, 1996; Neujahr et al., 1997a). During cytokinesis, MHCK C is localized in the cleavage furrows, where myosin II filaments accumulate; it likely phosphorylates the filaments driving myosin II turnover during and after cell division. MHCK A accumulates in the polar regions during cytokinesis, where actin filaments are enriched, and is probably involved in the exclusion of myosin II from those areas.
A key question raised by the aforementioned findings is, what determines the differential distributions of MHCK A and MHCK C. MHCK A contains a coiled-coil region at its N terminus (amino acid residues 1–504), whereas MHCK C does not. Recently, it was demonstrated that the coiled-coil domain of MHCK A alone was able to localize to F-actin–rich regions in vivo and to bind to actin filaments in vitro, whereas in vivo MHCK A lacking the coiled-coil domain diffuse in the cytosol (Steimle et al., 2002). The primary structure of MHCK A lacking the coiled-coil domain is similar to that of MHCK C, but unlike MHCK C, the truncation mutant of MHCK A did not localize to myosin II rich regions. How then does MHCK C recognize myosin II filaments in cells?
A feature unique to MHCK C is PPXYsequence, a putative WW domain binding motif, present in the catalytic domain of MHCK C. The WW domain is one of the most versatile of protein–protein interaction modules, and its ability to interact with a variety of proline-containing ligands yields a great deal of functional diversity (Sudol and Hunter, 2001; Ilsley et al., 2002, #345). From Dictyostelium, we have already cloned dwwA, a gene encoding a protein that contains two WW domains, and found that GFP-DWWA is also localized in the cortex of Dictyostelium cells (Nagasaki and Uyeda, unpublished data). However, we have not as yet been able to demonstrate a direct interaction between MHCK C and DWWA. Additional studies will be required before the molecular mechanism responsible for the differential targeting of MHCKs is completely understood.
Finally, using the fluorescent recovery after photobleaching technique, we have recently shown that turnover of myosin II filaments in the cell cortex is rapid and continuous (Yumura, 2001). As mentioned above, this shuttling of myosin II between the cortex and the cytoplasm is regulated by phosphorylation of the three threonine residues in the myosin II tail. MHCK C would seem to be a good candidate regulator of this dynamic process because, unlike other characterized kinases, MHCK C always colocalizes with myosin II filaments. In future studies, we plan to investigate this and related issues.
Note added in proof. The biochemical characterization of MHCKC was recently reported by W. Liang, et al. (BMC Cell Biol. [2002]. 3:19).
ACKNOWLEDGMENTS
We thank Dr. Tom Egelhoff for sharing unpublished data, and The Japanese cDNA Sequencing Project for the sequence data.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0228. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0228.
REFERENCES
- Chang WT, Newell PC, Gross JD. Identification of the cell fate gene stalky in Dictyostelium. Cell. 1996;87:471–481. doi: 10.1016/s0092-8674(00)81367-7. [DOI] [PubMed] [Google Scholar]
- Clancy C, Mendoza M, Naismith T, Kolman M, Egelhoff T. Identification of a protein kinase from Dictyostelium with homology to the novel catalytic domain of myosin heavy chain kinase A. J Biol Chem. 1997;272:11812–11815. doi: 10.1074/jbc.272.18.11812. [DOI] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Dembinsky A, Rubin H, Ravid S. Chemoattractant-mediated increases in cGMP induce changes in Dictyostelium myosin II heavy chain-specific protein kinase C activities. J Cell Biol. 1996;134:911–921. doi: 10.1083/jcb.134.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembinsky A, Rubin H, Ravid S. Autophosphorylation of Dictyostelium myosin II heavy chain-specific protein kinase C is required for its activation and membrane dissociation. J Biol Chem. 1997;272:828–834. doi: 10.1074/jbc.272.2.828. [DOI] [PubMed] [Google Scholar]
- Egelhoff TT, Brown SS, Spudich JA. Spatial and temporal control of nonmuscle myosin localization: identification of a domain that is necessary for myosin filament disassembly in vivo. J Cell Biol. 1991;112:677–688. doi: 10.1083/jcb.112.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Inoue S. Cell division in Dictyostelium with special emphasis on actomyosin organization in cytokinesis. Cell Motil Cytoskeleton. 1991;18:41–54. doi: 10.1002/cm.970180105. [DOI] [PubMed] [Google Scholar]
- Futey L, Medley Q, Cote G, Egelhoff T. Structural analysis of myosin heavy chain kinase A from Dictyostelium. Evidence for a highly divergent protein kinase domain, an amino-terminal coiled-coil domain, and a domain homologous to the beta-subunit of heterotrimeric G proteins. J Biol Chem. 1995;270:523–529. doi: 10.1074/jbc.270.2.523. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The mechanism and control of cytokinesis. Curr Opin Cell Biol. 1997;9:815–823. doi: 10.1016/s0955-0674(97)80082-8. [DOI] [PubMed] [Google Scholar]
- Ilsley J, Sudol M, Winder S. The WW domain: linking cell signaling to the membrane cytoskeleton. Cell Signal. 2002;14:183–189. doi: 10.1016/s0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- Kolman MF, Egelhoff TT. Dictyostelium myosin heavy chain kinase A subdomains. Coiled-coil and wd repeat roles in oligomerization and substrate targeting. J Biol Chem. 1997;272:16904–16910. doi: 10.1074/jbc.272.27.16904. [DOI] [PubMed] [Google Scholar]
- Kolman MF, Futey LM, Egelhoff TT. Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J Cell Biol. 1996;132:101–109. doi: 10.1083/jcb.132.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczmarski E, Spudich J. Regulation of myosin self-assembly: phosphorylation of Dictyostelium heavy chain inhibits formation of thick filaments. Proc Natl Acad Sci USA. 1980;77:7292–7296. doi: 10.1073/pnas.77.12.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ito K, Lee R, Uyeda T. Involvement of tail domains in regulation of Dictyostelium myosin II. Biochem Biophys Res Commun. 2000;271:75–81. doi: 10.1006/bbrc.2000.2582. [DOI] [PubMed] [Google Scholar]
- Luo X, Crawley S, Steimle P, Egelhoff T, Cote G. Specific phosphorylation of threonine by the Dictyostelium myosin II heavy chain kinase family. J Biol Chem. 2001;276:17836–17843. doi: 10.1074/jbc.M009366200. [DOI] [PubMed] [Google Scholar]
- Mabuchi I. Biochemical aspects of cytokinesis. Int Rev Cytol. 1986;101:175–213. doi: 10.1016/s0074-7696(08)60249-1. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein DJ, Titus MA, DeLozanne A, Spudich JA. Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki A, de Hostos EL, Uyeda TQ. Genetic and morphological evidence for two parallel pathways of cell cycle-coupled cytokinesis in Dictyostelium. J Cell Sci. 2002;115:2241–2251. doi: 10.1242/jcs.115.10.2241. [DOI] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Albrecht R, Ecke M, Schwartz JM, Weber I, Gerisch G. Three-dimensional patterns and redistribution of myosin II and actin in mitotic Dictyostelium cells. J Cell Biol. 1997a;139:1793–1804. doi: 10.1083/jcb.139.7.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Gerisch G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J Cell Sci. 1997b;110:123–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- Nock S, Liang W, Warrick H, Spudich J. Mutational analysis of phosphorylation sites in the Dictyostelium myosin II tail: disruption of myosin function by a single charge change. FEBS Lett. 2000;466:267–272. doi: 10.1016/s0014-5793(99)01796-2. [DOI] [PubMed] [Google Scholar]
- Ravid S, Spudich J. Membrane-bound Dictyostelium myosin heavy chain kinase: a developmentally regulated substrate-specific member of the protein kinase C family. Proc Natl Acad Sci USA. 1992;89:5877–5881. doi: 10.1073/pnas.89.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redowicz M. Regulation of nonmuscle myosins by heavy chain phosphorylation. J Muscle Res Cell Motil. 2001;22:163–173. doi: 10.1023/a:1010552929028. [DOI] [PubMed] [Google Scholar]
- Robinson D, Spudich J. Towards a molecular understanding of cytokinesis. Trends Cell Biol. 2000;10:228–237. doi: 10.1016/s0962-8924(00)01747-5. [DOI] [PubMed] [Google Scholar]
- Ruppel KM, Uyeda TQ, Spudich JA. Role of highly conserved lysine 130 of myosin motor domain. In vivo and in vitro characterization of site specifically mutated myosin. J Biol Chem. 1994;269:18773–18780. [PubMed] [Google Scholar]
- Sabry JH, Moores SL, Ryan S, Zang JH, Spudich JA. Myosin heavy chain phosphorylation sites regulate myosin localization during cytokinesis in live cells. Mol Biol Cell. 1997;8:2605–2615. doi: 10.1091/mbc.8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle P, Licate L, Cote G, Egelhoff TT. Lamellipodial localization of Dictyostelium myosin heavy chain kinase A is mediated via F-actin binding by the coiled-coil domain. FEBS Lett. 2002;516:58–62. doi: 10.1016/s0014-5793(02)02494-8. [DOI] [PubMed] [Google Scholar]
- Steimle P, Naismith T, Licate L, Egelhoff T. WD repeat domains target Dictyostelium myosin heavy chain kinases by binding directly to myosin filaments. J Biol Chem. 2001a;276:6853–6860. doi: 10.1074/jbc.M008992200. [DOI] [PubMed] [Google Scholar]
- Steimle P, Yumura S, Cote G, Medley Q, Polyakov M, Leppert B, Egelhoff T. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr Biol. 2001b;11:708–713. doi: 10.1016/s0960-9822(01)00182-8. [DOI] [PubMed] [Google Scholar]
- Sudol M, Hunter T. New wrinkles for an old domain. Cell. 2001;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. In: Spudich JA, editor. Dictyostelium discoideum: Molecular Approaches to Cell Biology. Vol. 28. Orlando, FL: Academic Press; 1987. pp. 9–29. [DOI] [PubMed] [Google Scholar]
- Uyeda TQ, Yumura S. Molecular biological approaches to study myosin functions in cytokinesis of Dictyostelium. Microsc Res Tech. 2000;49:136–144. doi: 10.1002/(SICI)1097-0029(20000415)49:2<136::AID-JEMT5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Vaillancourt J, Lyons C, Cote G. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J Biol Chem. 1988;263:10082–10087. [PubMed] [Google Scholar]
- Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz J, Marriott G, Gerisch G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- Yumura S. Spatial distribution of fluorescently labeled actin in living Dictyostelium amoebae. Cell Struct Funct. 1996;21:189–197. doi: 10.1247/csf.21.189. [DOI] [PubMed] [Google Scholar]
- Yumura S. Myosin II dynamics and cortical flow during contractile ring formation in Dictyostelium cells. J Cell Biol. 2001;154:137–146. doi: 10.1083/jcb.200011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S, Fukui Y. Reversible cyclic AMP-dependent change in distribution of myosin thick filaments in Dictyostelium. Nature. 1985;314:194–196. doi: 10.1038/314194a0. [DOI] [PubMed] [Google Scholar]
- Yumura S, Mori H, Fukui Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J Cell Biol. 1984;99:894–899. doi: 10.1083/jcb.99.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S, Uyeda TQ. Transport of myosin II to the equatorial region without its own motor activity in mitotic Dictyostelium cells. Mol Biol Cell. 1997;8:2089–2099. doi: 10.1091/mbc.8.10.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang JH, Cavet G, Sabry JH, Wagner P, Moores SL, Spudich JA. On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol Biol Cell. 1997;8:2617–2629. doi: 10.1091/mbc.8.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]